In silico reconstruction of an ancestral mammalian genome
















- Slides: 45

In silico reconstruction of an ancestral mammalian genome UQAM Seminaire de bioinformatique Mathieu Blanchette

CGACTGCATCAGACGACGATCAGACTACTATATCAGCAGATTACGGT GCATCGTATTTACGCATGACGATCAGACTACGCATAGA TGCATCAGACGACGATCAGACTACTATATCAGCAGATTACGGTCGAT • Sequence of ~3*109 nucleotides TATTTACGCATGACGATCAGACTACGCATAGAGCAATA CGACTGCATCAGACGACGATCAGACTACTATATCAGCAGATTACGGT • Complete sequence is known (2001) GCGTATTTACGCATGACGATCAGACTACGCATAGAGCA CGCATCAGACGACGATCAGACTACTATATCAGCAGATTACGGTCGTA ACGTTACGCATGACGATCAGACTACGCATAGAGCCGATCATCT CAGACGACGATCAGACTACTATATCAGCAGATTACGGTGGCATACTA ATCGTATTTACGCATGACGATCAGACTACGCATAGAAA CGACGATCAGACTACTATATCAGCAGATTACGGTGCGCGAATTCATA TATTTACGCATGACGATCAGACTACGCATAGATTGATA CATCAGACGACGATCAGACTACTATATCAGCAGATTACGGTGCATAT TTTACGCATGACGATCAGACTACGCATAGAGATCATCA TCAGACGACGATCAGACTACTATATCAGCAGATTACGGTAGCATTCT CGTATTTACGCATGACGATCAGACTACGCATAGAATGC ACGACGATCAGACTACTATATCAGCAGATTACGGTGATACGAT CGTATTTACGCATGACGATCAGACTACGCATAGAGATA The Human genome O H O D W T I ES O W ? ? RK

Comparative Genomics • Goal: Functional annotation of the genome – What is the role of each region of the genome? – Very hard to answer…. • Idea: Look not only at what our genome is now, but also at how it evolved – Different types of functional regions have different evolutionary signatures • Complete genomes are sequenced for: – Human, chimp, mouse, rat, house, chicken, zebrafish, pufferfish • Partial genomes are available for: – Dog, cow, rabbit, elephant, armadillo


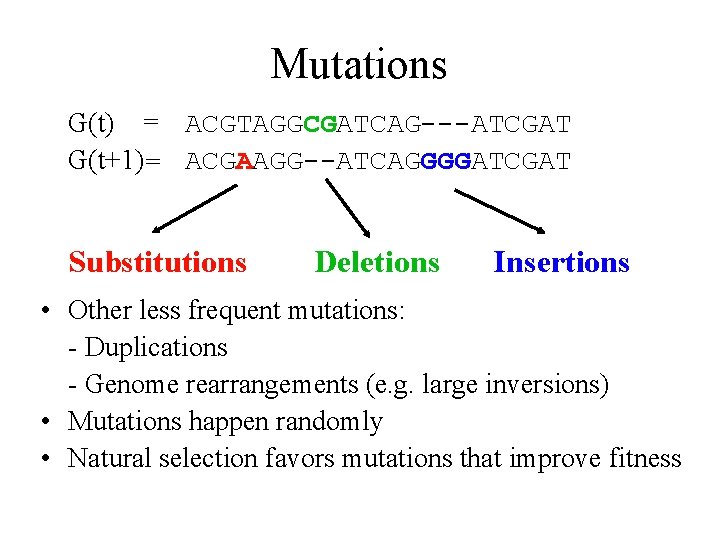
Mutations G(t) = ACGTAGGCGATCAG---ATCGAT G(t+1)= ACGAAGG--ATCAGGGGATCGAT Substitutions Deletions Insertions • Other less frequent mutations: - Duplications - Genome rearrangements (e. g. large inversions) • Mutations happen randomly • Natural selection favors mutations that improve fitness

A random walk in genome space

Mammalian evolution -Rapid radiation ~75 Myrs ago -Many nearly independent phyla -Many “noisy” copies of ancestor - Accurate reconstruction of ancestors may be feasible http: //www. broad. mit. edu/personal/jpvinson/phylogenetics/bigtree_1_0. jpg

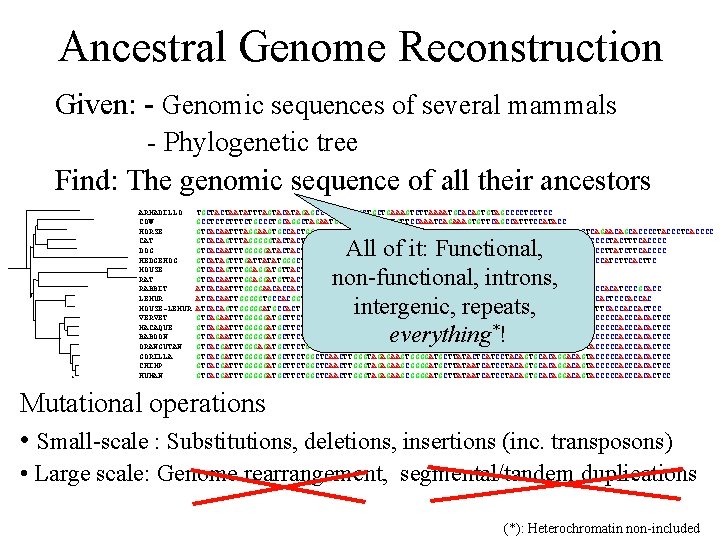
Ancestral Genome Reconstruction Given: - Genomic sequences of several mammals - Phylogenetic tree Find: The genomic sequence of all their ancestors ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSE-LEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN TGCTACTAATATTTAGTACATAGAGCCCAGGGGTGCTGCTGAAAGTCTTAAAATGCACAGTGTAGCCCCTCCTCC GCCTCTCTTTCTGCCCTGCAGGCTAGAATGTATCACTTAGATGTTCCAAATCAGAAAGTGTTCAGCCATTTCCATACC GTCACAATTTAGGAAGTGCCACTGGCCTCTAGAGGGTAGAAGACAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTACCCTCACCCC GTCACAGTTTAGGGGGTACTACTGGCATCTATCGGGTGGAGGATAGGGATACTGATAATCATTCTACAGTGCACAGGACAGTACCCCTACTTTCACCCC GTCACAATTTGGGGGATACTACTGGCATCTAATGGGTAGAGGACAGGGATACTGATAATTGCTTTACAGTGCACAGGACAGCACCCTTATCTTCACCCC GTCATAGTTTGATTATATGGGCTTCTTAGTAGACAAAGAAAAAGATGTTCTGGTAGTCATTCTGCTTTCCATATGATAGCACTCCCATCTTCACTTC GTCACAGTTTGGAGGATGTTACTGACATCTAGAGAGTAGACTTTAAAGATACTGATAGTCACCCCATTGTGCACCTCC GTCACAATTTGGAGGATGTTACTGGCATCTAGAGAGTAGACTTTAAGGACACTGATAATCATACTATGCTGCACTTCC ATCACAATTTGGGGAACACCACTGGCATCTCGGGTAGCAGGCATGCTGGTAATTATACTACAGTGCACAGTTCCCCACATCCCGCACC ATCACAATTGGGGGTGCCACGGTCCTCCAGTGGGTAGAGAACAGGGAGGCTGATAACCACCCTGCAGTGCACAGGGCAGTGCCCCACTCCCACCAC ATCACAGTTGGGGGATGCCACTGGCCTCAAGTGGGTAGAGAACAGGGAGGCTGAAAACCACCCTGCAGAGCACGGGGCAGTGCCTTCACCACCACTCC GTCAGAATTTGGGGGATGCTTCTGGCTCTACTTGGGTAGAGAAACAGGGATGCTTATAATCATCCTACAGTGCACAGGACAGTACCCCCACACTCC GTCAGAATTTGGGGGATGCTTCTGGCTCTACTTGGGTAGAGAAACAGGAATGCTTATAATCATCCTACAGTGCACAGGTCAGTACCCCCACACTCC GTCAGAATTTGGGGGATGCTTCTGGCTCTACTTGGGTAGAAAAACAGGGATGCTTATAATCATCCTACAGTGCACAGGACAGTACCCCCACACTCC GTCACGATTTGGGAGATGCTTCTGGCTCGACTTGGGTAGAGAAGCGGGGATGCTTATAATCATCCAACAGTGCACAGGACAGTACCCCCACACTCC GTCACGATTTGGGGGATGCTTCTGGCTCAACTTGGGTAGAGAAGTGGGGATGCTTATACTCATCCTACAGTGCACAGGACAGTACCCCCACCCACACTCC GTCACGATTTGGGGGATGCTTCTGGCTCAACTTGGGTAGAGAAGCGGGGATGCTTATAATCATCCTACAGTGCACAGGACAGTACCCCCACACTCC All of it: Functional, non-functional, introns, intergenic, repeats, everything*! Mutational operations • Small-scale : Substitutions, deletions, insertions (inc. transposons) • Large scale: Genome rearrangement, segmental/tandem duplications (*): Heterochromatin non-included

