Markov Chains Dependencies along the genome In previous









- Slides: 32

Markov Chains .

Dependencies along the genome In previous classes we assumed every letter in a sequence is sampled randomly from some distribution q( ) over the alpha bet {A, C, T, G}. This model could suffice for alignment scoring, but it is not the case in true genomes. 1. There are special subsequences in the genome, like TATA within the regulatory area, upstream a gene. 2. The pairs C followed by G is less common than expected for random sampling. We model such dependencies by Markov chains and hidden Markov model, which we define next. 2

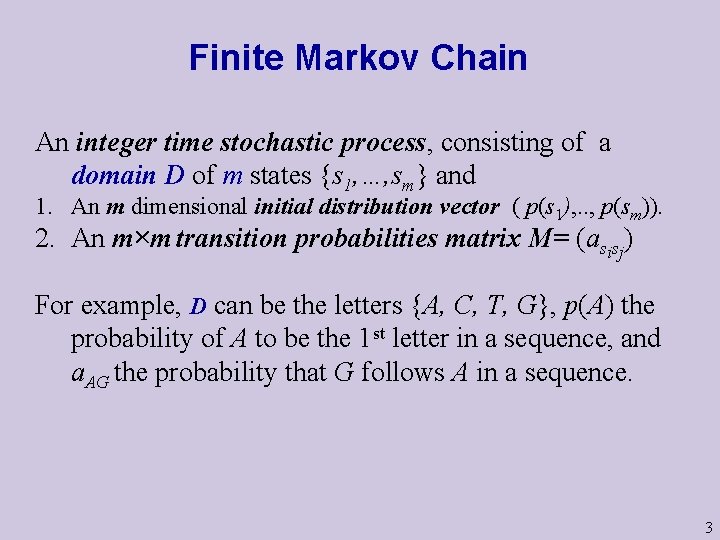
Finite Markov Chain An integer time stochastic process, consisting of a domain D of m states {s 1, …, sm} and 1. An m dimensional initial distribution vector ( p(s 1), . . , p(sm)). 2. An m×m transition probabilities matrix M= (asisj) For example, D can be the letters {A, C, T, G}, p(A) the probability of A to be the 1 st letter in a sequence, and a. AG the probability that G follows A in a sequence. 3

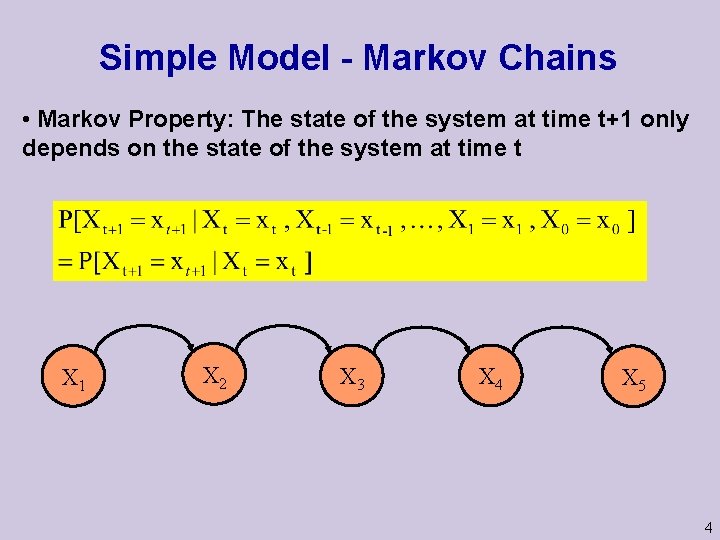
Simple Model - Markov Chains • Markov Property: The state of the system at time t+1 only depends on the state of the system at time t X 1 X 2 X 3 X 4 X 5 4

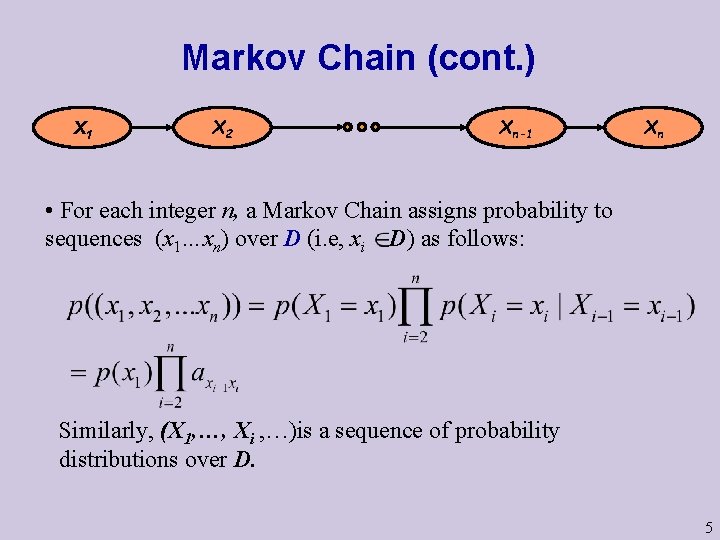
Markov Chain (cont. ) X 1 X 2 Xn-1 Xn • For each integer n, a Markov Chain assigns probability to sequences (x 1…xn) over D (i. e, xi D) as follows: Similarly, (X 1, …, Xi , …)is a sequence of probability distributions over D. 5

