Protein network analysis Network motifs Network clusters modules





- Slides: 39

Protein network analysis • • Network motifs Network clusters / modules Co-clustering networks & expression Network comparison (species, conditions) • Integration of genetic & physical nets • Network visualization

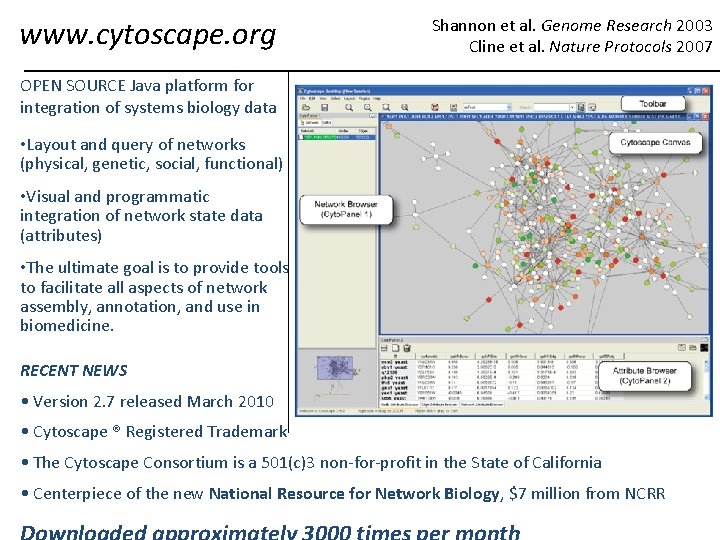
www. cytoscape. org Shannon et al. Genome Research 2003 Cline et al. Nature Protocols 2007 OPEN SOURCE Java platform for integration of systems biology data • Layout and query of networks (physical, genetic, social, functional) • Visual and programmatic integration of network state data (attributes) • The ultimate goal is to provide tools to facilitate all aspects of network assembly, annotation, and use in biomedicine. RECENT NEWS • Version 2. 7 released March 2010 • Cytoscape ® Registered Trademark • The Cytoscape Consortium is a 501(c)3 non-for-profit in the State of California • Centerpiece of the new National Resource for Network Biology, $7 million from NCRR

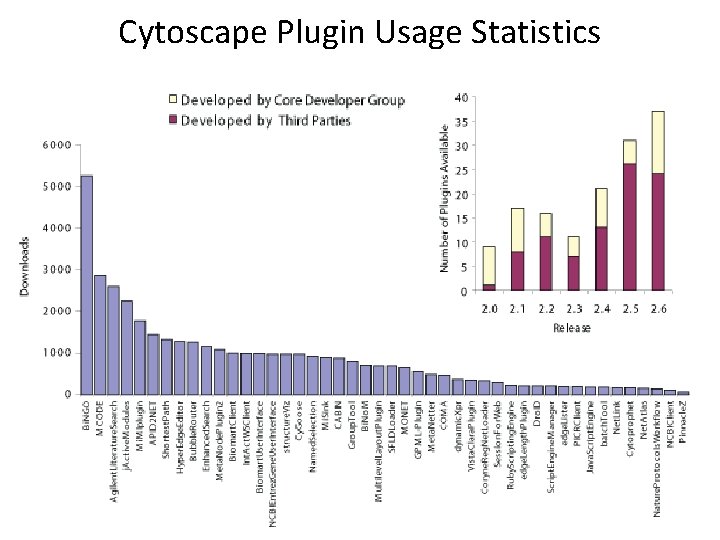
Cytoscape Plugin Usage Statistics

Integration of networks and expression

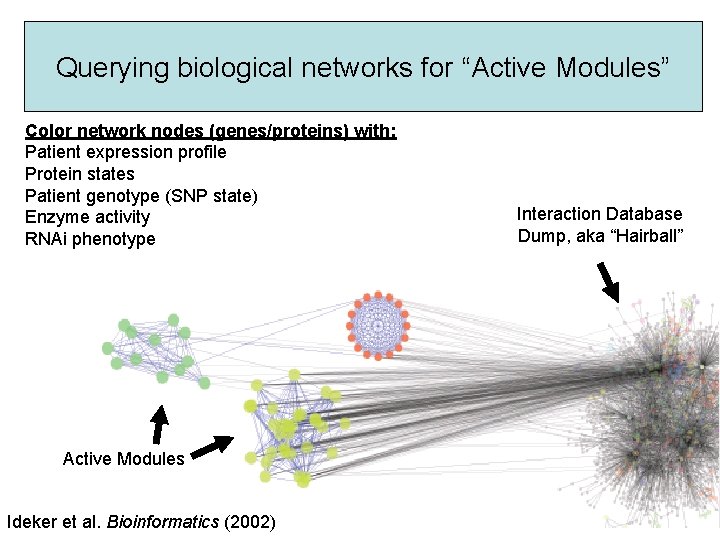
Querying biological networks for “Active Modules” Color network nodes (genes/proteins) with: Patient expression profile Protein states Patient genotype (SNP state) Enzyme activity RNAi phenotype Active Modules Ideker et al. Bioinformatics (2002) Interaction Database Dump, aka “Hairball”

