Selecting Genomes for Reconstruction of Ancestral Genomes Louxin





![Calculating the Reconstruction Accuracy of Fitch Method The unambiguous reconstruction accuracy: P[{1}|1]= the probability Calculating the Reconstruction Accuracy of Fitch Method The unambiguous reconstruction accuracy: P[{1}|1]= the probability](https://slidetodoc.com/presentation_image/8c811bf432665a412f49709692340881/image-13.jpg)














- Slides: 47

Selecting Genomes for Reconstruction of Ancestral Genomes Louxin Zhang Department of Mathematics National University of Singapore

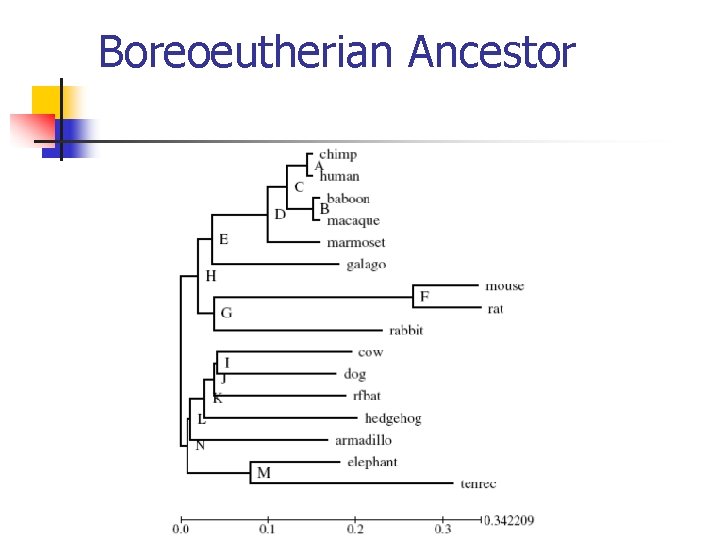
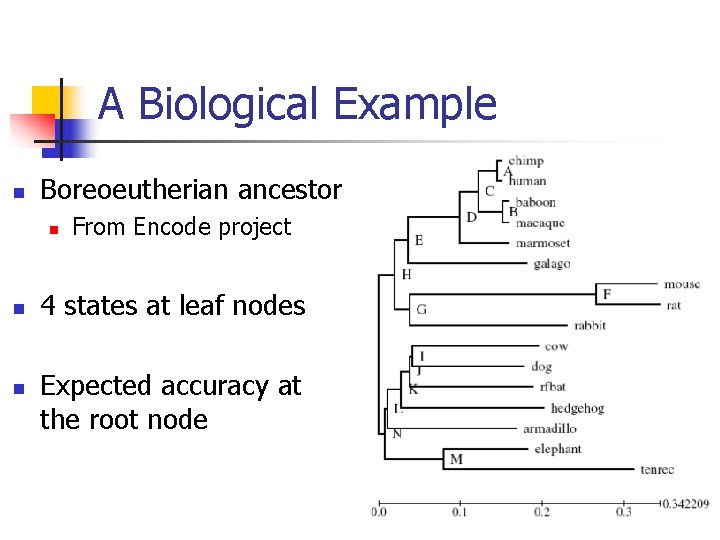
Boreoeutherian Ancestor

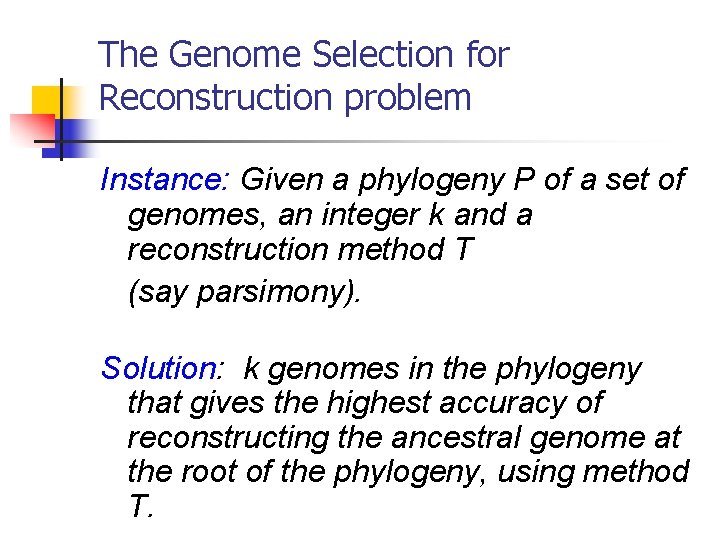
The Genome Selection for Reconstruction problem Instance: Given a phylogeny P of a set of genomes, an integer k and a reconstruction method T (say parsimony). Solution: k genomes in the phylogeny that gives the highest accuracy of reconstructing the ancestral genome at the root of the phylogeny, using method T.