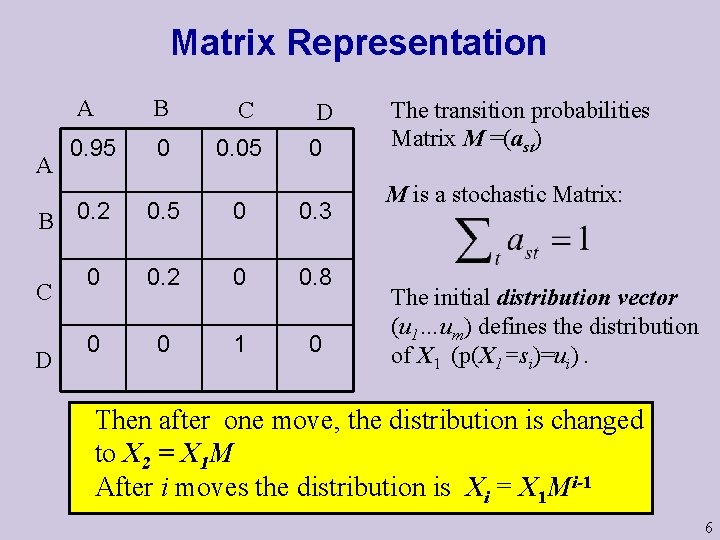
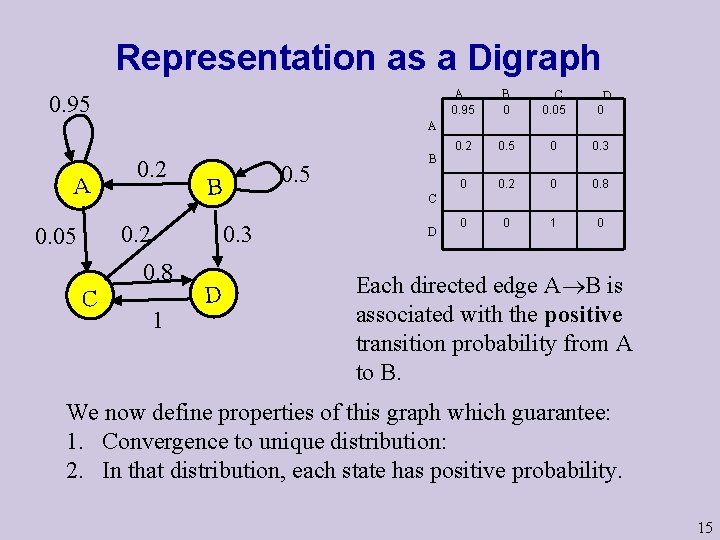
Matrix Representation A A B C 0. 95 0 0. 05 B 0. 2 C D D 0 0. 5 0 0. 3 0 0. 2 0 0. 8 0 0 1 0 The transition probabilities Matrix M =(ast) M is a stochastic Matrix: The initial distribution vector (u 1…um) defines the distribution of X 1 (p(X 1=si)=ui). Then after one move, the distribution is changed to X 2 = X 1 M After i moves the distribution is Xi = X 1 Mi-1 6

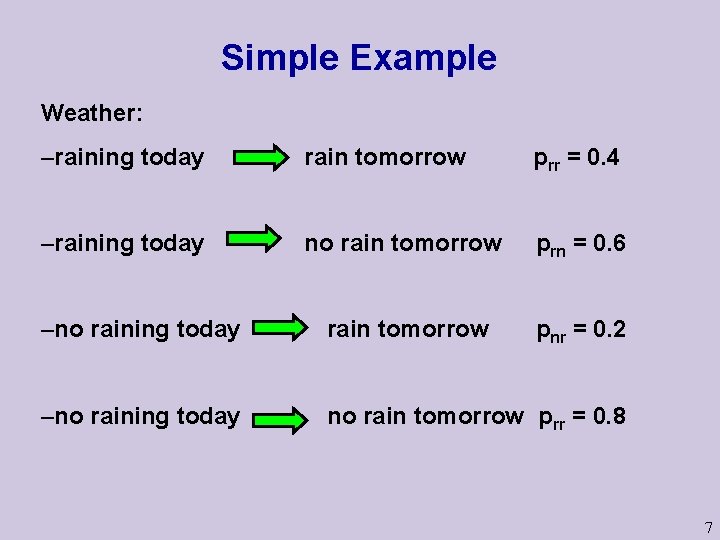
Simple Example Weather: –raining today rain tomorrow prr = 0. 4 –raining today no rain tomorrow prn = 0. 6 –no raining today rain tomorrow pnr = 0. 2 –no raining today no rain tomorrow prr = 0. 8 7