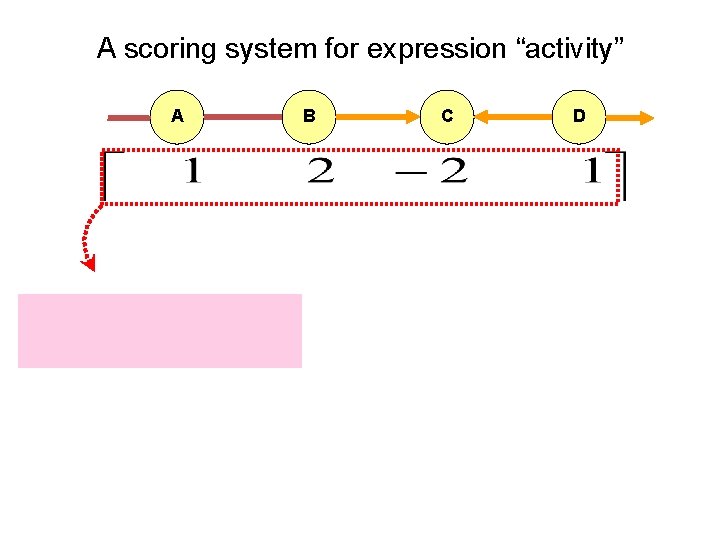
A scoring system for expression “activity” A B C D

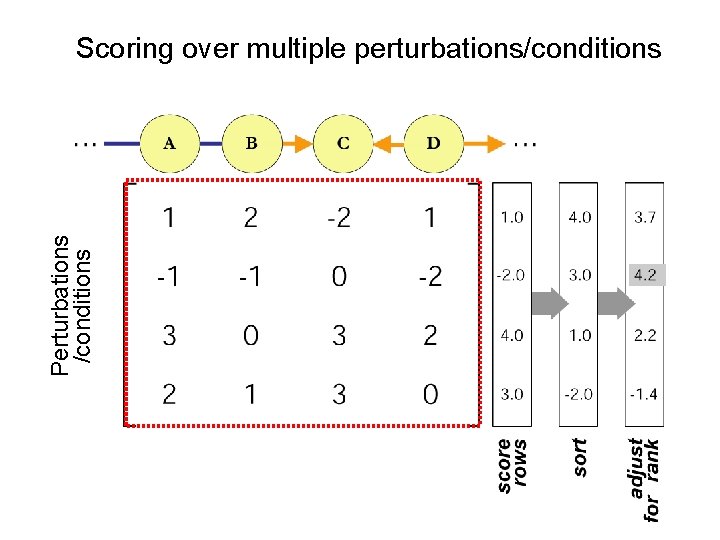
Perturbations /conditions Scoring over multiple perturbations/conditions

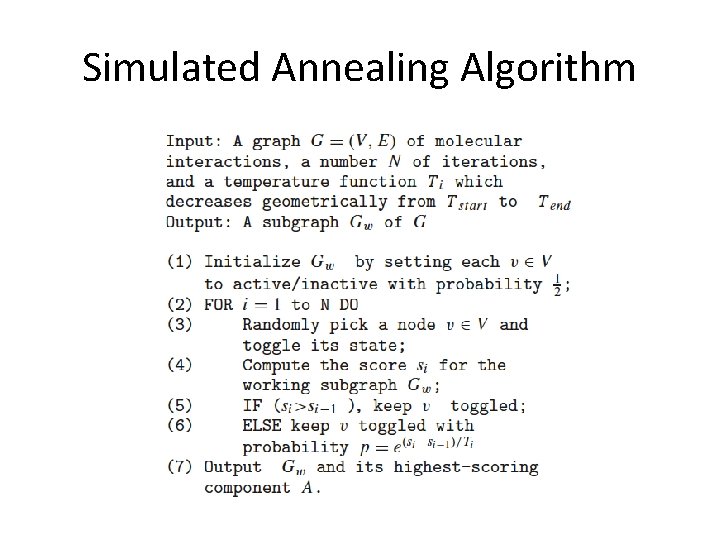
Searching for “active” pathways in a large network • Score subnetworks according to their overall amount of activity • Finding the highest scoring subnetworks is NP hard, so we use heuristic search algs. to identify a collection of high-scoring subnetworks (local optima) • Simulated annealing and/or greedy search starting from an initial subnetwork “seed” • During the search we must also worry about issues such as local topology and whether a subnetwork’s score is higher than would be expected at random

Simulated Annealing Algorithm

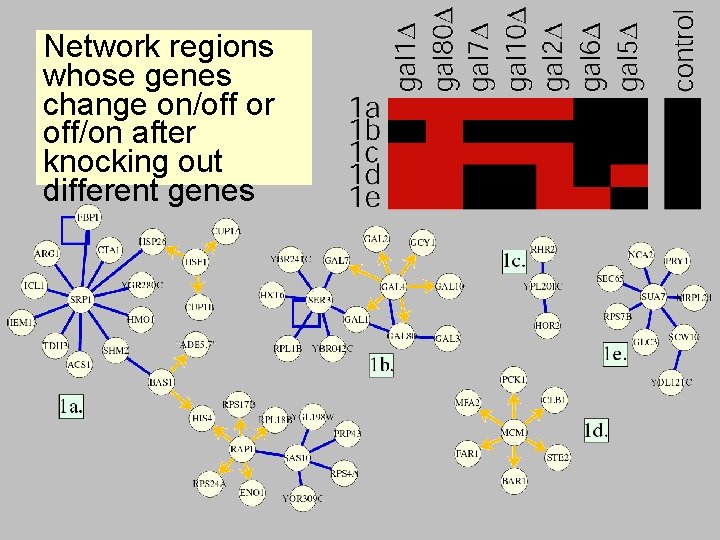
Network regions whose genes change on/off or off/on after knocking out different genes

Initial Application to Toxicity: Networks responding to DNA damage in yeast Tom Begley and Leona Samson; MIT Dept. of Bioengineering Systematic phenotyping of gene knockout strains in yeast Evaluation of growth of each strain in the presence of MMS (and other DNA damaging agents) Sensitive Not sensitive Not tested MMS sensitivity in ~25% of strains Screening against a network of protein interactions…

