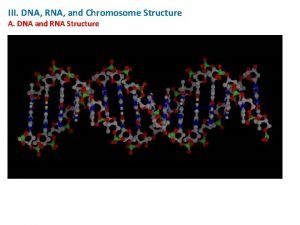
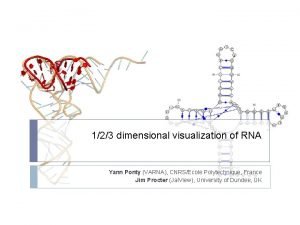
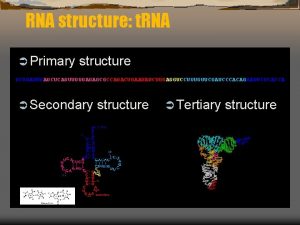
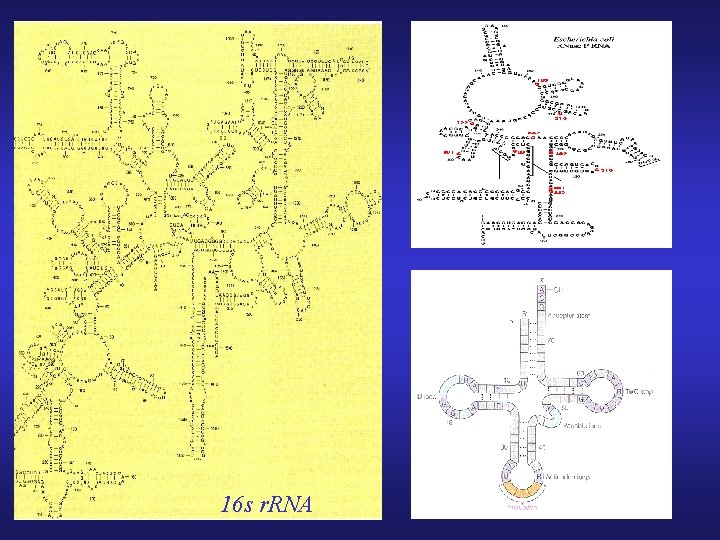
RNA Secondary Structure Prediction 16 s r RNA







- Slides: 16

RNA Secondary Structure Prediction


16 s r. RNA

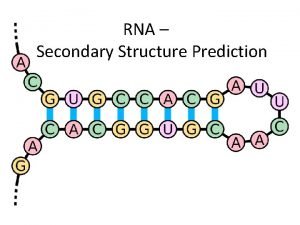
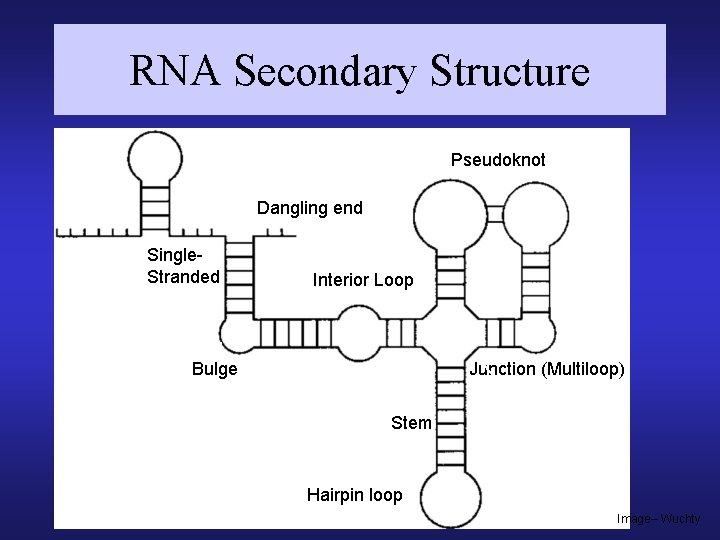
RNA Secondary Structure Pseudoknot Dangling end Single. Stranded Interior Loop Bulge Junction (Multiloop) Stem Hairpin loop Image– Wuchty