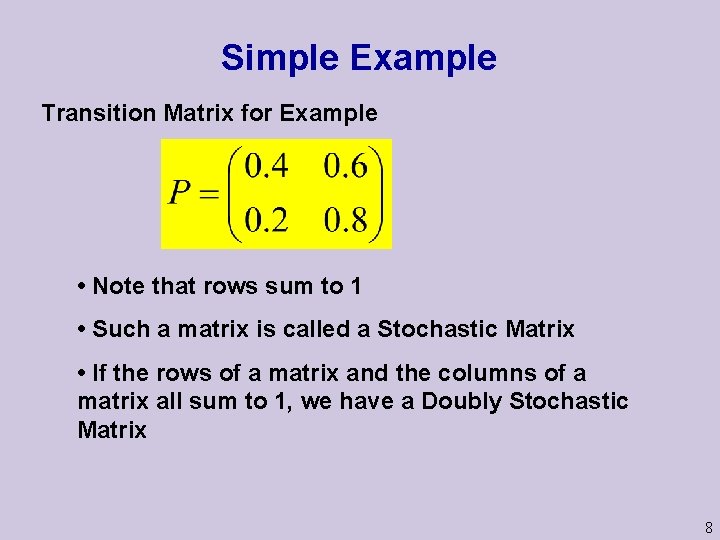
Simple Example Transition Matrix for Example • Note that rows sum to 1 • Such a matrix is called a Stochastic Matrix • If the rows of a matrix and the columns of a matrix all sum to 1, we have a Doubly Stochastic Matrix 8

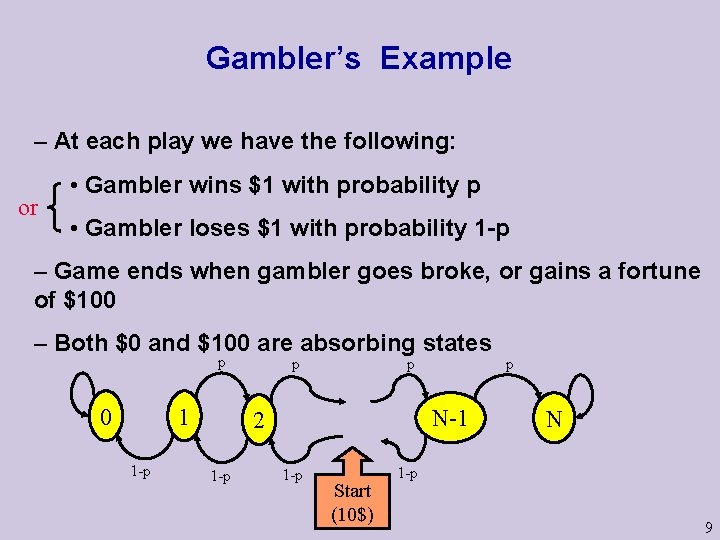
Gambler’s Example – At each play we have the following: or • Gambler wins $1 with probability p • Gambler loses $1 with probability 1 -p – Game ends when gambler goes broke, or gains a fortune of $100 – Both $0 and $100 are absorbing states p 0 1 1 -p p p N-1 2 1 -p Start (10$) p N 1 -p 9

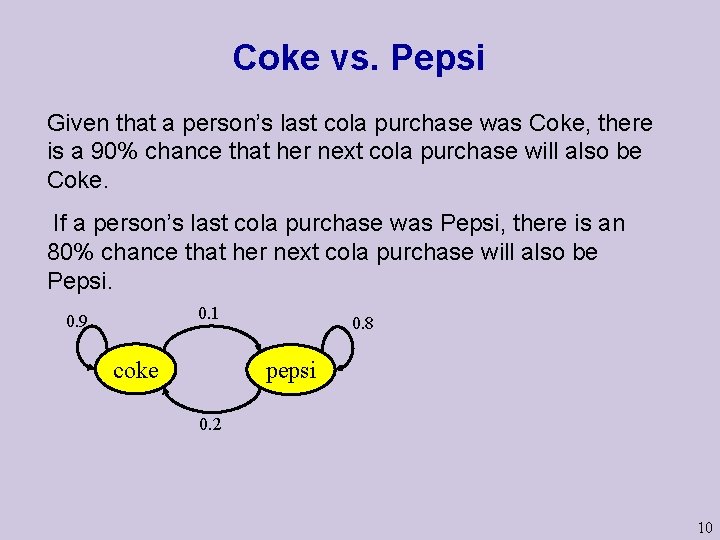
Coke vs. Pepsi Given that a person’s last cola purchase was Coke, there is a 90% chance that her next cola purchase will also be Coke. If a person’s last cola purchase was Pepsi, there is an 80% chance that her next cola purchase will also be Pepsi. 0. 1 0. 9 coke 0. 8 pepsi 0. 2 10