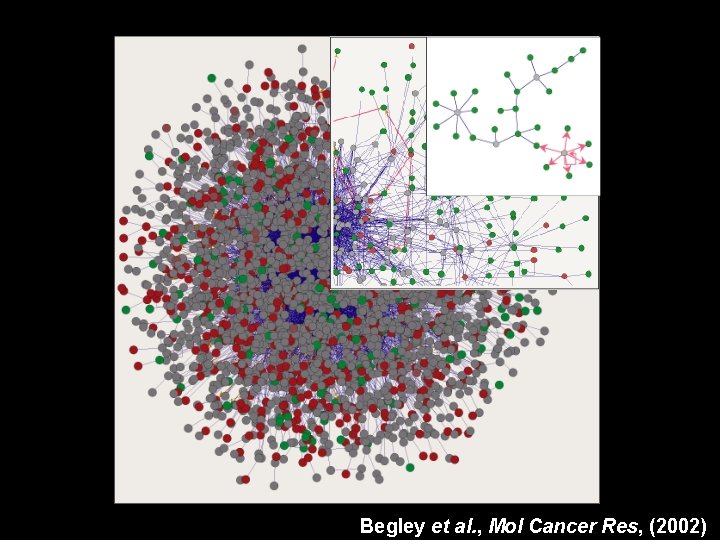
Begley et al. , Mol Cancer Res, (2002)

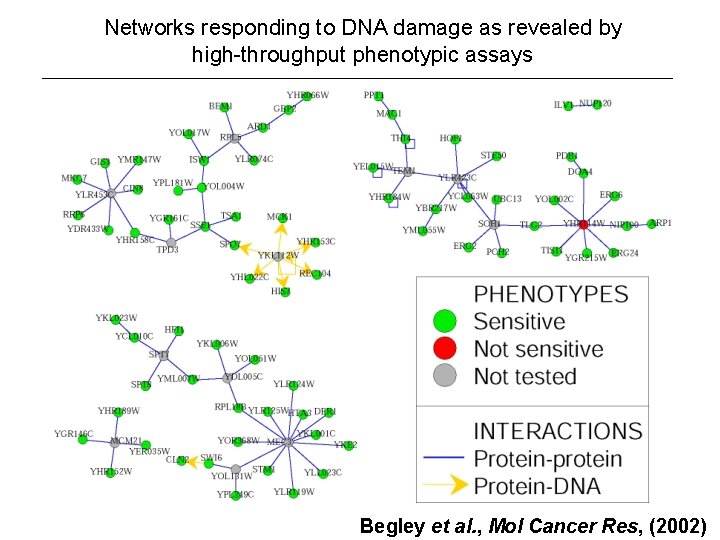
Networks responding to DNA damage as revealed by high-throughput phenotypic assays Begley et al. , Mol Cancer Res, (2002)

Host-pathogen interactions regulating early stage HIV-1 infection Genome-wide RNAi screens for genes required for infection utilizing a single cycle HIV-1 reporter virus engineered to encode luciferase and bearing the Vesicular Stomatitis Virus Glycoprotein (VSV-G) on its surface to facilitate efficient infection… Sumit Chanda

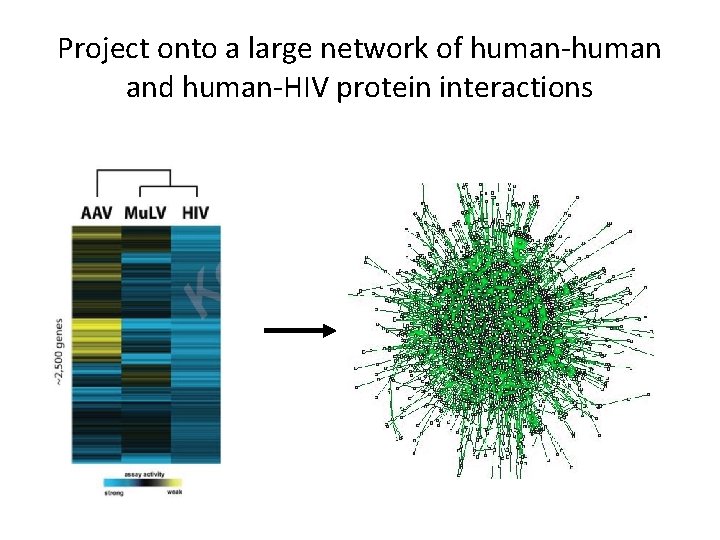
Project onto a large network of human-human and human-HIV protein interactions

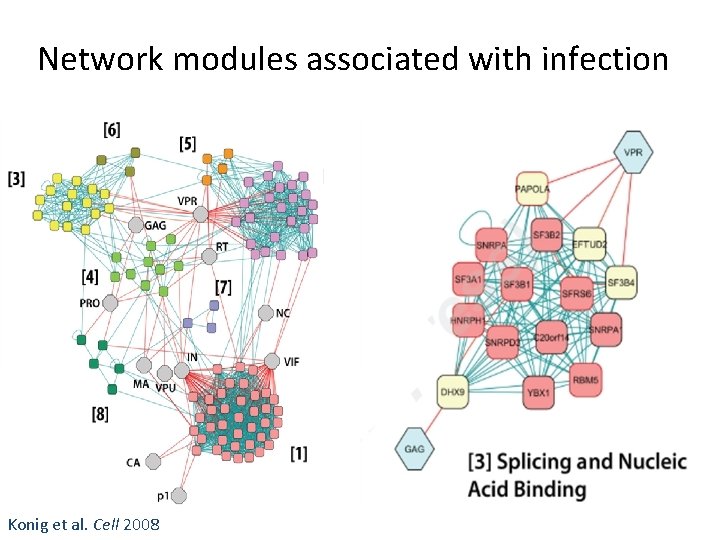
Network modules associated with infection Konig et al. Cell 2008