Two reasons n n It is often impossible to sequence all descendent genomes below an ancestor; More taxa do not necessarily give a higher accuracy for the reconstruction of ancestral character states in general (examples will be given below)

Outline n n n Introduction to reconstruction accuracy analysis More genomes are not necessarily better for reconstruction Greedy algorithms for genome selection A joint work with G. Li, J. Ma and M. Steel

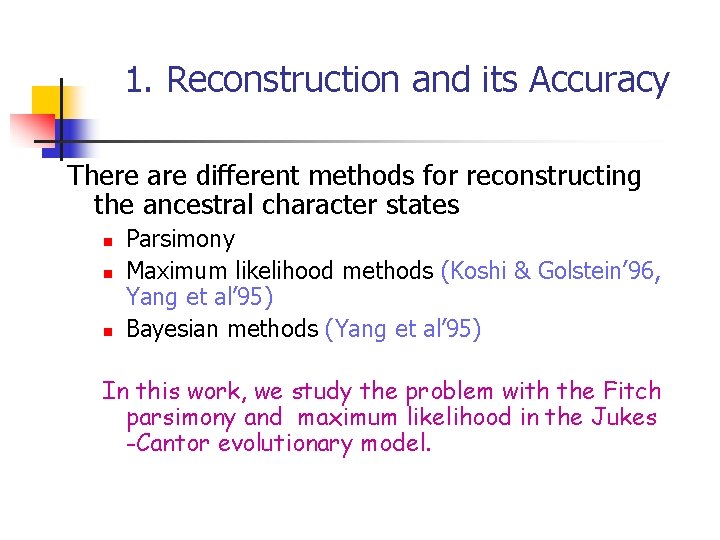
1. Reconstruction and its Accuracy There are different methods for reconstructing the ancestral character states n n n Parsimony Maximum likelihood methods (Koshi & Golstein’ 96, Yang et al’ 95) Bayesian methods (Yang et al’ 95) In this work, we study the problem with the Fitch parsimony and maximum likelihood in the Jukes -Cantor evolutionary model.

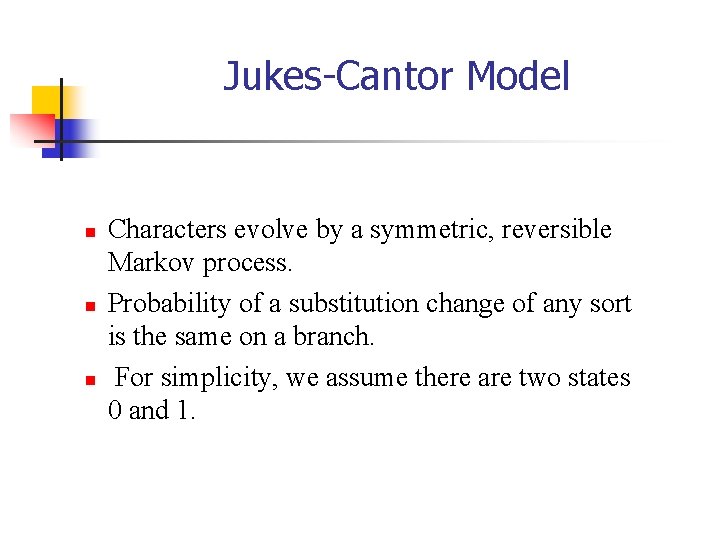
Jukes-Cantor Model n n n Characters evolve by a symmetric, reversible Markov process. Probability of a substitution change of any sort is the same on a branch. For simplicity, we assume there are two states 0 and 1.

