Introduction to Single Nucleotide Polymorphisms SNPs Zhongming Zhao









- Slides: 37

Introduction to Single Nucleotide Polymorphisms (SNPs) Zhongming Zhao Department of Psychiatry and Center for the Study of Biological Complexity June 28, 2004 Email: zzhao@vcu. edu

Organization § Introduction to single nucleotide polymorphism (SNPs) § An overview of mammalian genome projects § Online resource of SNPs and genome sequences

SNPs are DNA sequence variations that occur when a single nucleotide (A, T, C, or G) is altered (a single base variation).

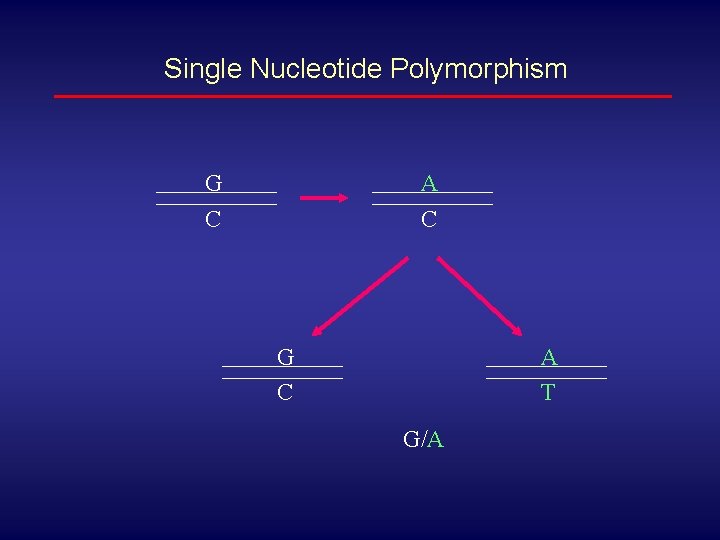
Single Nucleotide Polymorphism G A C C G C A T G/A

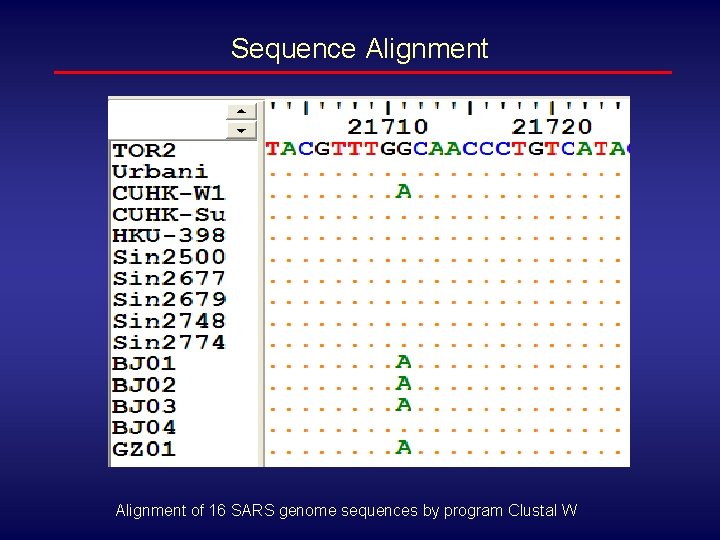
Sequence Alignment of 16 SARS genome sequences by program Clustal W

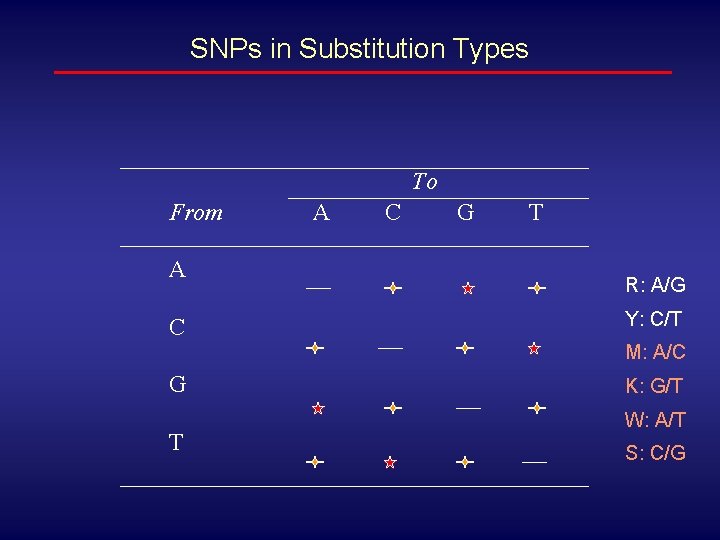
SNPs in Substitution Types To From A C A C G T R: A/G Y: C/T M: A/C G T K: G/T W: A/T S: C/G