Coke vs. Pepsi Given that a person is currently a Pepsi purchaser, what is the probability that she will purchase Coke two purchases from now? The transition matrix is: (Corresponding to one purchase ahead) 11

Coke vs. Pepsi Given that a person is currently a Coke drinker, what is the probability that she will purchase Pepsi three purchases from now? 12

Coke vs. Pepsi Assume each person makes one cola purchase per week. Suppose 60% of all people now drink Coke, and 40% drink Pepsi. What fraction of people will be drinking Coke three weeks from now? Let (Q 0, Q 1)=(0. 6, 0. 4) be the initial probabilities. P 00 We will regard Coke as 0 and Pepsi as 1 We want to find P(X 3=0) 13

“Good” Markov chains For certain Markov Chains, the distributions Xi , as i ∞: (1) converge to a unique distribution, independent of the initial distribution. (2) In that unique distribution, each state has a positive probability. Call these Markov Chain “good”. We describe these “good” Markov Chains by considering Graph representation of Stochastic matrices. 14

Representation as a Digraph 0. 95 A 0. 95 B 0 C 0. 05 0. 2 0. 5 0 0. 3 0 0. 2 0 0. 8 0 0 1 0 D 0 A A 0. 2 0. 05 C 0. 3 0. 8 C 0. 5 B 1 D B D Each directed edge A B is associated with the positive transition probability from A to B. We now define properties of this graph which guarantee: 1. Convergence to unique distribution: 2. In that distribution, each state has positive probability. 15

Examples of “Bad” Markov Chains Markov chains are not “good” if either : 1. They do not converge to a unique distribution. 2. They do converge to u. d. , but some states in this distribution have zero probability. 16

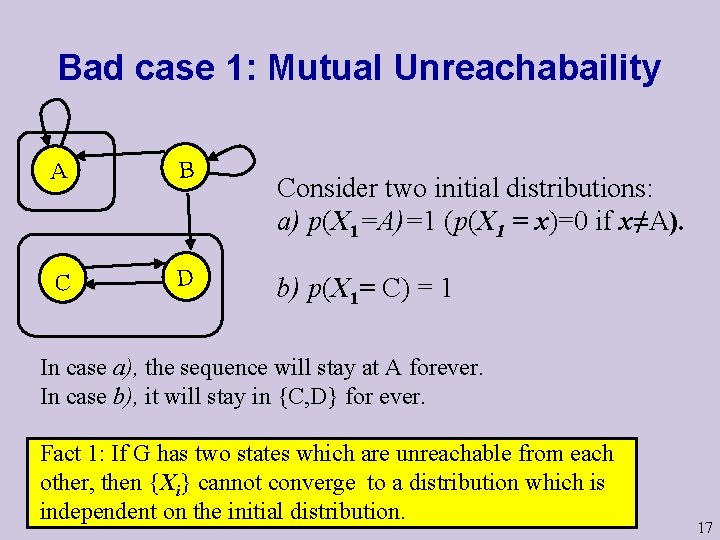
Bad case 1: Mutual Unreachabaility A B C D Consider two initial distributions: a) p(X 1=A)=1 (p(X 1 = x)=0 if x≠A). b) p(X 1= C) = 1 In case a), the sequence will stay at A forever. In case b), it will stay in {C, D} for ever. Fact 1: If G has two states which are unreachable from each other, then {Xi} cannot converge to a distribution which is independent on the initial distribution. 17