Reconstruction Accuracy In the symmetric Jukes-Cantor model, n the reconstruction accuracy of a method is independent of the prior distribution of the states at the root.

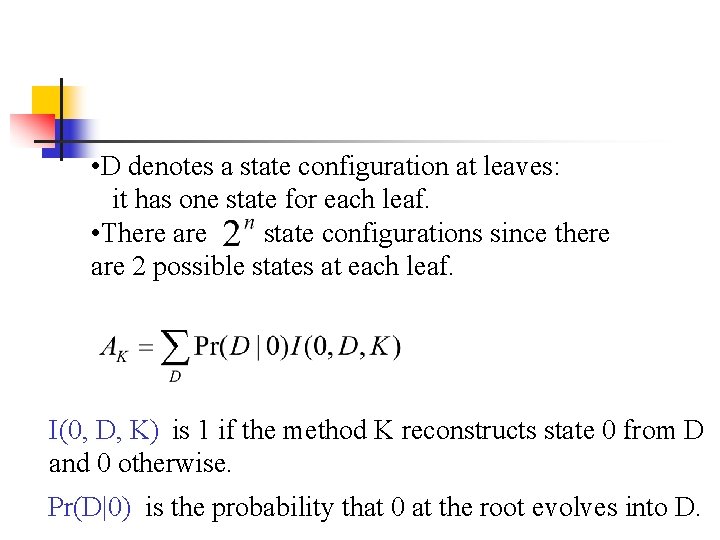
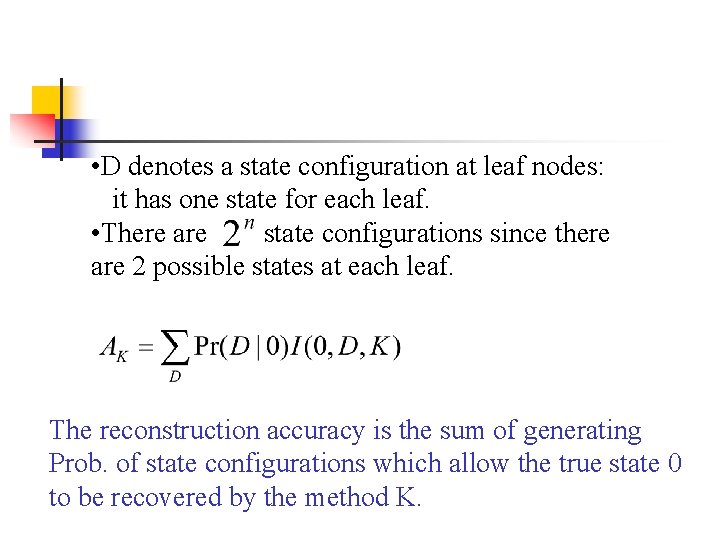
• D denotes a state configuration at leaves: it has one state for each leaf. • There are state configurations since there are 2 possible states at each leaf. I(0, D, K) is 1 if the method K reconstructs state 0 from D and 0 otherwise. Pr(D|0) is the probability that 0 at the root evolves into D.

• D denotes a state configuration at leaf nodes: it has one state for each leaf. • There are state configurations since there are 2 possible states at each leaf. The reconstruction accuracy is the sum of generating Prob. of state configurations which allow the true state 0 to be recovered by the method K.

Previous Analysis Works n n Simulation study (Martins’ 99, Mooers’ 04, Salisbury & Kim’ 01, Zhang & Nei’ 97, Yang et al’ 95); Theoretical study (Mossel’ 01, Lucena and Haussler’ 05, Maddison’ 95)