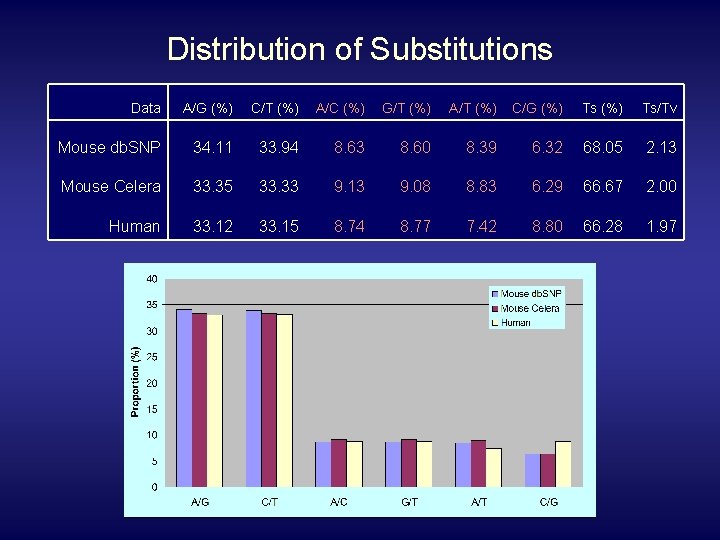
Distribution of Substitutions Data A/G (%) C/T (%) A/C (%) G/T (%) A/T (%) C/G (%) Ts/Tv Mouse db. SNP 34. 11 33. 94 8. 63 8. 60 8. 39 6. 32 68. 05 2. 13 Mouse Celera 33. 35 33. 33 9. 13 9. 08 8. 83 6. 29 66. 67 2. 00 Human 33. 12 33. 15 8. 74 8. 77 7. 42 8. 80 66. 28 1. 97

SNPs are Valuable Tools in Genetic Analysis § Disease Studies − Causes of genetic diseases − Association studies of complex diseases § Population Studies − Population structures and history − Haplotype analysis § Functional Analysis − Pharmacogenomics § Genome Mapping − Dense/fine marker set − Haplotype map § Comparative Genomics − Genome evolution − Mechanism of molecular evolution

SNP Databases Public: § NCBI db. SNP § TSC § Whitehead Institute SNP Database § HGMD § HGBase (now HGVD) § UCSC Genome Browser § Ensembl § Mouse Phenome Database Private § Celera Ref. SNP § Sequenom Real. SNP § Incyte SNP Program

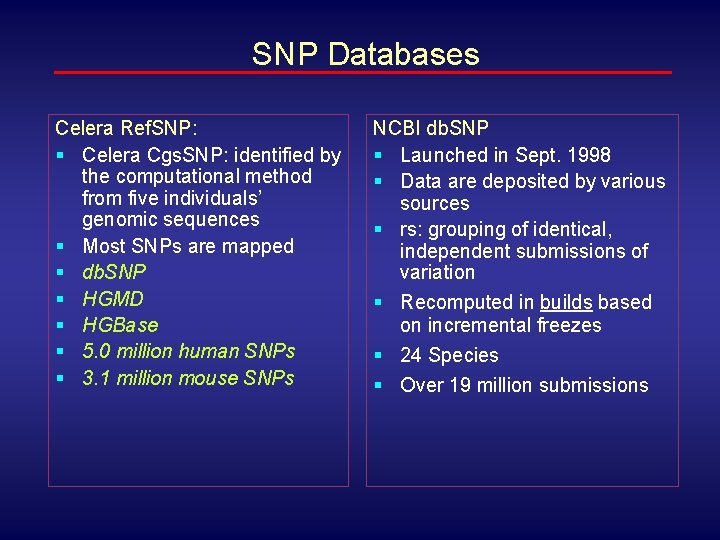
SNP Databases Celera Ref. SNP: § Celera Cgs. SNP: identified by the computational method from five individuals’ genomic sequences § Most SNPs are mapped § db. SNP § HGMD § HGBase § 5. 0 million human SNPs § 3. 1 million mouse SNPs NCBI db. SNP § Launched in Sept. 1998 § Data are deposited by various sources § rs: grouping of identical, independent submissions of variation § Recomputed in builds based on incremental freezes § 24 Species § Over 19 million submissions