Reconstruction algorithm 1) Identify syntenic regions in each species • Blastz (Schwartz et al. ) and Chaining/netting program (Kent et al. ) • In ENCODE case: targeted BAC sequencing

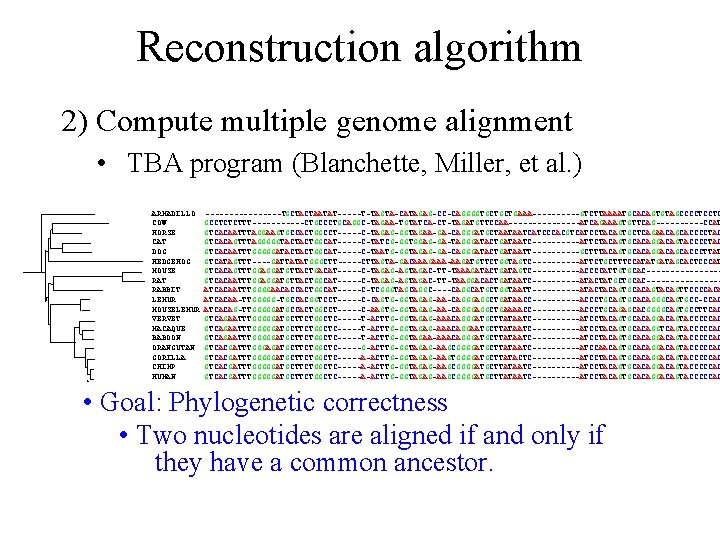
Reconstruction algorithm 2) Compute multiple genome alignment • TBA program (Blanchette, Miller, et al. ) ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSELEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN --------TGCTACTAATAT-----T-TAGTA-CATAGAG-CC-CAGGGGTGCTGCTGAAA-----GTCTTAAAATGCACAGTGTAGCCCCTCCTC GCCTCTCTTT------CTGCCCTGCAGGC-TAGAA-TGTATCA-CT-TAGATGTTCCAA--------ATCAGAAAGTGTTCAG-----CCAT GTCACAATTTAGGAAGTGCCACTGGCCT-----C-TAGAG-GGTAGAA-GA-CAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTAC GTCACAGTTTAGGGGGTACTACTGGCAT-----C-TATCG-GGTGGAG-GA-TAGGGATACTGATAATC-----ATTCTACAGTGCACAGGACAGTACCCCTAC GTCACAATTTGGGGGATACTACTGGCAT-----C-TAATG-GGTAGAG-GA-CAGGGATACTGATAATT-----GCTTTACAGTGCACAGGACAGCACCCTTAT GTCATAGTTT----GATTATATGGGCTT-----CTTAGTA-GACAAAGAAA-AAGATGTTCTGGTAGTC-----ATTCTGCTTTCCATATGATAGCACTCCCAT GTCACAGTTTGGAGGATGTTACTGACAT-----C-TAGAG-AGTAGAC-TT-TAAAGATACTGATAGTC-----ACCCCATTGTGCAC--------GTCACAATTTGGAGGATGTTACTGGCAT-----C-TAGAG-AGTAGAC-TT-TAAGGACACTGATAATC-----ATACTATGCTGCAC--------ATCACAATTTGGGGAACACCACTGGCAT-----C-TCGGGTAGCAGGC----CAGGCATGCTGGTAATT-----ATACTACAGTGCACAGTTCCCCACA ATCACAA-TTGGGGG-TGCCACGGTCCT-----C-CAGTG-GGTAGAG-AA-CAGGGAGGCTGATAACC-----ACCCTGCAGTGCACAGGGCAGTGCC-CCAC ATCACAG-TTGGGGGATGCCACTGGCCT-----C-AAGTG-GGTAGAG-AA-CAGGGAGGCTGAAAACC-----ACCCTGCAGAGCACGGGGCAGTGCCTTCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGAATGCTTATAATC-----ATCCTACAGTGCACAGGTCAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAA-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGAGATGCTTCTGGCTC-----G-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCAACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGTGGGGATGCTTATACTC----------ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC • Goal: Phylogenetic correctness • Two nucleotides are aligned if and only if they have a common ancestor.