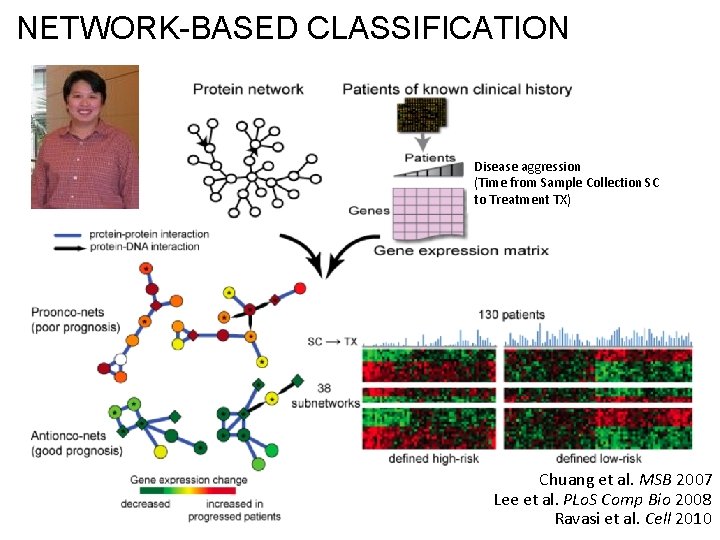
Network-based classification

NETWORK-BASED CLASSIFICATION Disease aggression (Time from Sample Collection SC to Treatment TX) Chuang et al. MSB 2007 Lee et al. PLo. S Comp Bio 2008 Ravasi et al. Cell 2010

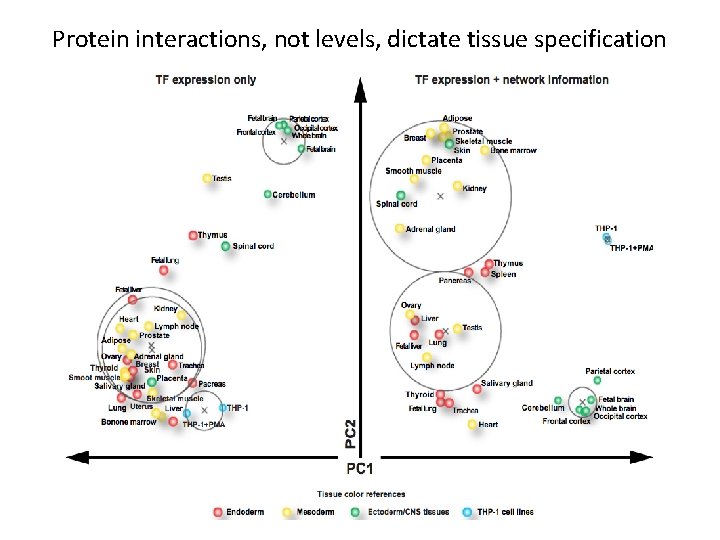
The Mammalian Cell Fate Map: Can we classify tissue type using expression, networks, etc? Gilbert Developmental Biology 4 th Edition

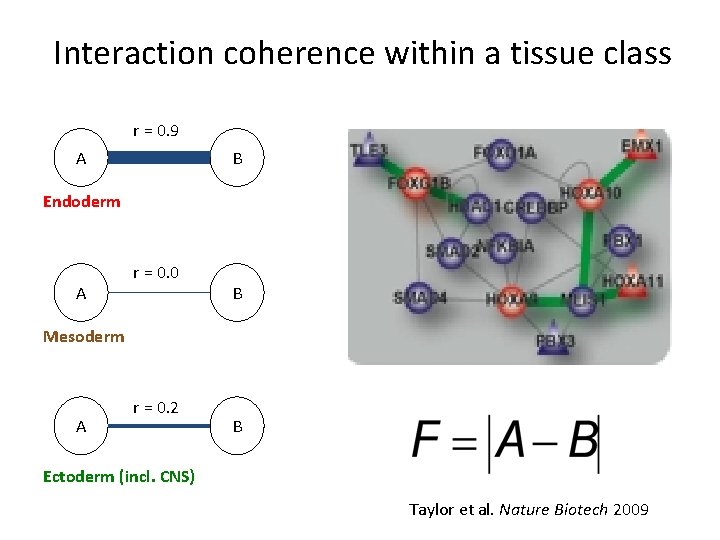
Interaction coherence within a tissue class r = 0. 9 A B Endoderm A r = 0. 0 B Mesoderm A r = 0. 2 B Ectoderm (incl. CNS) Taylor et al. Nature Biotech 2009