NCBI db. SNP

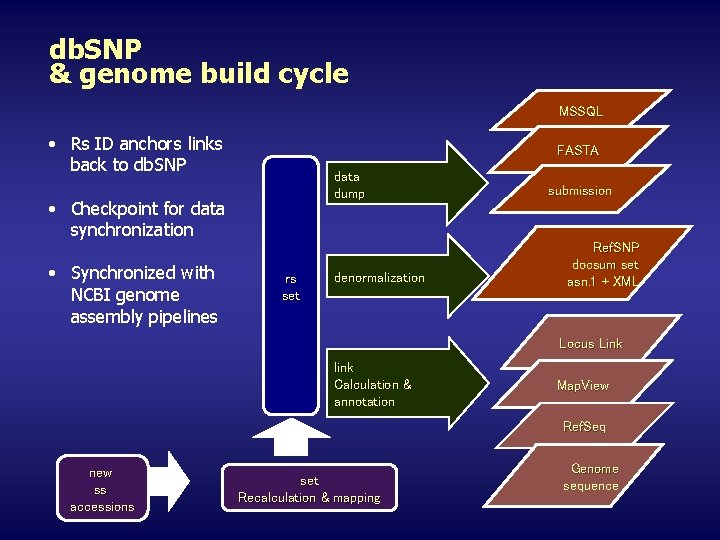
db. SNP & genome build cycle MSSQL • Rs ID anchors links back to db. SNP FASTA data dump • Checkpoint for data synchronization • Synchronized with NCBI genome assembly pipelines rs set denormalization submission Ref. SNP docsum set asn. 1 + XML Locus Link link Calculation & annotation Map. View Ref. Seq new ss accessions set Recalculation & mapping Genome sequence

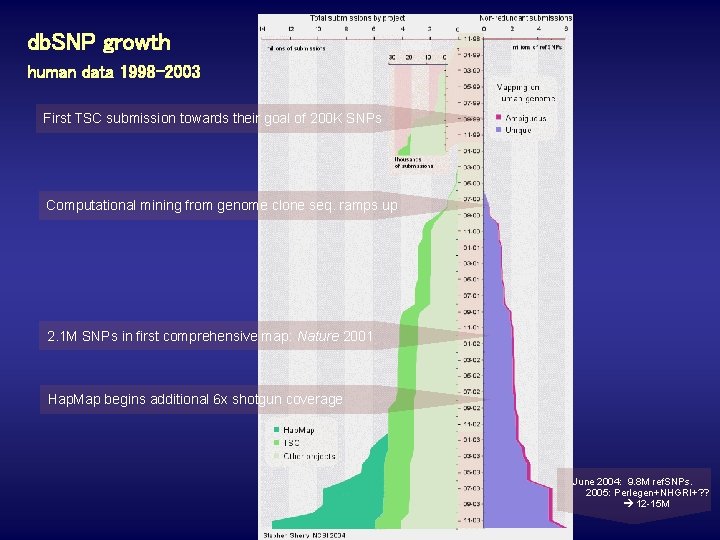
db. SNP growth human data 1998 -2003 First TSC submission towards their goal of 200 K SNPs Computational mining from genome clone seq. ramps up 2. 1 M SNPs in first comprehensive map: Nature 2001 Hap. Map begins additional 6 x shotgun coverage June 2004: 9. 8 M ref. SNPs. 2005: Perlegen+NHGRI+? ? 12 -15 M