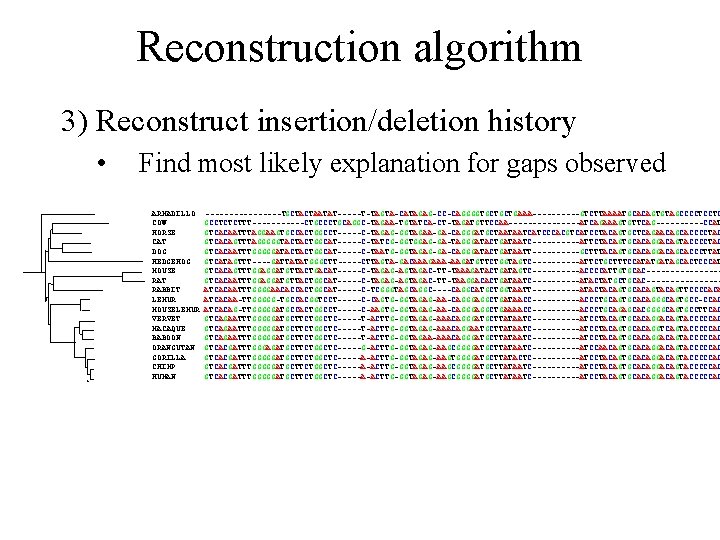
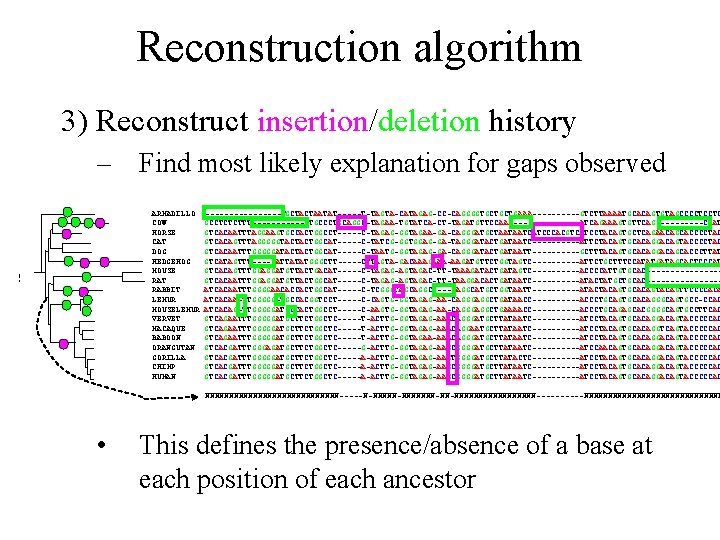
Reconstruction algorithm 3) Reconstruct insertion/deletion history • Find most likely explanation for gaps observed ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSELEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN --------TGCTACTAATAT-----T-TAGTA-CATAGAG-CC-CAGGGGTGCTGCTGAAA-----GTCTTAAAATGCACAGTGTAGCCCCTCCTC GCCTCTCTTT------CTGCCCTGCAGGC-TAGAA-TGTATCA-CT-TAGATGTTCCAA--------ATCAGAAAGTGTTCAG-----CCAT GTCACAATTTAGGAAGTGCCACTGGCCT-----C-TAGAG-GGTAGAA-GA-CAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTAC GTCACAGTTTAGGGGGTACTACTGGCAT-----C-TATCG-GGTGGAG-GA-TAGGGATACTGATAATC-----ATTCTACAGTGCACAGGACAGTACCCCTAC GTCACAATTTGGGGGATACTACTGGCAT-----C-TAATG-GGTAGAG-GA-CAGGGATACTGATAATT-----GCTTTACAGTGCACAGGACAGCACCCTTAT GTCATAGTTT----GATTATATGGGCTT-----CTTAGTA-GACAAAGAAA-AAGATGTTCTGGTAGTC-----ATTCTGCTTTCCATATGATAGCACTCCCAT GTCACAGTTTGGAGGATGTTACTGACAT-----C-TAGAG-AGTAGAC-TT-TAAAGATACTGATAGTC-----ACCCCATTGTGCAC--------GTCACAATTTGGAGGATGTTACTGGCAT-----C-TAGAG-AGTAGAC-TT-TAAGGACACTGATAATC-----ATACTATGCTGCAC--------ATCACAATTTGGGGAACACCACTGGCAT-----C-TCGGGTAGCAGGC----CAGGCATGCTGGTAATT-----ATACTACAGTGCACAGTTCCCCACA ATCACAA-TTGGGGG-TGCCACGGTCCT-----C-CAGTG-GGTAGAG-AA-CAGGGAGGCTGATAACC-----ACCCTGCAGTGCACAGGGCAGTGCC-CCAC ATCACAG-TTGGGGGATGCCACTGGCCT-----C-AAGTG-GGTAGAG-AA-CAGGGAGGCTGAAAACC-----ACCCTGCAGAGCACGGGGCAGTGCCTTCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGAATGCTTATAATC-----ATCCTACAGTGCACAGGTCAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAA-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGAGATGCTTCTGGCTC-----G-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCAACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGTGGGGATGCTTATACTC----------ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC

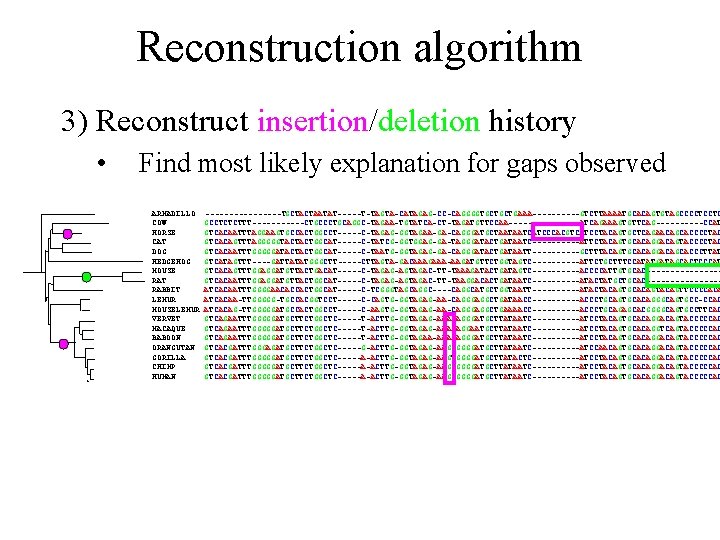
Reconstruction algorithm 3) Reconstruct insertion/deletion history • Find most likely explanation for gaps observed ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSELEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN --------TGCTACTAATAT-----T-TAGTA-CATAGAG-CC-CAGGGGTGCTGCTGAAA-----GTCTTAAAATGCACAGTGTAGCCCCTCCTC GCCTCTCTTT------CTGCCCTGCAGGC-TAGAA-TGTATCA-CT-TAGATGTTCCAA--------ATCAGAAAGTGTTCAG-----CCAT GTCACAATTTAGGAAGTGCCACTGGCCT-----C-TAGAG-GGTAGAA-GA-CAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTAC GTCACAGTTTAGGGGGTACTACTGGCAT-----C-TATCG-GGTGGAG-GA-TAGGGATACTGATAATC-----ATTCTACAGTGCACAGGACAGTACCCCTAC GTCACAATTTGGGGGATACTACTGGCAT-----C-TAATG-GGTAGAG-GA-CAGGGATACTGATAATT-----GCTTTACAGTGCACAGGACAGCACCCTTAT GTCATAGTTT----GATTATATGGGCTT-----CTTAGTA-GACAAAGAAA-AAGATGTTCTGGTAGTC-----ATTCTGCTTTCCATATGATAGCACTCCCAT GTCACAGTTTGGAGGATGTTACTGACAT-----C-TAGAG-AGTAGAC-TT-TAAAGATACTGATAGTC-----ACCCCATTGTGCAC--------GTCACAATTTGGAGGATGTTACTGGCAT-----C-TAGAG-AGTAGAC-TT-TAAGGACACTGATAATC-----ATACTATGCTGCAC--------ATCACAATTTGGGGAACACCACTGGCAT-----C-TCGGGTAGCAGGC----CAGGCATGCTGGTAATT-----ATACTACAGTGCACAGTTCCCCACA ATCACAA-TTGGGGG-TGCCACGGTCCT-----C-CAGTG-GGTAGAG-AA-CAGGGAGGCTGATAACC-----ACCCTGCAGTGCACAGGGCAGTGCC-CCAC ATCACAG-TTGGGGGATGCCACTGGCCT-----C-AAGTG-GGTAGAG-AA-CAGGGAGGCTGAAAACC-----ACCCTGCAGAGCACGGGGCAGTGCCTTCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGAATGCTTATAATC-----ATCCTACAGTGCACAGGTCAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAA-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGAGATGCTTCTGGCTC-----G-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCAACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGTGGGGATGCTTATACTC----------ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC