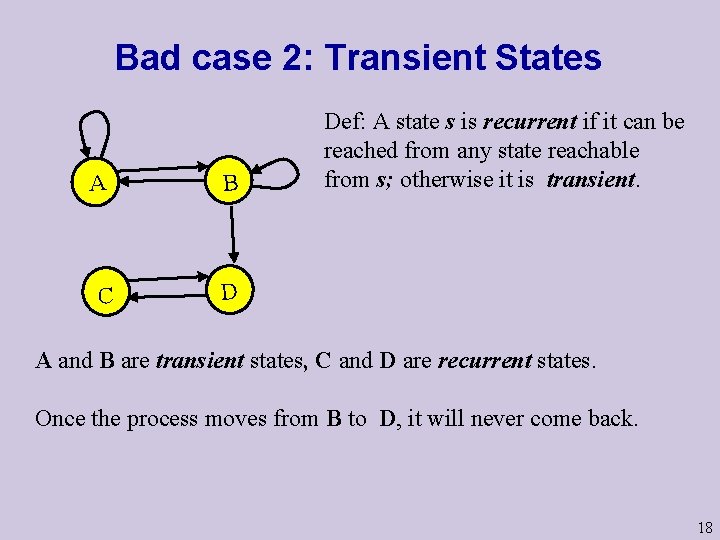
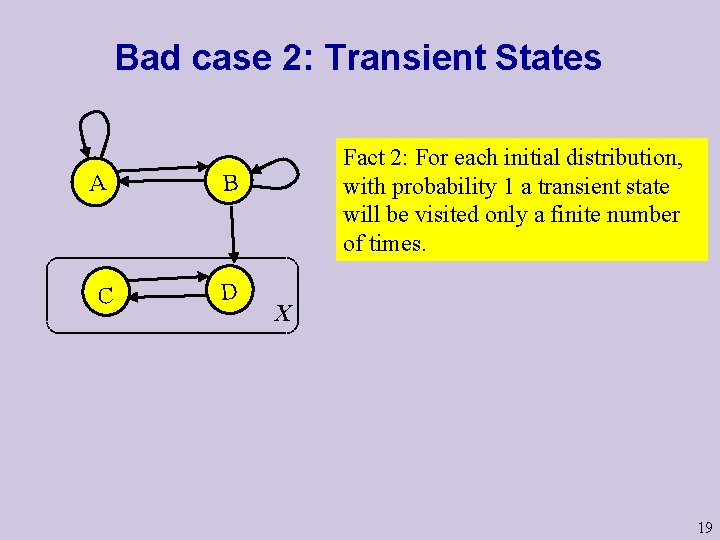
Bad case 2: Transient States A B C D Def: A state s is recurrent if it can be reached from any state reachable from s; otherwise it is transient. A and B are transient states, C and D are recurrent states. Once the process moves from B to D, it will never come back. 18

Bad case 2: Transient States A B C D Fact 2: For each initial distribution, with probability 1 a transient state will be visited only a finite number of times. X 19

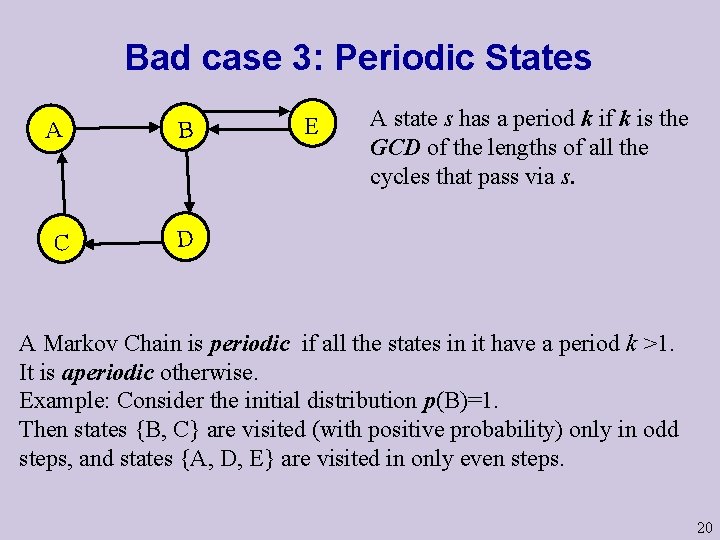
Bad case 3: Periodic States A B C D E A state s has a period k if k is the GCD of the lengths of all the cycles that pass via s. A Markov Chain is periodic if all the states in it have a period k >1. It is aperiodic otherwise. Example: Consider the initial distribution p(B)=1. Then states {B, C} are visited (with positive probability) only in odd steps, and states {A, D, E} are visited in only even steps. 20

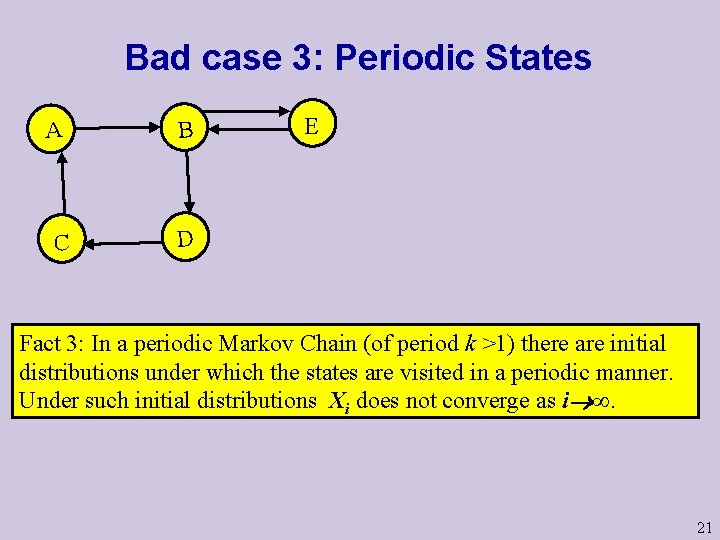
Bad case 3: Periodic States A B C D E Fact 3: In a periodic Markov Chain (of period k >1) there are initial distributions under which the states are visited in a periodic manner. Under such initial distributions Xi does not converge as i ∞. 21