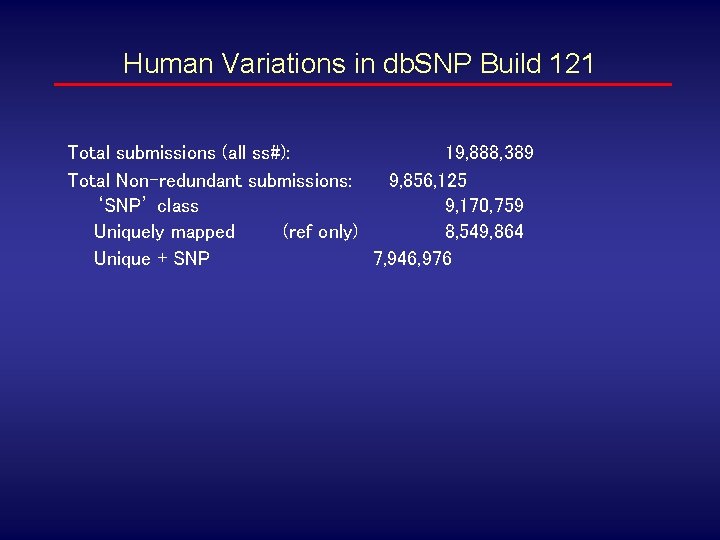
Human Variations in db. SNP Build 121 Total submissions (all ss#): 19, 888, 389 Total Non-redundant submissions: 9, 856, 125 ‘SNP’ class 9, 170, 759 Uniquely mapped (ref only) 8, 549, 864 Unique + SNP 7, 946, 976

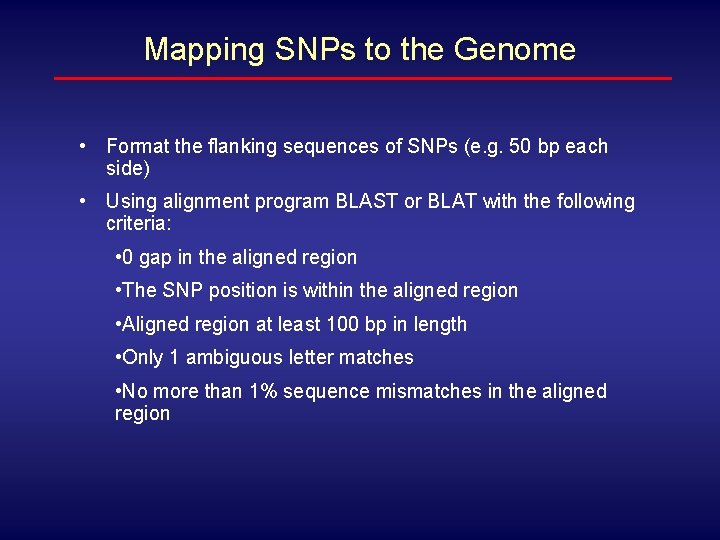
Mapping SNPs to the Genome • Format the flanking sequences of SNPs (e. g. 50 bp each side) • Using alignment program BLAST or BLAT with the following criteria: • 0 gap in the aligned region • The SNP position is within the aligned region • Aligned region at least 100 bp in length • Only 1 ambiguous letter matches • No more than 1% sequence mismatches in the aligned region

Most SNPs Map Uniquely during Genome Annotation

FASTA Format and Data Structure for a rs Record define for FASTA records start with ">" | object-type=general | | database name | | | offset tax. ID list of | | | rs# | length | SNP class alleles | | | | | define: >gnl|db. SNP|rs 271_allele. Pos=51 totallen=101|taxid=9606|snp. Class=1|alleles='G/A' 5' sequence: CTGCATCACA TGTACTGATT CTGTCCATTG GAACAGAGAT GATGACTGGT variation: R 3' sequence: TTACTAAACC CTGAGCCCTG GTGTTTCTGT TGATAGGGGG TTGCATTGAT http: //www. ncbi. nlm. nih. gov/SNP/snp_ref. cgi? rs=rs 271

The SNP Consortium (TSC)

The SNP Consortium (TSC) • The SNP Consortium (TSC) is a public/private collaboration that has to date discovered and characterized nearly 1. 8 million SNPs • The TSC was funded by 11 corporate members and the Wellcome Trust. • Started in April 1999 and that time its mission is to develop up to 300, 000 SNPs distributed evenly throughout the human genome. Finally, in 2001, it finished by 1. 5 million SNPs • Well designed. Good quality of SNP data and allele frequencies.