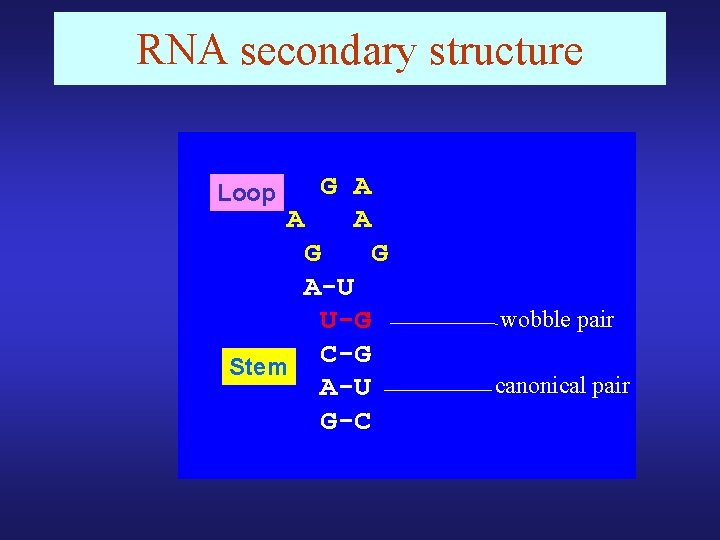
RNA secondary structure G A A A G G A-U U-G C-G Stem A-U G-C Loop wobble pair canonical pair

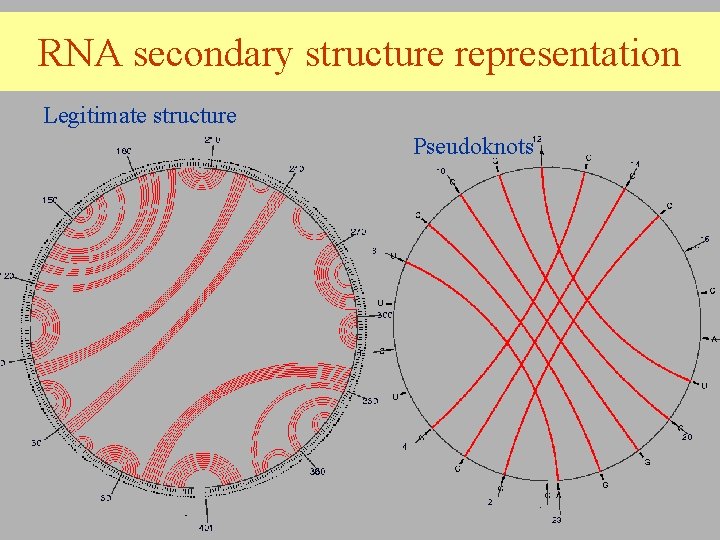
RNA secondary structure representation Legitimate structure Pseudoknots

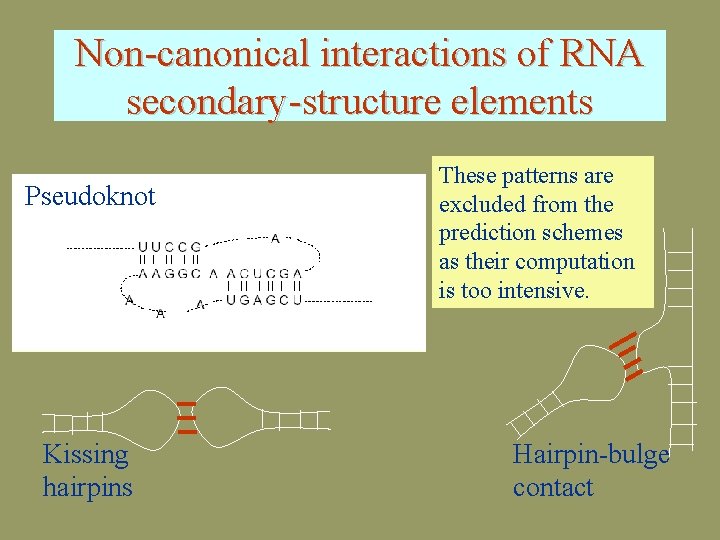
Non-canonical interactions of RNA secondary-structure elements Pseudoknot Kissing hairpins These patterns are excluded from the prediction schemes as their computation is too intensive. Hairpin-bulge contact

“Rules for 2 D RNA prediction” • Base Pairs in stems: GOOD • Additional possible assumptions: e. g. , G: C better than A: T • Bulges, Loops: BAD • Canonical Interactions (base pairs, stems, bulges, loops): OK • Non canonical interactions (pseudoknots, kissing hairpins): Forbidden • The more interactions: The better