Reconstruction algorithm 3) Reconstruct insertion/deletion history – Find most likely explanation for gaps observed ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSELEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN --------TGCTACTAATAT-----T-TAGTA-CATAGAG-CC-CAGGGGTGCTGCTGAAA-----GTCTTAAAATGCACAGTGTAGCCCCTCCTC GCCTCTCTTT------CTGCCCTGCAGGC-TAGAA-TGTATCA-CT-TAGATGTTCCAA--------ATCAGAAAGTGTTCAG-----CCAT GTCACAATTTAGGAAGTGCCACTGGCCT-----C-TAGAG-GGTAGAA-GA-CAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTAC GTCACAGTTTAGGGGGTACTACTGGCAT-----C-TATCG-GGTGGAG-GA-TAGGGATACTGATAATC-----ATTCTACAGTGCACAGGACAGTACCCCTAC GTCACAATTTGGGGGATACTACTGGCAT-----C-TAATG-GGTAGAG-GA-CAGGGATACTGATAATT-----GCTTTACAGTGCACAGGACAGCACCCTTAT GTCATAGTTT----GATTATATGGGCTT-----CTTAGTA-GACAAAGAAA-AAGATGTTCTGGTAGTC-----ATTCTGCTTTCCATATGATAGCACTCCCAT GTCACAGTTTGGAGGATGTTACTGACAT-----C-TAGAG-AGTAGAC-TT-TAAAGATACTGATAGTC-----ACCCCATTGTGCAC--------GTCACAATTTGGAGGATGTTACTGGCAT-----C-TAGAG-AGTAGAC-TT-TAAGGACACTGATAATC-----ATACTATGCTGCAC--------ATCACAATTTGGGGAACACCACTGGCAT-----C-TCGGGTAGCAGGC----CAGGCATGCTGGTAATT-----ATACTACAGTGCACAGTTCCCCACA ATCACAA-TTGGGGG-TGCCACGGTCCT-----C-CAGTG-GGTAGAG-AA-CAGGGAGGCTGATAACC-----ACCCTGCAGTGCACAGGGCAGTGCC-CCAC ATCACAG-TTGGGGGATGCCACTGGCCT-----C-AAGTG-GGTAGAG-AA-CAGGGAGGCTGAAAACC-----ACCCTGCAGAGCACGGGGCAGTGCCTTCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGAATGCTTATAATC-----ATCCTACAGTGCACAGGTCAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAA-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGAGATGCTTCTGGCTC-----G-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCAACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGTGGGGATGCTTATACTC----------ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC NNNNNNNNNNNNNN-----N-NNNNNNN-NN-NNNNNNNNN-----NNNNNNNNNNNNNNN • This defines the presence/absence of a base at each position of each ancestor

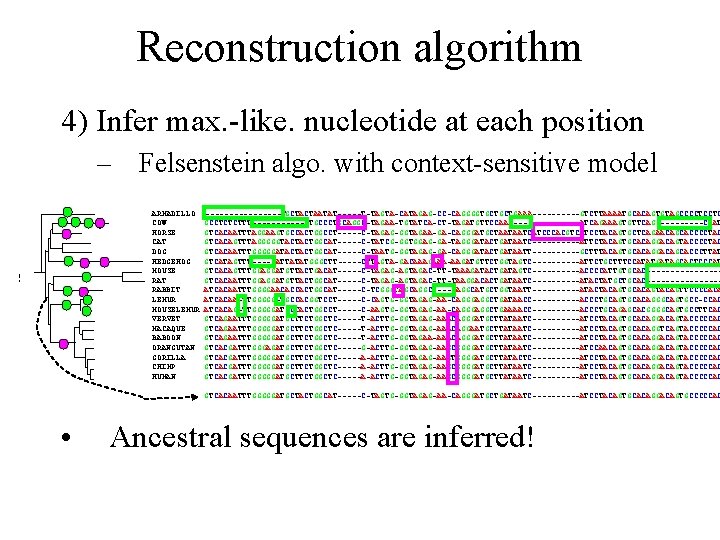
Reconstruction algorithm 4) Infer max. -like. nucleotide at each position – Felsenstein algo. with context-sensitive model ARMADILLO COW HORSE CAT DOG HEDGEHOG MOUSE RAT RABBIT LEMUR MOUSELEMUR VERVET MACAQUE BABOON ORANGUTAN GORILLA CHIMP HUMAN --------TGCTACTAATAT-----T-TAGTA-CATAGAG-CC-CAGGGGTGCTGCTGAAA-----GTCTTAAAATGCACAGTGTAGCCCCTCCTC GCCTCTCTTT------CTGCCCTGCAGGC-TAGAA-TGTATCA-CT-TAGATGTTCCAA--------ATCAGAAAGTGTTCAG-----CCAT GTCACAATTTAGGAAGTGCCACTGGCCT-----C-TAGAG-GGTAGAA-GA-CAGGGATGCTAATAATCATCCCACGTCATCCTACAGTGCTCAGAACAGCACCCCTAC GTCACAGTTTAGGGGGTACTACTGGCAT-----C-TATCG-GGTGGAG-GA-TAGGGATACTGATAATC-----ATTCTACAGTGCACAGGACAGTACCCCTAC GTCACAATTTGGGGGATACTACTGGCAT-----C-TAATG-GGTAGAG-GA-CAGGGATACTGATAATT-----GCTTTACAGTGCACAGGACAGCACCCTTAT GTCATAGTTT----GATTATATGGGCTT-----CTTAGTA-GACAAAGAAA-AAGATGTTCTGGTAGTC-----ATTCTGCTTTCCATATGATAGCACTCCCAT GTCACAGTTTGGAGGATGTTACTGACAT-----C-TAGAG-AGTAGAC-TT-TAAAGATACTGATAGTC-----ACCCCATTGTGCAC--------GTCACAATTTGGAGGATGTTACTGGCAT-----C-TAGAG-AGTAGAC-TT-TAAGGACACTGATAATC-----ATACTATGCTGCAC--------ATCACAATTTGGGGAACACCACTGGCAT-----C-TCGGGTAGCAGGC----CAGGCATGCTGGTAATT-----ATACTACAGTGCACAGTTCCCCACA ATCACAA-TTGGGGG-TGCCACGGTCCT-----C-CAGTG-GGTAGAG-AA-CAGGGAGGCTGATAACC-----ACCCTGCAGTGCACAGGGCAGTGCC-CCAC ATCACAG-TTGGGGGATGCCACTGGCCT-----C-AAGTG-GGTAGAG-AA-CAGGGAGGCTGAAAACC-----ACCCTGCAGAGCACGGGGCAGTGCCTTCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAG-AAACAGGAATGCTTATAATC-----ATCCTACAGTGCACAGGTCAGTACCCCCAC GTCAGAATTTGGGGGATGCTTCTGGCTC-----T-ACTTG-GGTAGAA-AAACAGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGAGATGCTTCTGGCTC-----G-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCAACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGTGGGGATGCTTATACTC----------ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACGATTTGGGGGATGCTTCTGGCTC-----A-ACTTG-GGTAGAG-AAGCGGGGATGCTTATAATC-----ATCCTACAGTGCACAGGACAGTACCCCCAC GTCACAATTTGGGGGATGCTACTGGCAT-----C-TAGTG-GGTAGAG-AA-CAGGGATGCTGATAATC-----ATCCTACAGTGCACAGGACAGTGCCCCCAC • Ancestral sequences are inferred!