Celera CDS

The Sequenom’s Real. SNP • Aims to develop assays for Sequenom’s Mass Spec Genotyping machine. • Most candidate SNPs were obtained from db. SNPs, some were from Incyte’s proprietary SNPs • Started in 2002 • Over 5. 4 M designed SNP assays • Over 400, 000 working assays • Over 220, 000 confirmed polymorphic SNPs

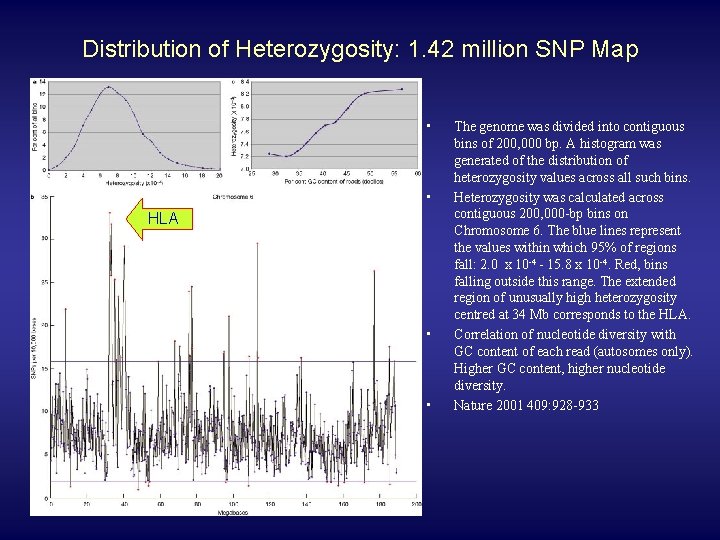
Distribution of Heterozygosity: 1. 42 million SNP Map • • HLA • • The genome was divided into contiguous bins of 200, 000 bp. A histogram was generated of the distribution of heterozygosity values across all such bins. Heterozygosity was calculated across contiguous 200, 000 -bp bins on Chromosome 6. The blue lines represent the values within which 95% of regions fall: 2. 0 x 10 -4 - 15. 8 x 10 -4. Red, bins falling outside this range. The extended region of unusually high heterozygosity centred at 34 Mb corresponds to the HLA. Correlation of nucleotide diversity with GC content of each read (autosomes only). Higher GC content, higher nucleotide diversity. Nature 2001 409: 928 -933

• To develop a haplotype map of the human genome • To describe the common patterns of human DNA sequence variation • U. S. A. , Japan, the U. K. , Canada, China, and Nigeria • Over A total of 270 people • Yoruba, Nigeria (30 both-parent-and-adult-child trios) • Japanese (45 unrelated individuals) • Han Chinese (45 unrelated individuals) • CEPH (30 trios) • Genotyped for at least 1 million SNPs evenly across the human genome

The Human Genome & Variation Science February 2001 Nature February 2001

The Rodent Genome & Variation December 5, 2002 Nature April 1, 2004

Human Genome Sequencing Project § International Human Genome Sequencing Consortium (IHGSC) − A collaboration of 20 groups from the USA, the United Kingdom, Japan, France, Germany, and China − Goals: DNA sequence, genetic map, physical map, genetic variation, functional analysis, etc. − A 15 -year $3 billion project (1990 -2005, finished 2001) − Hierarchical shotgun sequencing strategy § Celera Human Genome Project − Compete IHGSC from the biotech industry − Whole-genome shotgun sequencing (WGS) strategy − DNA samples from five individuals, mainly from Craig Venter § Many follow-up studies Chromosome 6, 7, 9, 10, 13, 14, 16, 19, 20, 21, 22 Comparative genomics Nature 2001 409: 860 -921 Science 2001 291: 1304 -1351 Science 2003 300: 286 -290

The Automatic Production Line at the Whitehead Genome Sequencing Center

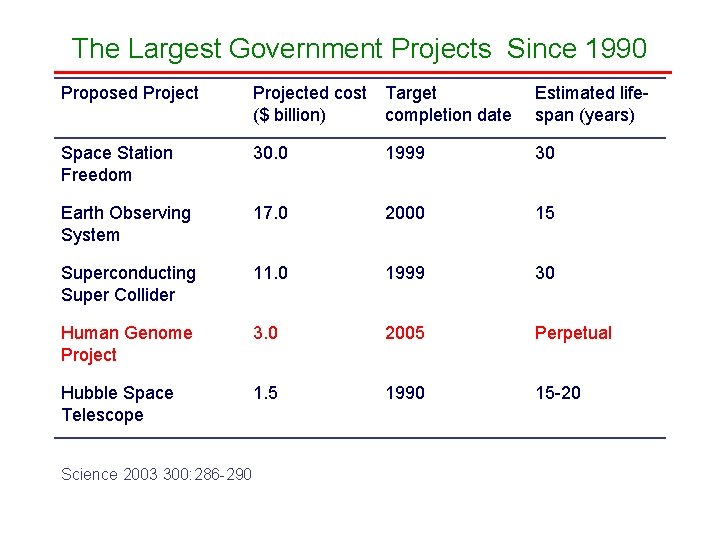
The Largest Government Projects Since 1990 Proposed Projected cost ($ billion) Target completion date Estimated lifespan (years) Space Station Freedom 30. 0 1999 30 Earth Observing System 17. 0 2000 15 Superconducting Super Collider 11. 0 1999 30 Human Genome Project 3. 0 2005 Perpetual Hubble Space Telescope 1. 5 1990 15 -20 Science 2003 300: 286 -290