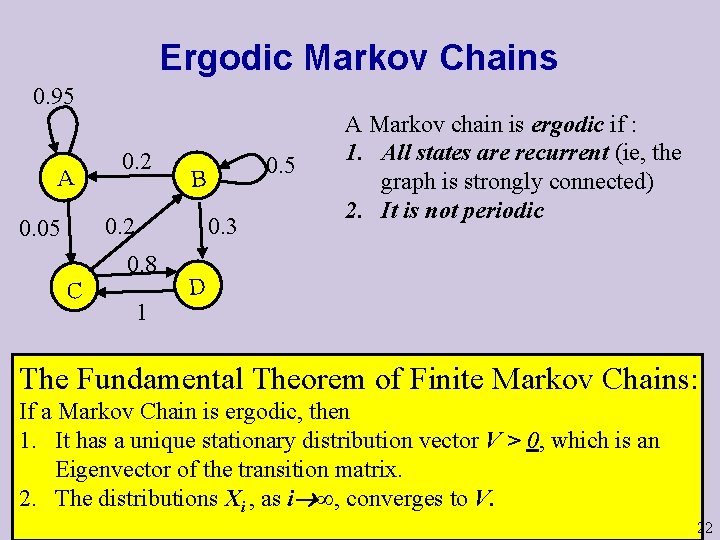
Ergodic Markov Chains 0. 95 A 0. 2 0. 05 0. 3 0. 8 C 0. 5 B 1 A Markov chain is ergodic if : 1. All states are recurrent (ie, the graph is strongly connected) 2. It is not periodic D The Fundamental Theorem of Finite Markov Chains: If a Markov Chain is ergodic, then 1. It has a unique stationary distribution vector V > 0, which is an Eigenvector of the transition matrix. 2. The distributions Xi , as i ∞, converges to V. 22

Use of Markov Chains in Genome search: Modeling Cp. G Islands In human genomes the pair CG often transforms to (methyl-C) G which often transforms to TG. Hence the pair CG appears less than expected from what is expected from the independent frequencies of C and G alone. Due to biological reasons, this process is sometimes suppressed in short stretches of genomes such as in the start regions of many genes. These areas are called Cp. G islands (p denotes “pair”). 23

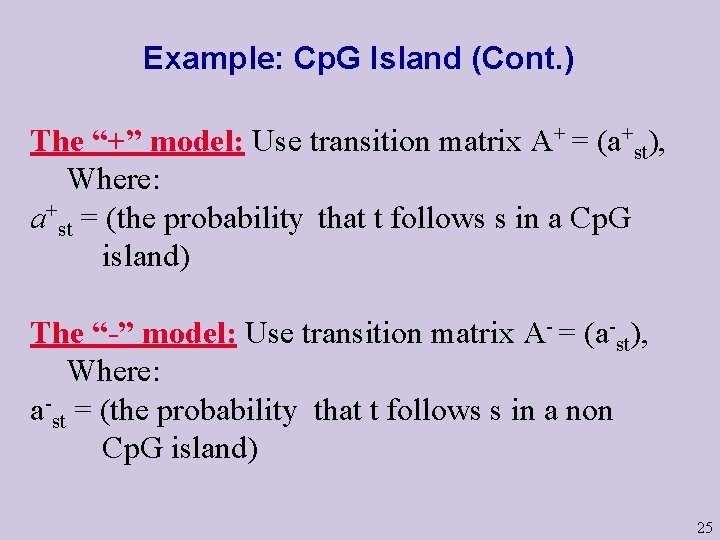
Example: Cp. G Island (Cont. ) We consider two questions (and some variants): Question 1: Given a short stretch of genomic data, does it come from a Cp. G island ? Question 2: Given a long piece of genomic data, does it contain Cp. G islands in it, where, what length ? We “solve” the first question by modeling strings with and without Cp. G islands as Markov Chains over the same states {A, C, G, T} but different transition probabilities: 24

Example: Cp. G Island (Cont. ) The “+” model: Use transition matrix A+ = (a+st), Where: a+st = (the probability that t follows s in a Cp. G island) The “-” model: Use transition matrix A- = (a-st), Where: a-st = (the probability that t follows s in a non Cp. G island) 25