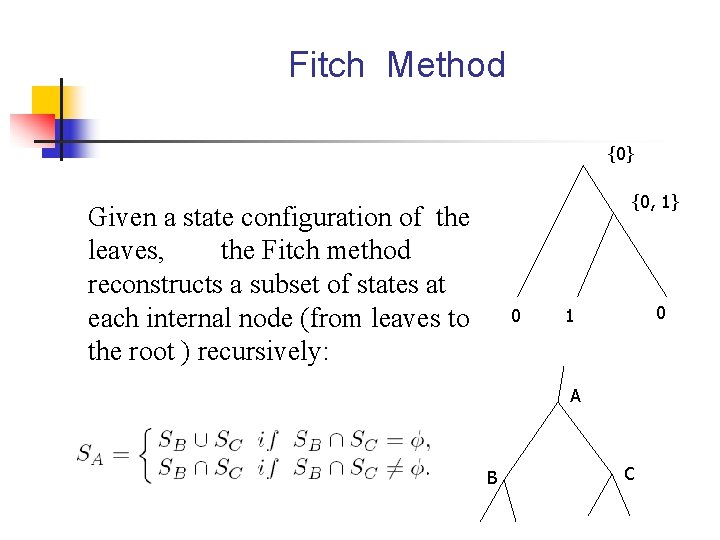
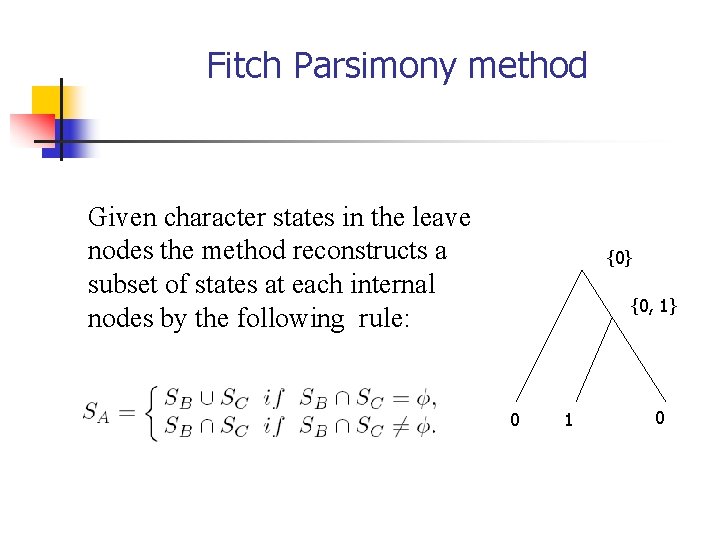
Fitch Method {0} {0, 1} Given a state configuration of the leaves, the Fitch method reconstructs a subset of states at each internal node (from leaves to the root ) recursively: 0 0 1 A B C
![Calculating the Reconstruction Accuracy of Fitch Method The unambiguous reconstruction accuracy P11 the probability Calculating the Reconstruction Accuracy of Fitch Method The unambiguous reconstruction accuracy: P[{1}|1]= the probability](https://slidetodoc.com/presentation_image/8c811bf432665a412f49709692340881/image-13.jpg)
Calculating the Reconstruction Accuracy of Fitch Method The unambiguous reconstruction accuracy: P[{1}|1]= the probability that Fitch method outputs PAccuracy = P[{1}|1]=P[{0}|0] true state at the root. and the reconstruction accuracy P[{0}|1], P[{1}|1], and P[{0, 1}|1] can be calculated by a dynamic approach (Maddison, 1995)

Outline n n n Introduction to reconstruction accuracy analysis More genomes are not necessarily better for reconstruction accuracy Greedy algorithms for genome selection

2. Reconstruction accuracy is not a monotone function of the size of taxon sampling • There is a large clade with a long stem • A short single sister lineage umbalanced tree Such a phylogeny is used when both fossil record and data at extant species are used for reconstruction (Finarelli and Flynn, 2006)