Protein interactions, not levels, dictate tissue specification

Functional Enrichment

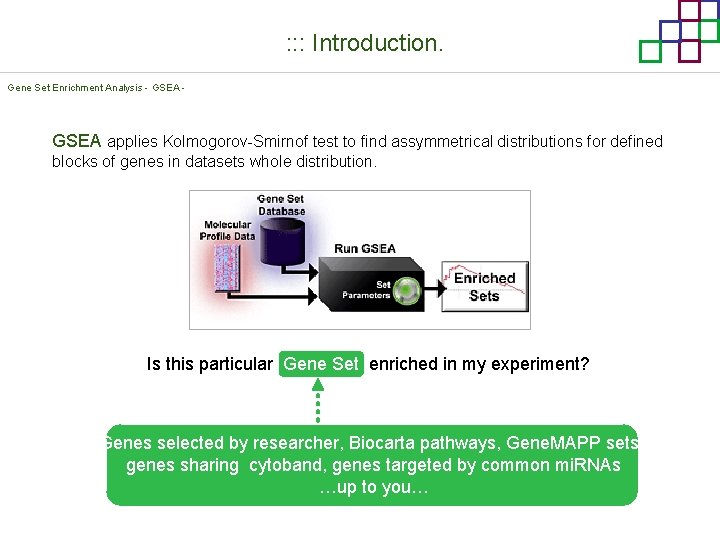
: : : Introduction. Gene Set Enrichment Analysis - GSEA MIT Broad Institute v 2. 0 available since Jan 2007 v 2. 0. 1 available since Feb 16 th 2007 Version 2. 0 includes Biocarta, Broad Institute, Gene. MAPP, KEGG annotations and more. . . Platforms: Affymetrix, Agilent, Code. Link, custom. . . (Subramanian et al. PNAS. 2005. )

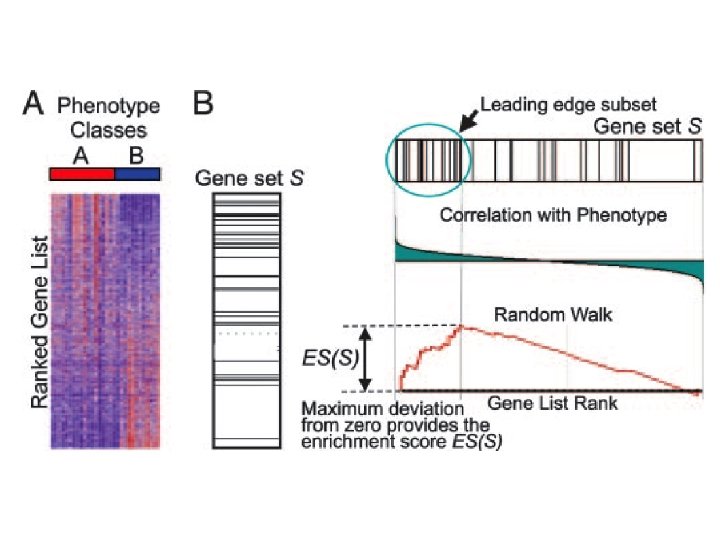
: : : Introduction. Gene Set Enrichment Analysis - GSEA applies Kolmogorov-Smirnof test to find assymmetrical distributions for defined blocks of genes in datasets whole distribution. Is this particular Gene Set enriched in my experiment? Genes selected by researcher, Biocarta pathways, Gene. MAPP sets, genes sharing cytoband, genes targeted by common mi. RNAs …up to you…

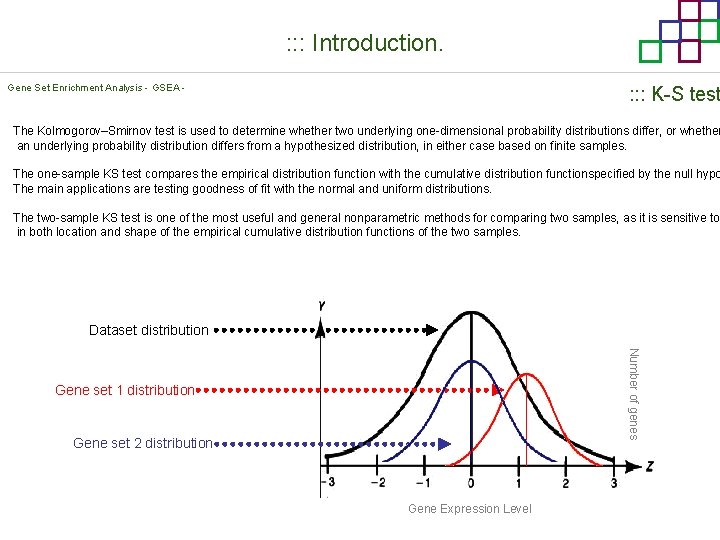
: : : Introduction. Gene Set Enrichment Analysis - GSEA - : : : K-S test The Kolmogorov–Smirnov test is used to determine whether two underlying one-dimensional probability distributions differ, or whether an underlying probability distribution differs from a hypothesized distribution, in either case based on finite samples. The one-sample KS test compares the empirical distribution function with the cumulative distribution functionspecified by the null hypo The main applications are testing goodness of fit with the normal and uniform distributions. The two-sample KS test is one of the most useful and general nonparametric methods for comparing two samples, as it is sensitive to in both location and shape of the empirical cumulative distribution functions of the two samples. Dataset distribution Number of genes Gene set 1 distribution Gene set 2 distribution Gene Expression Level

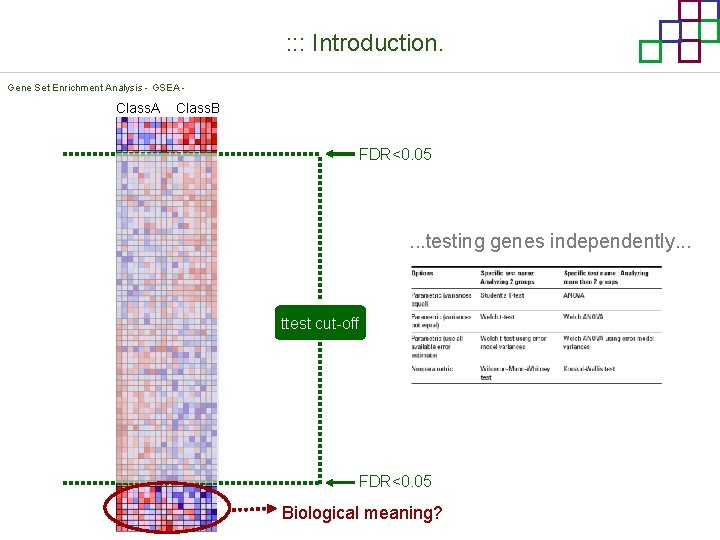
: : : Introduction. Gene Set Enrichment Analysis - GSEA - Class. A Class. B FDR<0. 05 . . . testing genes independently. . . ttest cut-off FDR<0. 05 Biological meaning?