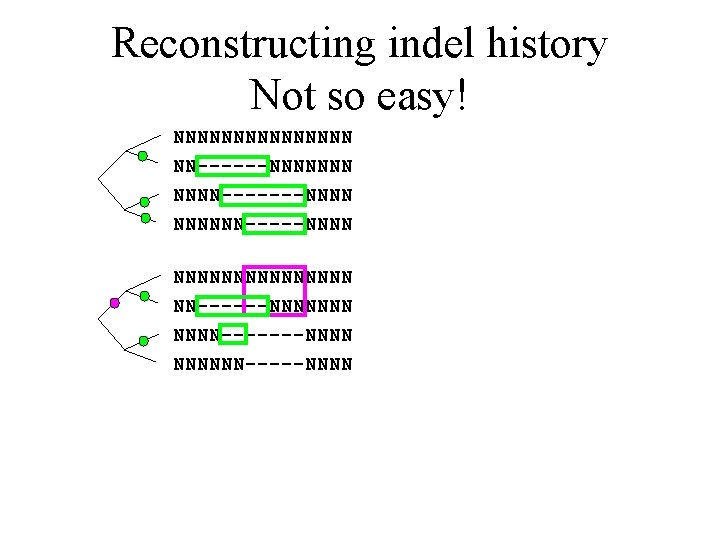
Optimal indel reconstruction Not so easy! NNNNNNNN NN------NNNNNNN-------NNNNNN-----NNNN

Reconstructing indel history Not so easy! NNNNNNNN NN------NNNNNNN-------NNNNNN-----NNNN

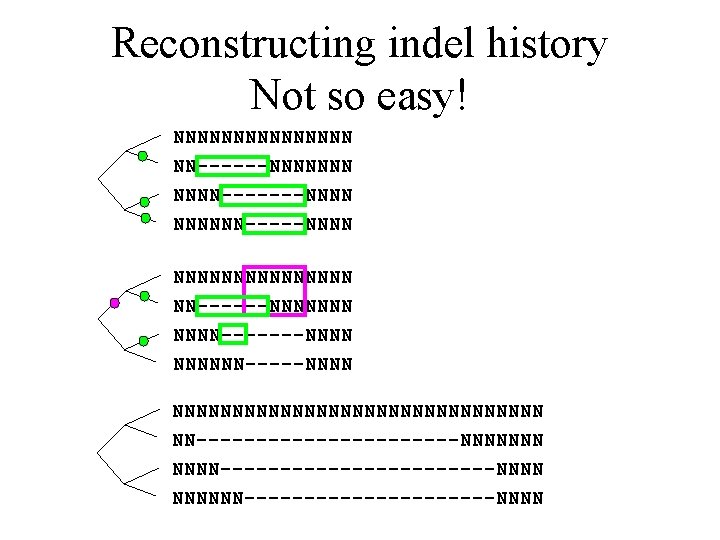
Reconstructing indel history Not so easy! NNNNNNNNNNNNNNN NN------NNNNNNN-------NNNNNN-----NNNN

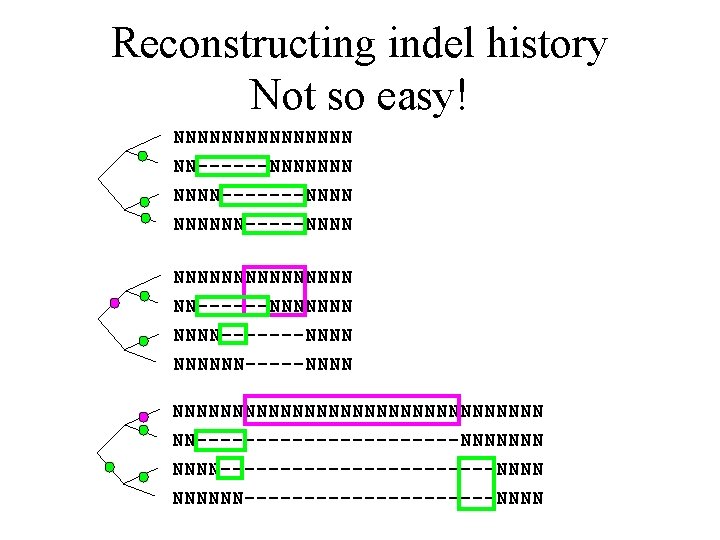
Reconstructing indel history Not so easy! NNNNNNNNNNNNNNN NN------NNNNNNN-------NNNNNN-----NNNNNNNNNNNNNNNN NN-----------NNNNNNN------------NNNNNN-----------NNNN

Reconstructing indel history Not so easy! NNNNNNNNNNNNNNN NN------NNNNNNN-------NNNNNN-----NNNNNNNNNNNNNNNN NN-----------NNNNNNN------------NNNNNN-----------NNNN

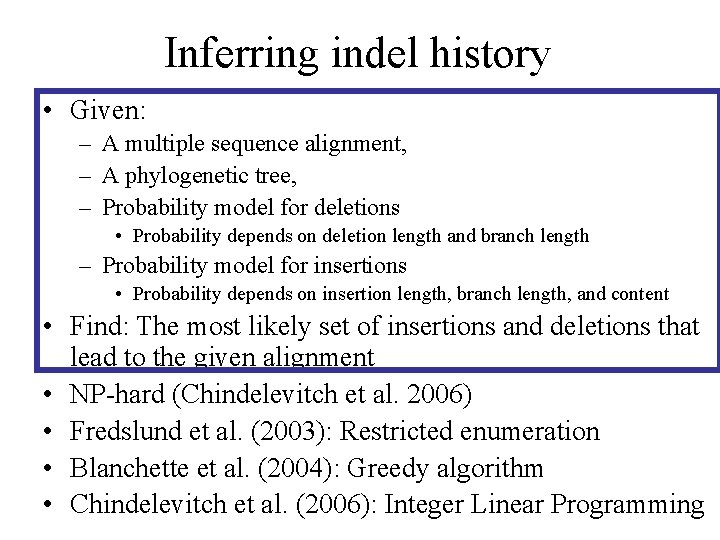
Inferring indel history • Given: – A multiple sequence alignment, – A phylogenetic tree, – Probability model for deletions • Probability depends on deletion length and branch length – Probability model for insertions • Probability depends on insertion length, branch length, and content • Find: The most likely set of insertions and deletions that lead to the given alignment • NP-hard (Chindelevitch et al. 2006) • Fredslund et al. (2003): Restricted enumeration • Blanchette et al. (2004): Greedy algorithm • Chindelevitch et al. (2006): Integer Linear Programming