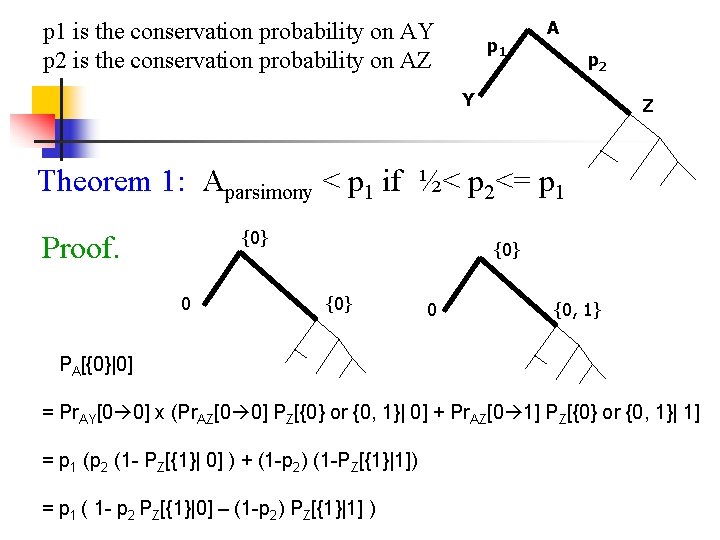
p 1 is the conservation probability on AY p 2 is the conservation probability on AZ p 1 A p 2 Y Z Theorem 1: Aparsimony < p 1 if ½< p 2<= p 1 {0} Proof. 0 {0} 0 {0, 1} PA[{0}|0] = Pr. AY[0 0] x (Pr. AZ[0 0] PZ[{0} or {0, 1}| 0] + Pr. AZ[0 1] PZ[{0} or {0, 1}| 1] = p 1 (p 2 (1 - PZ[{1}| 0] ) + (1 -p 2) (1 -PZ[{1}|1]) = p 1 ( 1 - p 2 PZ[{1}|0] – (1 -p 2) PZ[{1}|1] )

p 1 is the conservation probability on AY p 2 is the conservation probability on AZ p 2 Y ½ < p 2 <= p 1 {0, 1} 0 p 1 A Z {0, 1} {1} 1 {0} PA[{0, 1}|0] = [p 1 p 2+(1 -p 1)(1 -p 2)] x PZ[{1}|0] + [ p 1(1 -p 2)+p 2(1 -p 1)] X PZ[{0}|0] Aparsimony = PA[{0}|0] + ½ PA[{0, 1}|1] = p 1 + ½ (1 -p 2) PZ[{1}|0] + ½(p 2 -p 1) PZ[{0}|0] < p 1

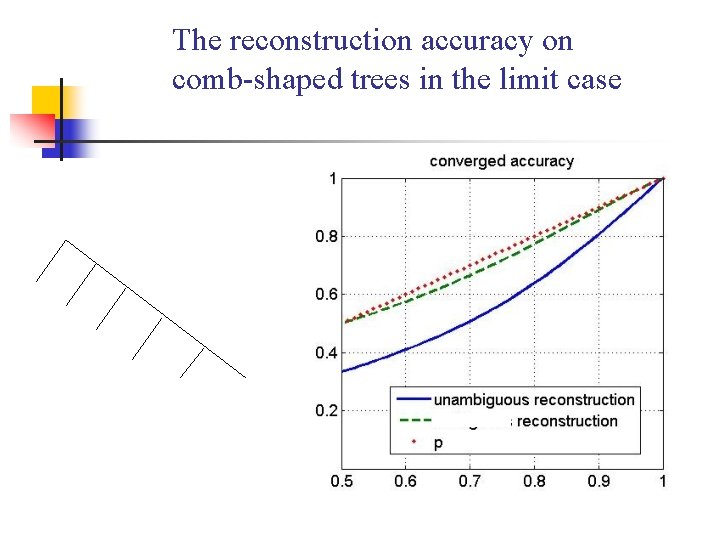
The reconstruction accuracy on comb-shaped trees in the limit case