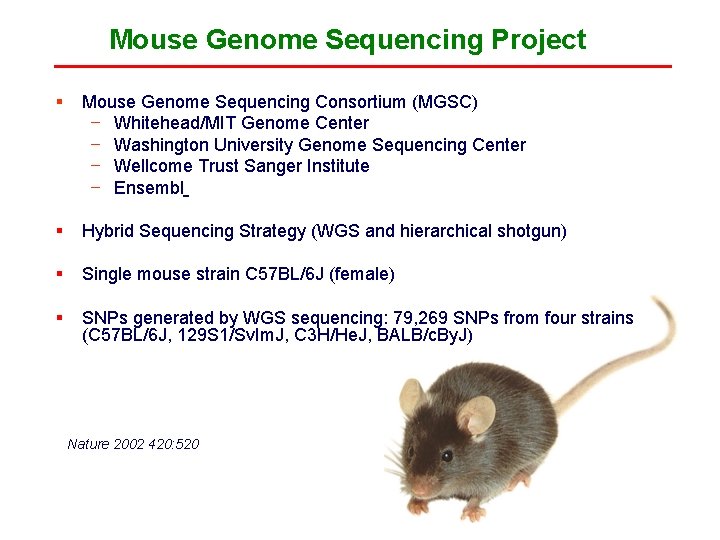
Mouse Genome Sequencing Project § Mouse Genome Sequencing Consortium (MGSC) − Whitehead/MIT Genome Center − Washington University Genome Sequencing Center − Wellcome Trust Sanger Institute − Ensembl § Hybrid Sequencing Strategy (WGS and hierarchical shotgun) § Single mouse strain C 57 BL/6 J (female) § SNPs generated by WGS sequencing: 79, 269 SNPs from four strains (C 57 BL/6 J, 129 S 1/Sv. Im. J, C 3 H/He. J, BALB/c. By. J) Nature 2002 420: 520