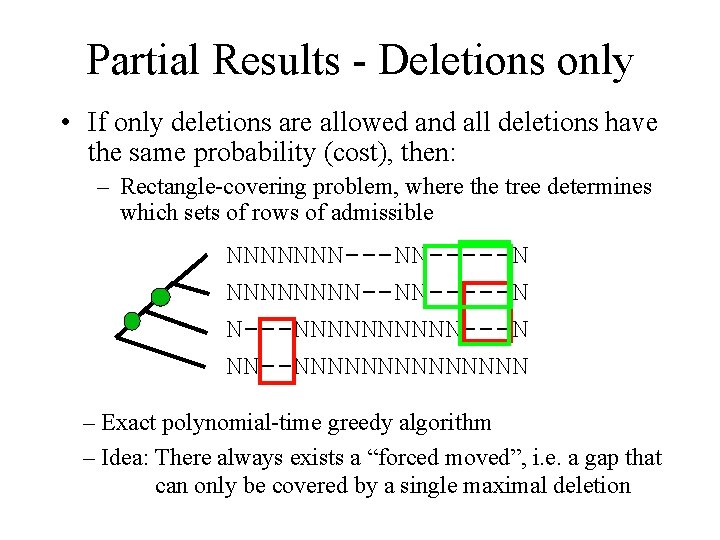
Partial Results - Deletions only • If only deletions are allowed and all deletions have the same probability (cost), then: – Rectangle-covering problem, where the tree determines which sets of rows of admissible NNNNNNN-----N NNNN--NN-----N N---NNNNN---N NN--NNNNNNN – Exact polynomial-time greedy algorithm – Idea: There always exists a “forced moved”, i. e. a gap that can only be covered by a single maximal deletion

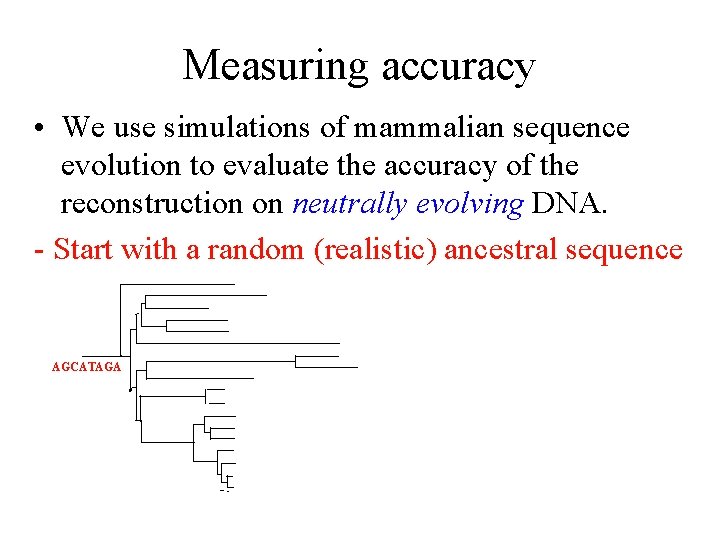
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. - Start with a random (realistic) ancestral sequence AGCATAGA

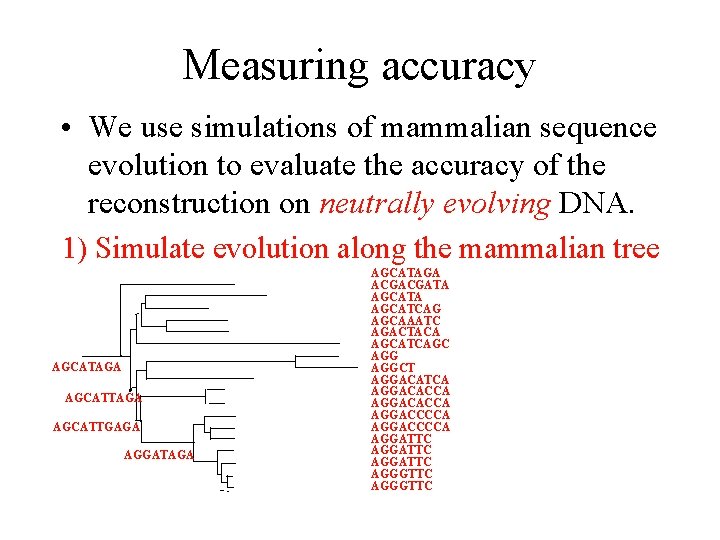
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. 1) Simulate evolution along the mammalian tree AGCATAGA AGCATTGAGA AGGATAGA AGCATAGA ACGACGATA AGCATCAG AGCAAATC AGACTACA AGCATCAGC AGGCT AGGACATCA AGGACACCA AGGACCCCA AGGATTC AGGGTTC

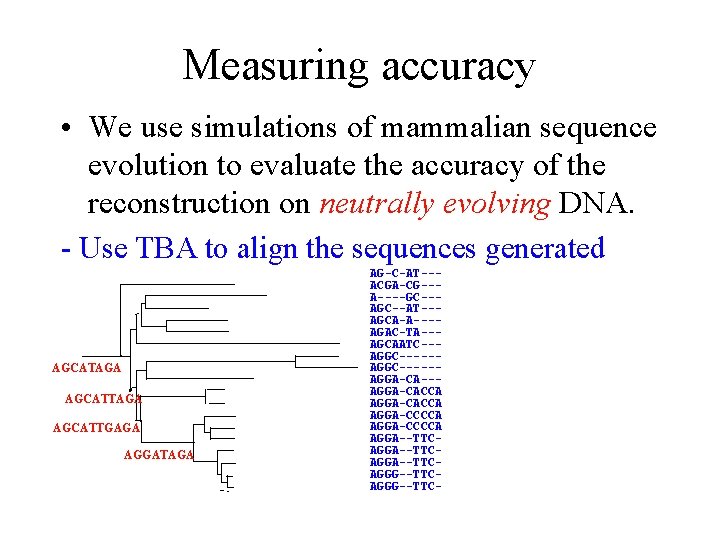
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. - Use TBA to align the sequences generated AGCATAGA AGCATTGAGA AGGATAGA AG-C-AT--ACGA-CG--A----GC--AT--AGCA-A---AGAC-TA--AGCAATC--AGGC-----AGGA-CACCA AGGA-CCCCA AGGA--TTCAGGA--TTCAGGG--TTC-