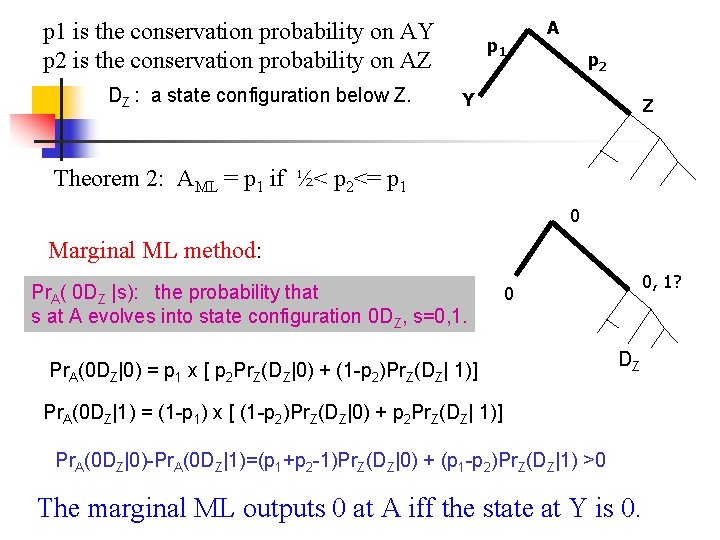
p 1 is the conservation probability on AY p 2 is the conservation probability on AZ DZ : a state configuration below Z. p 1 A p 2 Y Z Theorem 2: AML = p 1 if ½< p 2<= p 1 0 Marginal ML method: Pr. A( 0 DZ |s): the probability that s at A evolves into state configuration 0 DZ, s=0, 1. 0, 1? 0 Pr. A(0 DZ|0) = p 1 x [ p 2 Pr. Z(DZ|0) + (1 -p 2)Pr. Z(DZ| 1)] DZ Pr. A(0 DZ|1) = (1 -p 1) x [ (1 -p 2)Pr. Z(DZ|0) + p 2 Pr. Z(DZ| 1)] Pr. A(0 DZ|0)-Pr. A(0 DZ|1)=(p 1+p 2 -1)Pr. Z(DZ|0) + (p 1 -p 2)Pr. Z(DZ|1) >0 The marginal ML outputs 0 at A iff the state at Y is 0.

Another Example showing the Non-monotone Property of Reconstruction Accuracy

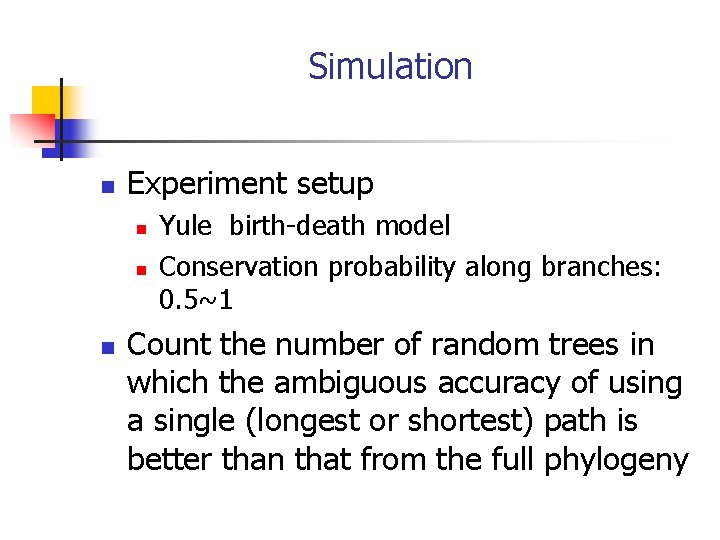
Simulation n Experiment setup n n n Yule birth-death model Conservation probability along branches: 0. 5~1 Count the number of random trees in which the ambiguous accuracy of using a single (longest or shortest) path is better than that from the full phylogeny

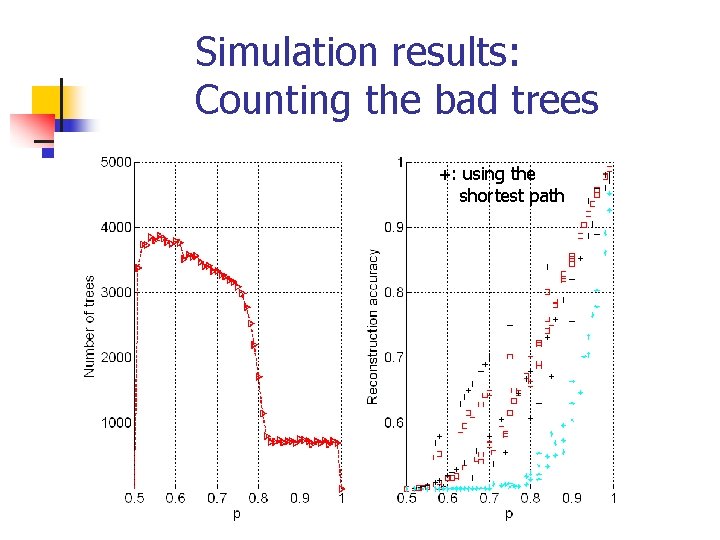
Simulation results: Counting the bad trees +: using the shortest path