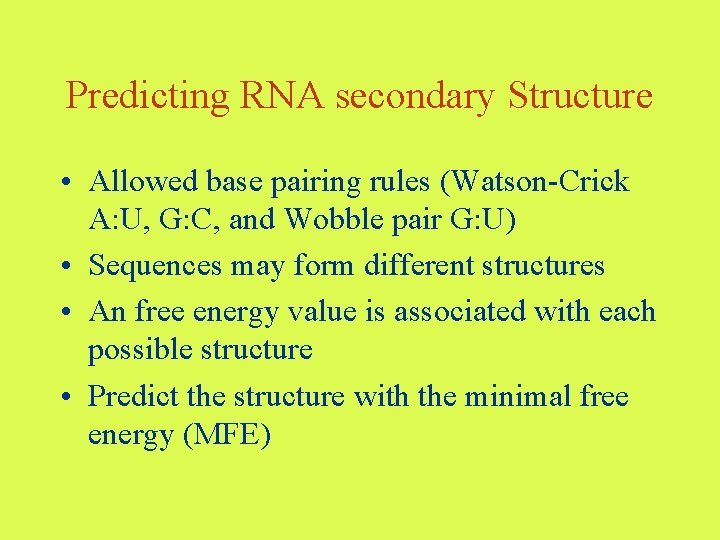
Predicting RNA secondary Structure • Allowed base pairing rules (Watson-Crick A: U, G: C, and Wobble pair G: U) • Sequences may form different structures • An free energy value is associated with each possible structure • Predict the structure with the minimal free energy (MFE)

Simplifying Assumptions for Structure Prediction • RNA folds into one minimum free-energy structure. • There are no non-canonical interactions. • The energy of a particular base pair in a double stranded regions is sequence independent – Neighbors have no influence. Was solved by dynamic programming Zucker and Steigler 1981

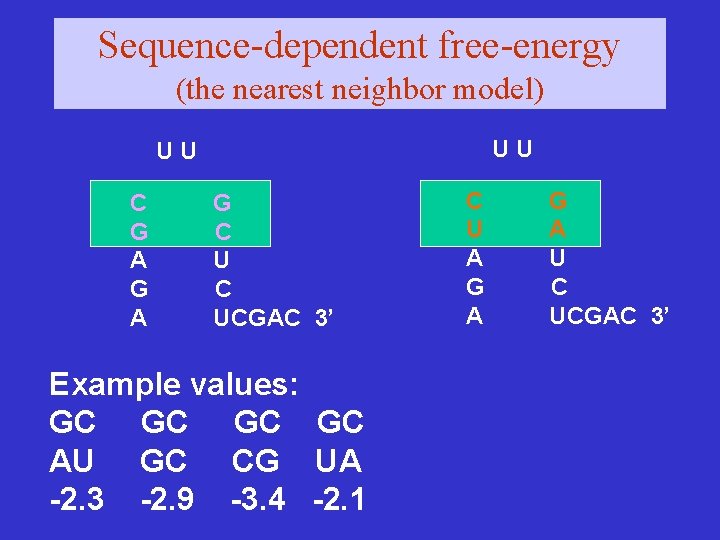
Sequence-dependent free-energy (the nearest neighbor model) UU UU C G A G C UCGAC 3’ Example values: GC GC AU GC CG UA -2. 3 -2. 9 -3. 4 -2. 1 C U A G A U C UCGAC 3’

Free energy computation +5. 9 (4 nt loop) U U A G C +3. 3 (1 nt bulge) G U A C A A A 5’ -1. 1 mismatch of hairpin -2. 9 stacking A 5’ dangling -0. 3 A C A U G U -1. 8 stacking -0. 9 stacking -1. 8 stacking -2. 1 stacking 3’ G= -4. 6 KCAL/MOL

Prediction Programs • Mfold http: //www. bioinfo. rpi. edu/applications/mfold/rna/form 1. cgi • Vienna RNA Secondary Structure Prediction http: //rna. tbi. univie. ac. at/cgi-bin/RNAfold. cgi

Mfold - Suboptimal Folding • For any sequence of N nucleotides, the expected number of structures is greater than 1. 8 N • A sequence of 100 nucleotides has ~3 1025 possible folds. If a computer can calculate 1000 folds/second, it would take 1015 years (age of universe = ~1010 years)! • Mfold generates suboptimal folds whose free energy fall within a certain range of values. Many of these structures are different in trivial ways. These suboptimal folds can still be useful for designing experiments.

Example:

Output:
 Rna secondary structure prediction
Rna secondary structure prediction Protein structure
Protein structure Phd secondary structure prediction
Phd secondary structure prediction Rna secondary structure dynamic programming
Rna secondary structure dynamic programming Jacobson algorithm
Jacobson algorithm Rna secondary structure dynamic programming
Rna secondary structure dynamic programming Frederick griffith transformation
Frederick griffith transformation Structure of chromosome
Structure of chromosome Varna rna
Varna rna Predictions may might will
Predictions may might will Championship branch prediction
Championship branch prediction Corner prediction
Corner prediction The hunger games questions
The hunger games questions Variance in regression
Variance in regression Meritsprediction
Meritsprediction Penggorengan keripik
Penggorengan keripik Make a prediction about kenny and franchesca
Make a prediction about kenny and franchesca