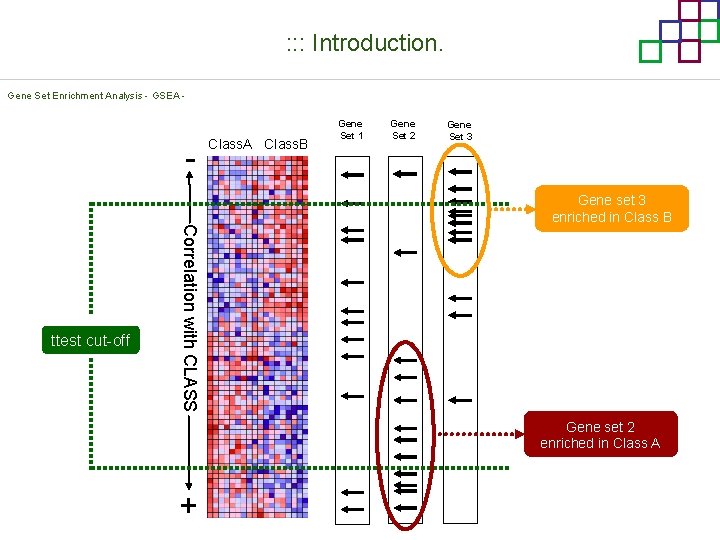
: : : Introduction. Gene Set Enrichment Analysis - GSEA - Correlation with CLASS ttest cut-off Class. A Class. B Gene Set 1 Gene Set 2 Gene Set 3 Gene set 3 enriched in Class B Gene set 2 enriched in Class A +


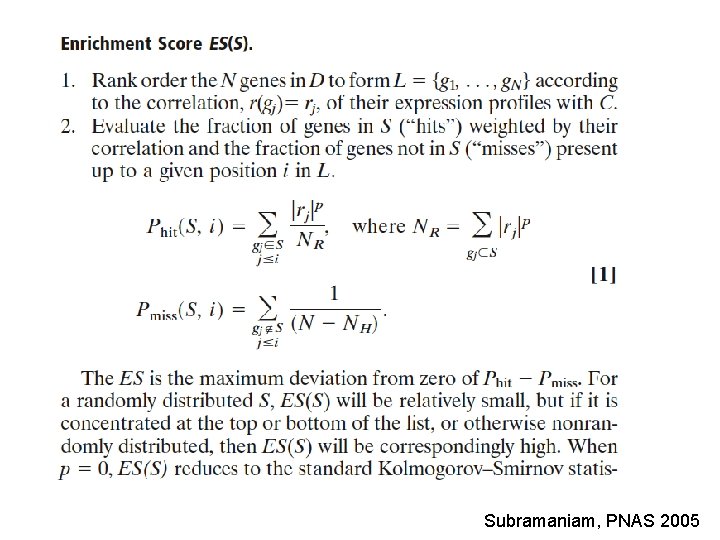
Subramaniam, PNAS 2005

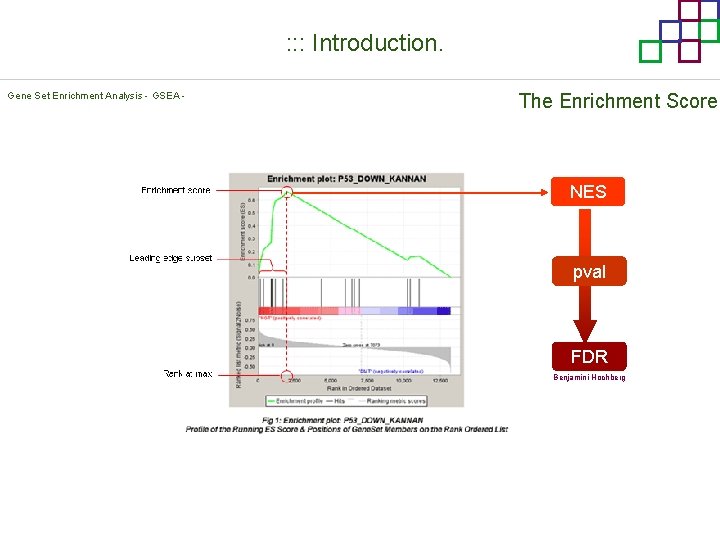
: : : Introduction. Gene Set Enrichment Analysis - GSEA - The Enrichment Score NES pval FDR Benjamini-Hochberg