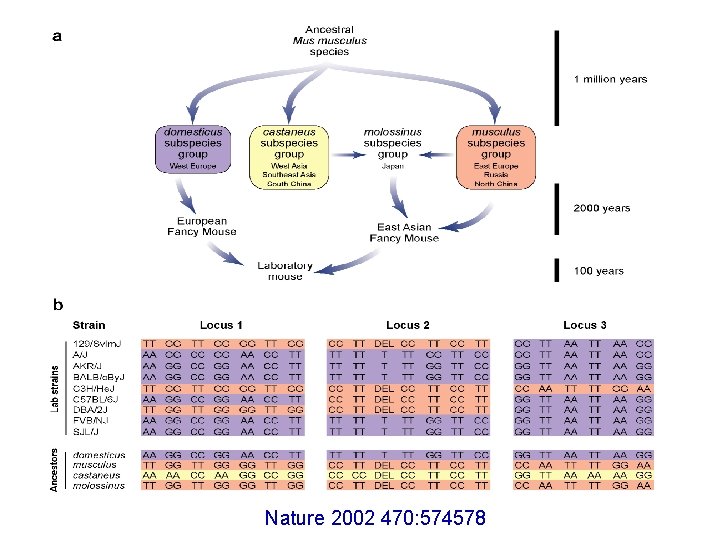
Nature 2002 470: 574578

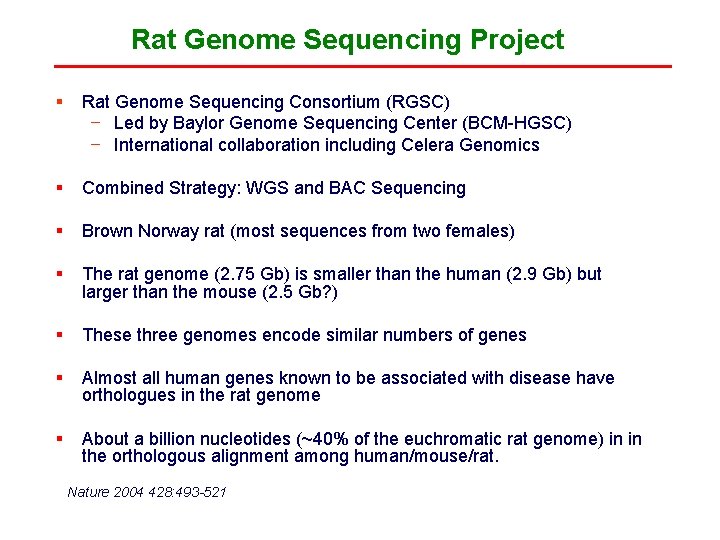
Rat Genome Sequencing Project § Rat Genome Sequencing Consortium (RGSC) − Led by Baylor Genome Sequencing Center (BCM-HGSC) − International collaboration including Celera Genomics § Combined Strategy: WGS and BAC Sequencing § Brown Norway rat (most sequences from two females) § The rat genome (2. 75 Gb) is smaller than the human (2. 9 Gb) but larger than the mouse (2. 5 Gb? ) § These three genomes encode similar numbers of genes § Almost all human genes known to be associated with disease have orthologues in the rat genome § About a billion nucleotides (~40% of the euchromatic rat genome) in in the orthologous alignment among human/mouse/rat. Nature 2004 428: 493 -521

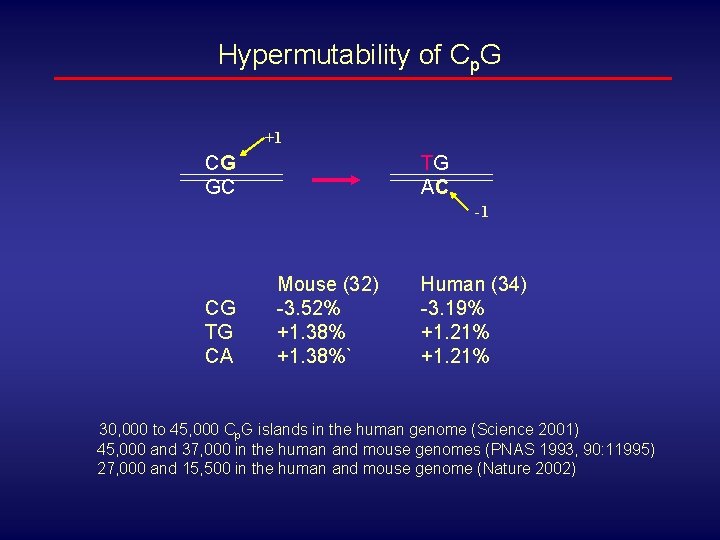
Hypermutability of Cp. G +1 CG GC TG AC -1 CG TG CA Mouse (32) -3. 52% +1. 38%` Human (34) -3. 19% +1. 21% 30, 000 to 45, 000 Cp. G islands in the human genome (Science 2001) 45, 000 and 37, 000 in the human and mouse genomes (PNAS 1993, 90: 11995) 27, 000 and 15, 500 in the human and mouse genome (Nature 2002)

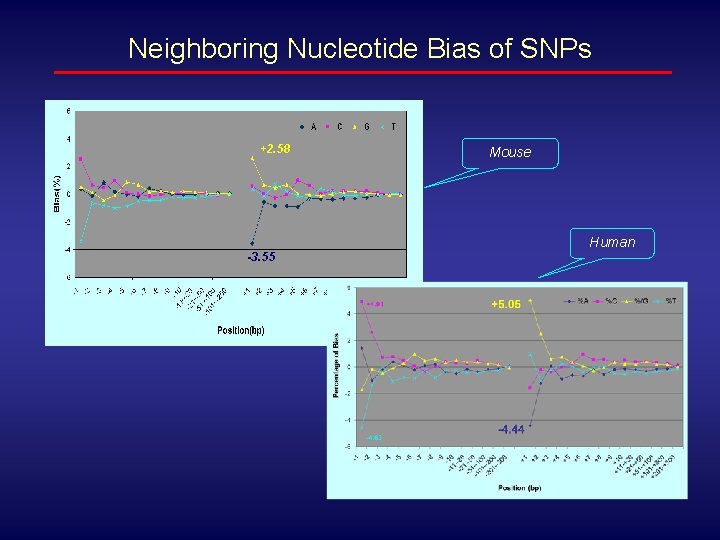
Neighboring Nucleotide Bias of SNPs +2. 58 Mouse Human -3. 55 -4. 44