Example: Cp. G Island (Cont. ) With this model, to solve Question 1 we need to decide whether a given short sequence of letters is more likely to come from the “+” model or from the “–” model. This is done by using the definitions of Markov Chain. [to solve Question 2 we need to decide which parts of a given long sequence of letters is more likely to come from the “+” model, and which parts are more likely to come from the “–” model. This is done by using the Hidden Markov Model, to be defined later. ] We start with Question 1: 26

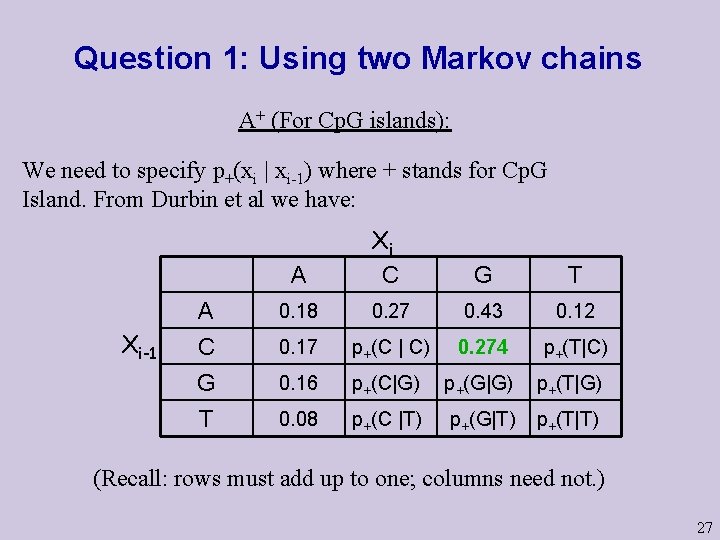
Question 1: Using two Markov chains A+ (For Cp. G islands): We need to specify p+(xi | xi-1) where + stands for Cp. G Island. From Durbin et al we have: Xi-1 A Xi C G T A C G 0. 18 0. 27 0. 43 0. 12 0. 17 p+(C | C) 0. 274 p+(T|C) 0. 16 p+(C|G) p+(G|G) p+(T|G) T 0. 08 p+(C |T) p+(G|T) p+(T|T) (Recall: rows must add up to one; columns need not. ) 27

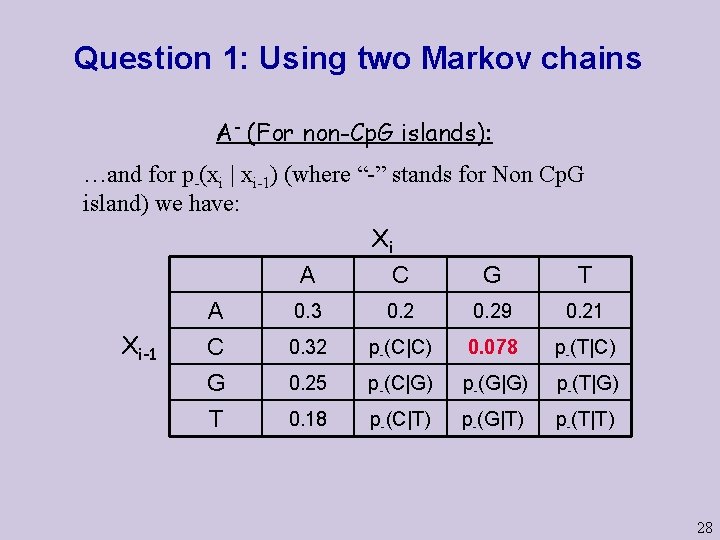
Question 1: Using two Markov chains A- (For non-Cp. G islands): …and for p-(xi | xi-1) (where “-” stands for Non Cp. G island) we have: Xi Xi-1 A C G T A C G 0. 3 0. 29 0. 21 0. 32 p-(C|C) 0. 078 p-(T|C) 0. 25 p-(C|G) p-(G|G) p-(T|G) T 0. 18 p-(C|T) p-(G|T) p-(T|T) 28