Network Alignment Species 1 vs. species 2 Physical vs. genetic

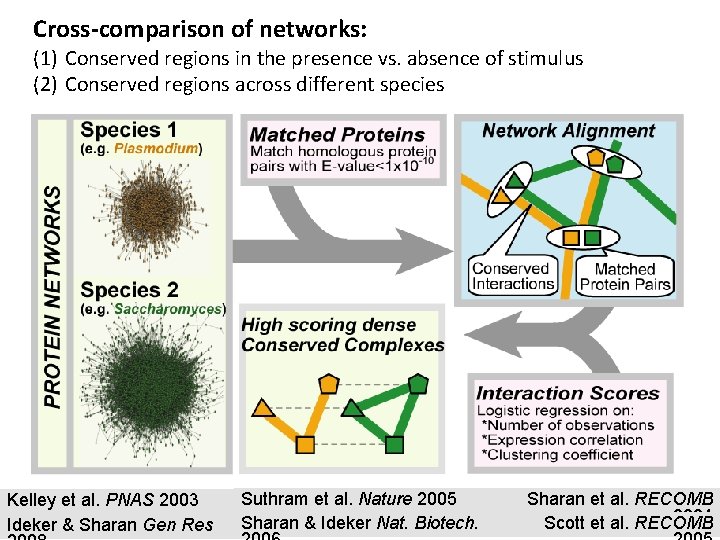
Cross-comparison of networks: (1) Conserved regions in the presence vs. absence of stimulus (2) Conserved regions across different species Kelley et al. PNAS 2003 Ideker & Sharan Gen Res Suthram et al. Nature 2005 Sharan & Ideker Nat. Biotech. Sharan et al. RECOMB 2004 Scott et al. RECOMB

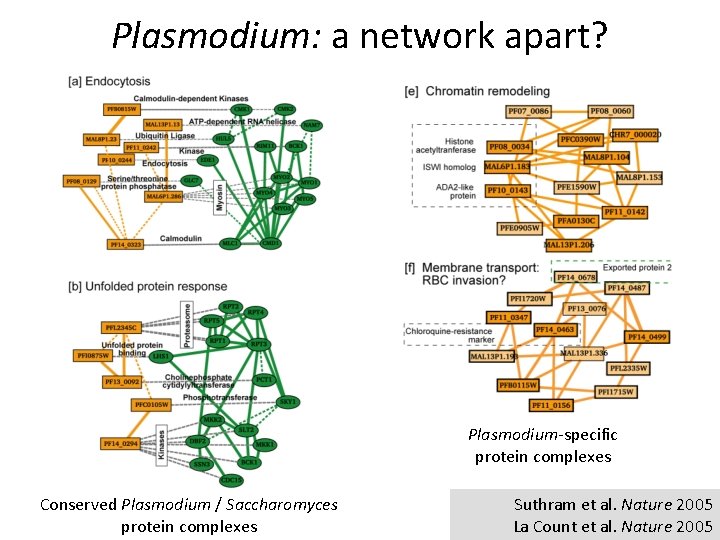
Plasmodium: a network apart? Plasmodium-specific protein complexes Conserved Plasmodium / Saccharomyces protein complexes Suthram et al. Nature 2005 La Count et al. Nature 2005

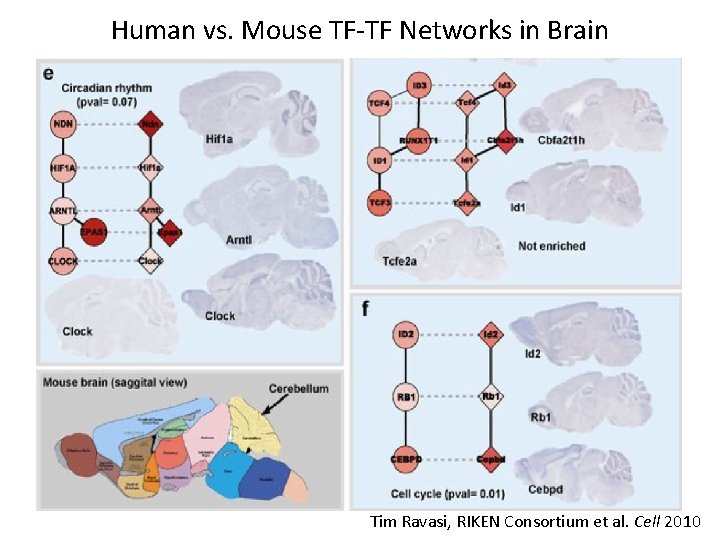
Human vs. Mouse TF-TF Networks in Brain Tim Ravasi, RIKEN Consortium et al. Cell 2010

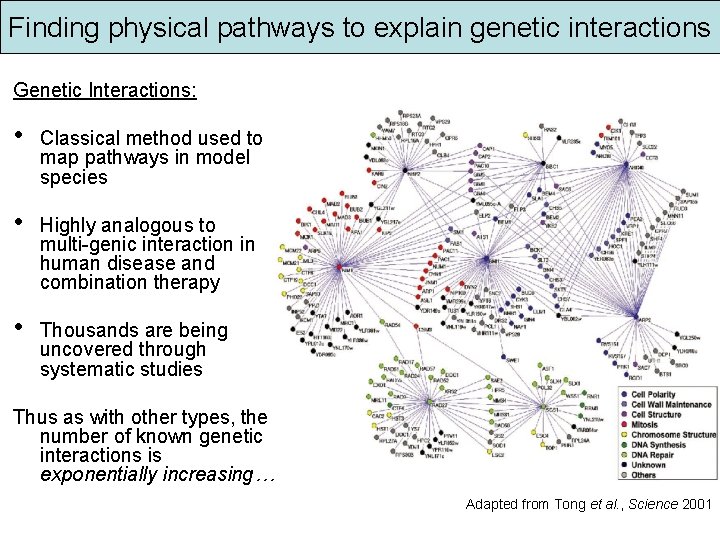
Finding physical pathways to explain genetic interactions Genetic Interactions: • Classical method used to map pathways in model species • Highly analogous to multi-genic interaction in human disease and combination therapy • Thousands are being uncovered through systematic studies Thus as with other types, the number of known genetic interactions is exponentially increasing… Adapted from Tong et al. , Science 2001