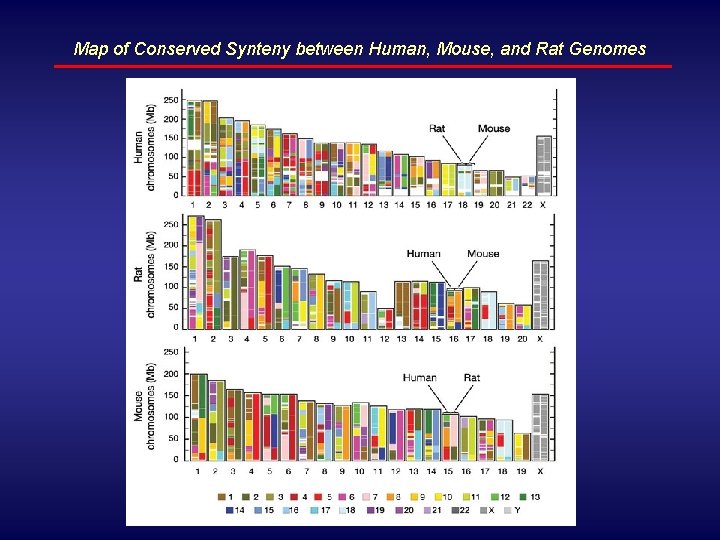
Map of Conserved Synteny between Human, Mouse, and Rat Genomes

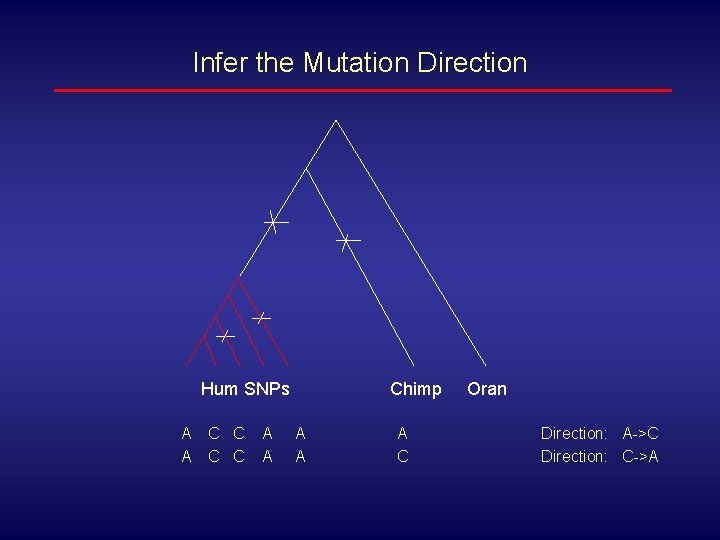
Infer the Mutation Direction • We have human SNPs with outgroup chimpanzee sequences (divergence time is about 4 -6 million years, sequence difference is about 1. 2%) • We have mouse SNPs with outgroup rat sequences (divergence time is about 12 -24 million years, sequence diversity is unknown )

Infer the Mutation Direction Hum SNPs A A C C A A Chimp A A A C Oran Direction: A->C Direction: C->A

Web Resources § NCBI db. SNP www. ncbi. nlm. nih. gov/SNP ftp. ncbi. nlm. nih. gov/snp §Celera Genomics: www. celera. com §The SNP Consortium (TSC): http: //snp. cshl. org §UCSC Genome Browser: http: //genome. ucsc. edu/ §The Human Gene Mutation Database (HGMD): http: //archive. uwcm. ac. uk/uwcm/mg/hgmd 0. html §Human Genome Variation Database (HGVD): http: //hgvbase. cgb. ki. se/ §MIT SNP database: Human: http: //www. broad. mit. edu/snp/human/ Mouse: http: //www. broad. mit. edu/snp/mouse/ §Sequenom Real. SNP: https: //www. realsnp. com/default. asp §Ensembl Genome Browser: http: //www. ensembl. org/ §The Hap. Map Project: http: //www. hapmap. org/ §Mouse Phenome Database: http: //aretha. jax. org/pub-cgi/phenome/mpdcgi? rtn=projects/details&sym=Mpd 1