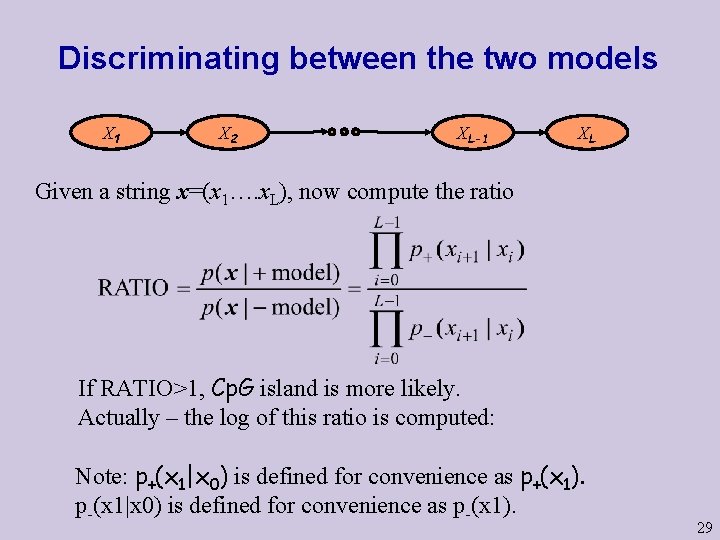
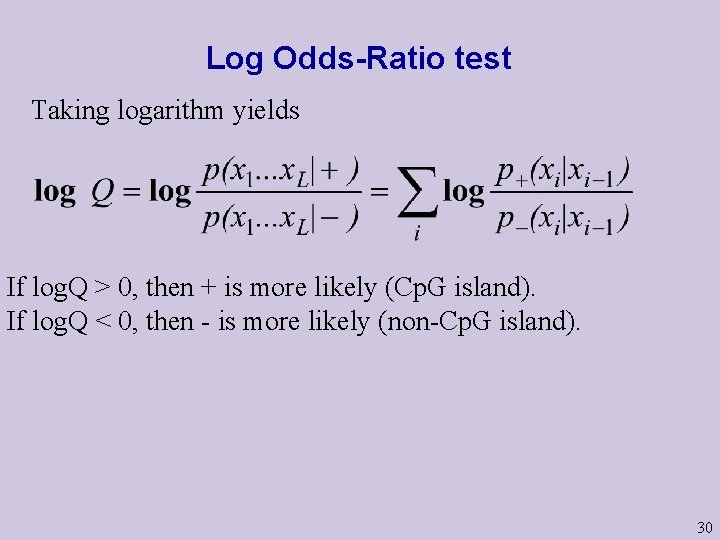
Discriminating between the two models X 1 X 2 XL-1 XL Given a string x=(x 1…. x. L), now compute the ratio If RATIO>1, Cp. G island is more likely. Actually – the log of this ratio is computed: Note: p+(x 1|x 0) is defined for convenience as p+(x 1). p-(x 1|x 0) is defined for convenience as p-(x 1). 29

Log Odds-Ratio test Taking logarithm yields If log. Q > 0, then + is more likely (Cp. G island). If log. Q < 0, then - is more likely (non-Cp. G island). 30

Where do the parameters (transitionprobabilities) come from ? Learning from complete data, namely, when the label is given and every xi is measured: Source: A collection of sequences from Cp. G islands, and a collection of sequences from non-Cp. G islands. Input: Tuples of the form (x 1, …, x. L, h), where h is + or Output: Maximum Likelihood parameters (MLE) Count all pairs (Xi=a, Xi-1=b) with label +, and with label -, say the numbers are Nba, + and Nba, -. 31

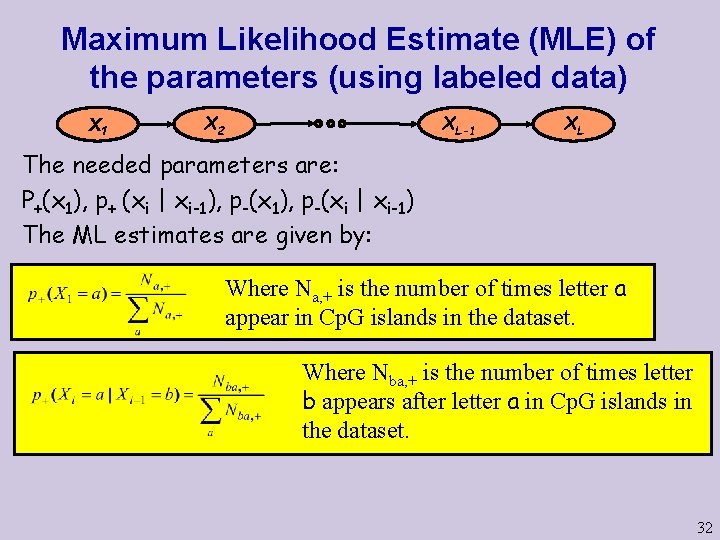
Maximum Likelihood Estimate (MLE) of the parameters (using labeled data) X 1 X 2 XL-1 XL The needed parameters are: P+(x 1), p+ (xi | xi-1), p-(xi | xi-1) The ML estimates are given by: Where Na, + is the number of times letter a appear in Cp. G islands in the dataset. Where Nba, + is the number of times letter b appears after letter a in Cp. G islands in the dataset. 32