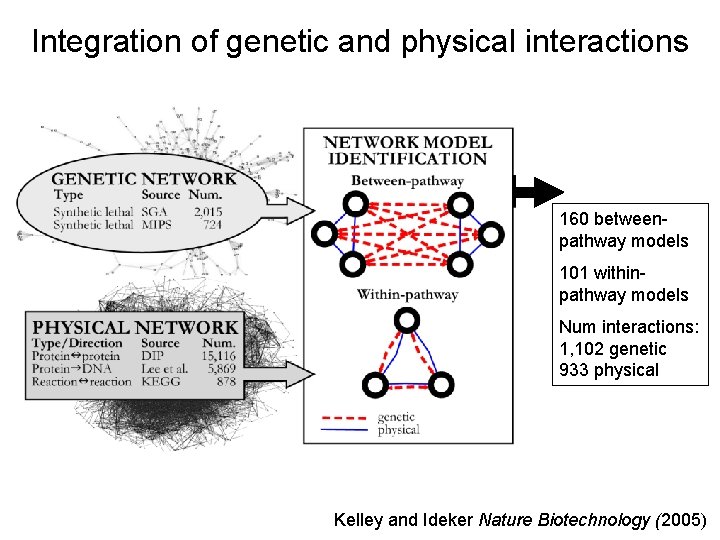
Integration of genetic and physical interactions 160 betweenpathway models 101 withinpathway models Num interactions: 1, 102 genetic 933 physical Kelley and Ideker Nature Biotechnology (2005)

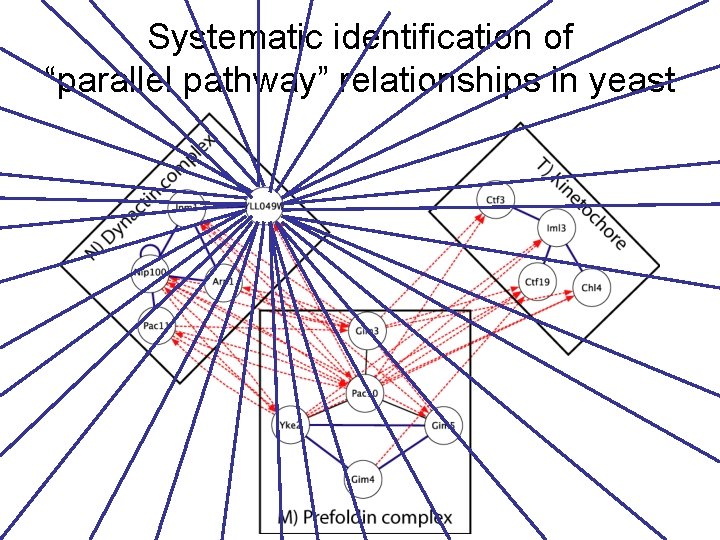
Systematic identification of “parallel pathway” relationships in yeast

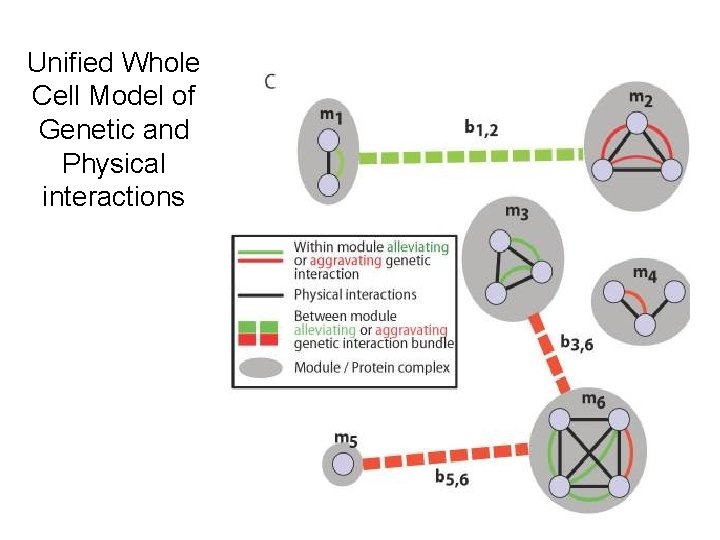
Unified Whole Cell Model of Genetic and Physical interactions

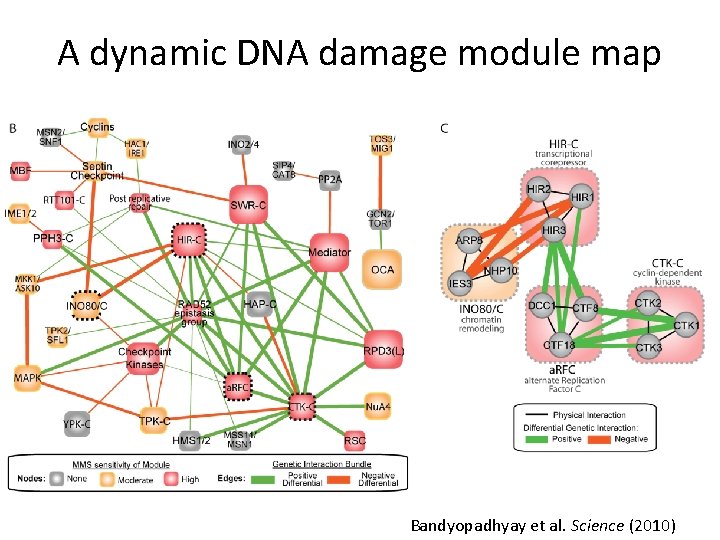
A dynamic DNA damage module map Bandyopadhyay et al. Science (2010)