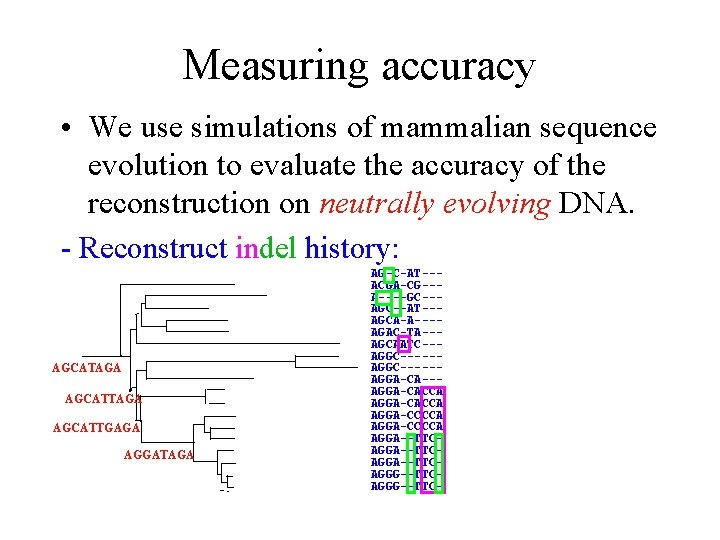
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. - Reconstruct indel history: AGCATAGA AGCATTGAGA AGGATAGA AG-C-AT--ACGA-CG--A----GC--AT--AGCA-A---AGAC-TA--AGCAATC--AGGC-----AGGA-CACCA AGGA-CCCCA AGGA--TTCAGGA--TTCAGGG--TTC-

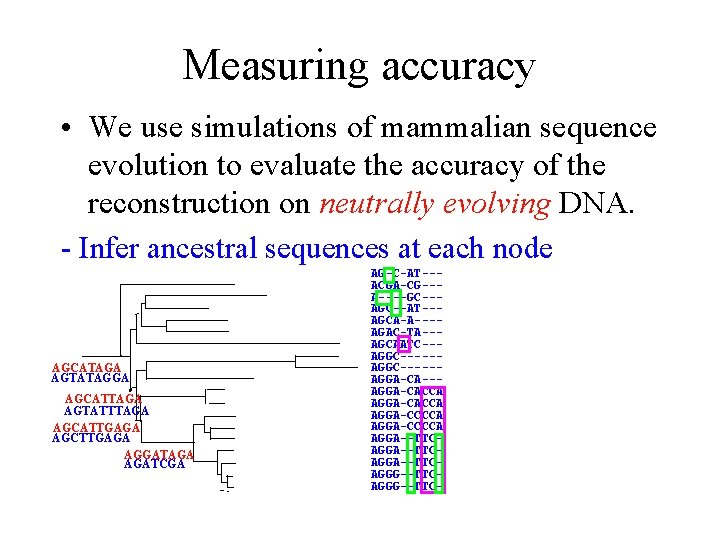
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. - Infer ancestral sequences at each node AGCATAGA AGTATAGGA AGCATTAGA AGTATTTAGA AGCATTGAGA AGCTTGAGA AGGATAGA AGATCGA AG-C-AT--ACGA-CG--A----GC--AT--AGCA-A---AGAC-TA--AGCAATC--AGGC-----AGGA-CACCA AGGA-CCCCA AGGA--TTCAGGA--TTCAGGG--TTC-

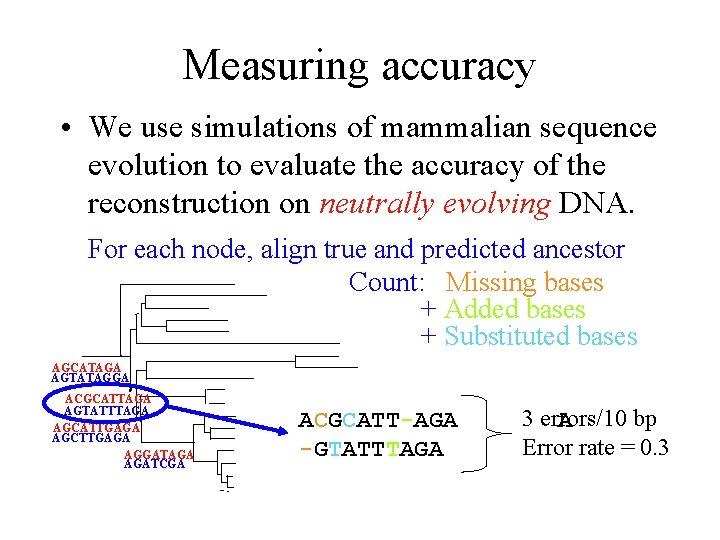
Measuring accuracy • We use simulations of mammalian sequence evolution to evaluate the accuracy of the reconstruction on neutrally evolving DNA. For each node, align true and predicted ancestor Count: Missing bases + Added bases + Substituted bases AGCATAGA AGTATAGGA ACGCATTAGA AGTATTTAGA AGCATTGAGA AGCTTGAGA AGGATAGA AGATCGA ACGCATT-AGA -GTATTTAGA 3 errors/10 bp A Error rate = 0. 3

Simulation details • We simulate neutrally evolving regions of 50 kb • We model: - Lineage-specific neutral mutation rates - Insertions and deletions based on empirical frequency and length distributions - Insertion of transposable elements - Cp. G effect • We don’t model: - DNA polymerase slippage - Positive selection - Genome rearrangement, duplications • Sanity checks: Simulated sequences are similar to actual mammalian sequences: – Same pair-wise percent identity – Same frequency and length distribution of insertions and deletions – Same repetitive content and age distribution of repeats

Guess which ancestor can be best reconstructed? Eizirik et al. 2001

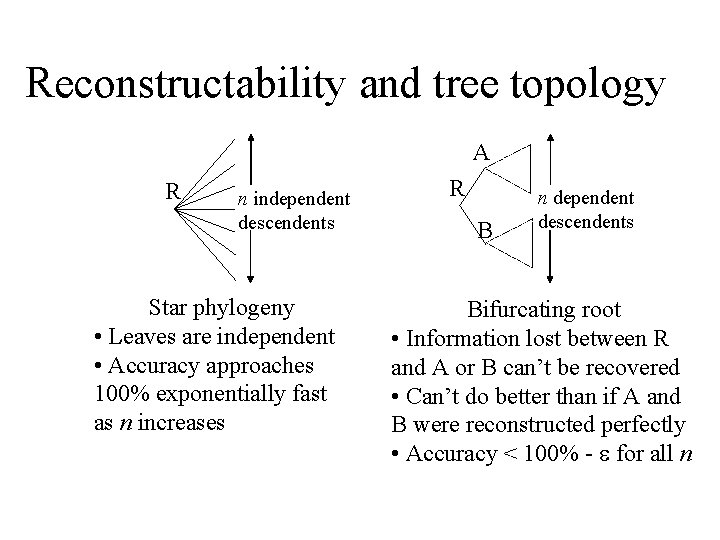
Reconstructability and tree topology A R n independent descendents Star phylogeny • Leaves are independent • Accuracy approaches 100% exponentially fast as n increases R B n dependent descendents Bifurcating root • Information lost between R and A or B can’t be recovered • Can’t do better than if A and B were reconstructed perfectly • Accuracy < 100% - for all n

Eizirik et al. 2001

How many species do we need? Best choice of species: - Sample many taxa - Choose slowly evolving species

What if the fast-radiation model is wrong?

Reconstructing real ancestors

COW For this set of species, simulations predict: - Expected accuracy ~95% RAT CHIMP, GORILLA, ORANGUTAN, MACAQUE, VERVET, BABOON MOUSE-LEMUR

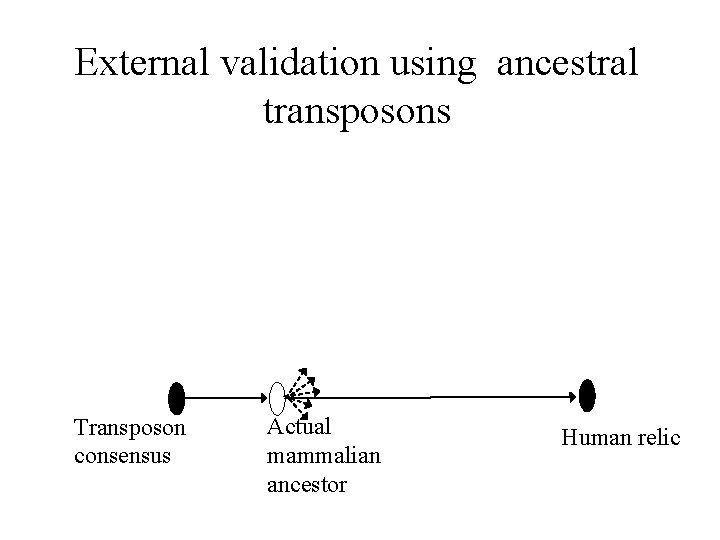
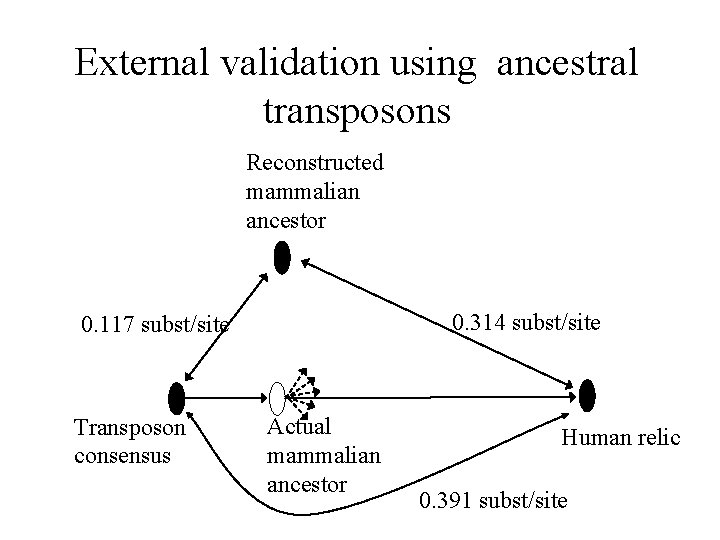
External validation using ancestral transposons Transposon consensus Actual mammalian ancestor Human relic

External validation using ancestral transposons Reconstructed mammalian ancestor 0. 314 subst/site 0. 117 subst/site Transposon consensus Actual mammalian ancestor Human relic 0. 391 subst/site

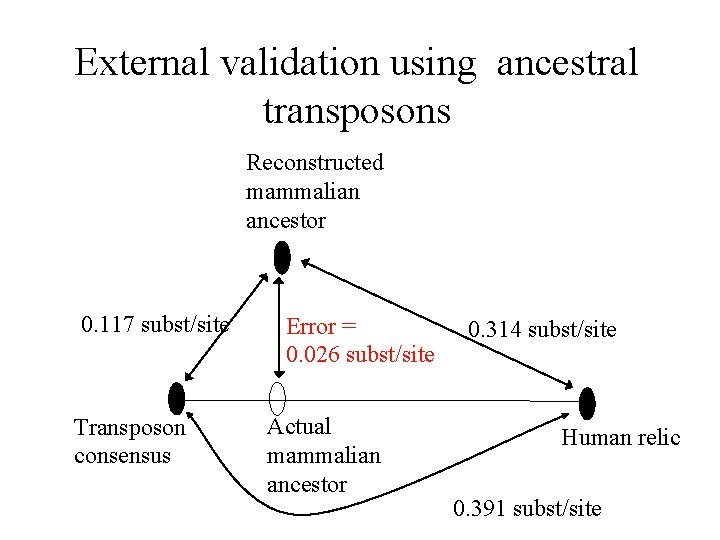
External validation using ancestral transposons Reconstructed mammalian ancestor 0. 117 subst/site Transposon consensus Error = 0. 026 subst/site Actual mammalian ancestor 0. 314 subst/site Human relic 0. 391 subst/site


What’s next? Whole genome! • Data available – Whole genomes: Human, chimp, mouse, rat, dog – Unassembled/ low coverage genomes: Cow, rabbit, armadillo, elephant • Challenges: – – Fewer species Unassembled contigs Genome rearrangements Recombination hotspots We expect that 90% of the Boreoeutherian genome can be reconstructed with ~90% accuracy


Why should we care? • Ancestral genome allows to see what and when changes happened in our genome – Allows detection and “dating” of lineage specific innovations (e. g. FOXP 2). • Allows a better understanding of the forces driving genome evolution • New model organism? – Human genome is 4 times closer to the ancestral genome than to the mouse genome: better model for human phenotypes?


Even if we had the full genomes of all living mammalian species: • Technological problem: – We can’t synthesize large regions of DNA • Many regions can’t be reconstructed at all: – Heterochromatin – Regions with high recombination rates • 99% base-by-base accuracy is not enough – One mistake may be enough to make life impossible

Acknowledgements • David Haussler, Brian Raney UC Santa Cruz • Webb Miller Penn State Univ. • Eric Green NHGRI • UC Santa Cruz group: – Adam Siepel, Robert Baertsch, Gill Bejerano, Jim Kent • Mc. Gill group: – Leonid Chindelevitch, Zhentao Li, Eric Blais
 Preoge
Preoge Semi-global alignment
Semi-global alignment Mesquite ancestral state reconstruction
Mesquite ancestral state reconstruction Epithelium of trachea
Epithelium of trachea The mammalian excretory system
The mammalian excretory system Mammalian excretory system
Mammalian excretory system Diving reflex
Diving reflex Tricodont
Tricodont Mammalian biomanufacturing
Mammalian biomanufacturing Glycolyaia
Glycolyaia Mammalian lung diagram
Mammalian lung diagram Chapter 30 section 2 diversity of mammals
Chapter 30 section 2 diversity of mammals Mammalian excretory system
Mammalian excretory system Tendinous cords
Tendinous cords Mammalian excretory system
Mammalian excretory system National human genome research institute
National human genome research institute Human genome project source code
Human genome project source code Genome definition
Genome definition Yac vector
Yac vector Genome.gov
Genome.gov What is genome
What is genome Tirmarker
Tirmarker National human genome research institute
National human genome research institute Genome klick
Genome klick Raspberry pi 3 model b specifications pdf
Raspberry pi 3 model b specifications pdf Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Genome project
Genome project Human genome size
Human genome size Igv genome browser
Igv genome browser Ap biol
Ap biol Human genome project
Human genome project Genome modification ustaz auni
Genome modification ustaz auni Human genome structure
Human genome structure What is a human genome
What is a human genome Plant genome research program
Plant genome research program Genome research limited
Genome research limited Genome sequencing
Genome sequencing Genome.ucsc.edu tutorial
Genome.ucsc.edu tutorial History of sequencing
History of sequencing Sickle cell karyotype
Sickle cell karyotype Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Maker genome annotation
Maker genome annotation Mash bioinformatics
Mash bioinformatics Img genome
Img genome 1000 genome project
1000 genome project