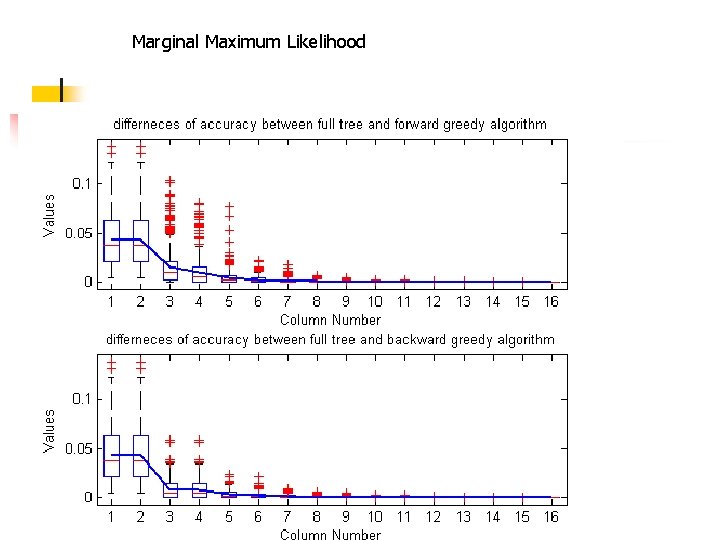
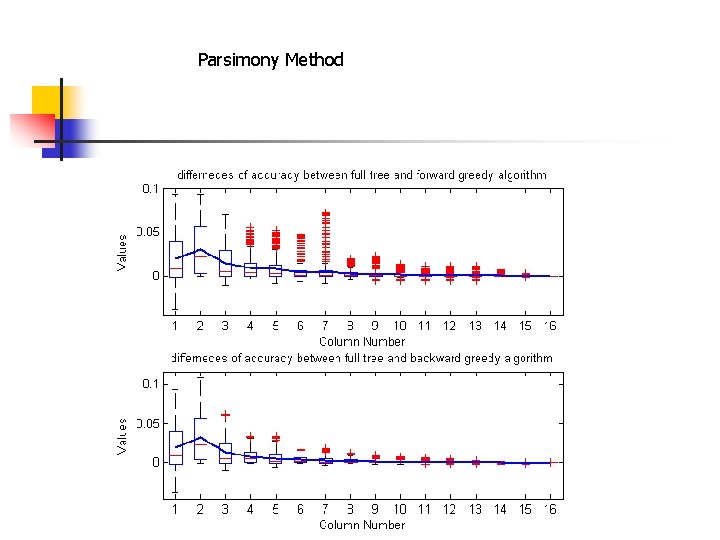
Comparison of Parsimony, Joint ML and Marginal ML 500 random trees with 12 leaves generated: • Yule birth-death model • branch length is uniform from 0 to 1 MML outperforms JML, MP. • In 80% of instances, MML is strictly better than JML • In 99% of instances, JML is strictly better than MP. •

Outline n n n Introduction to reconstruction accuracy analysis More genomes are not necessarily better for reconstruction accuracy Greedy algorithms for genome selection problem

Genome selection for reconstruction the problem n n Instance: A phylogeny P over n genomes, integer k and a reconstruction method T Question: Find k genomes that allows the ancestral genome of the root of P to be reconstructed with the maximum accuracy, using method T.

Our approaches n n The genome selection problem is unlikely polynomial-time solvable (no hardness proof yet) As a result, we propose two greedy algorithms for the problem: Forward greedy algorithm & Backward greedy algorithm

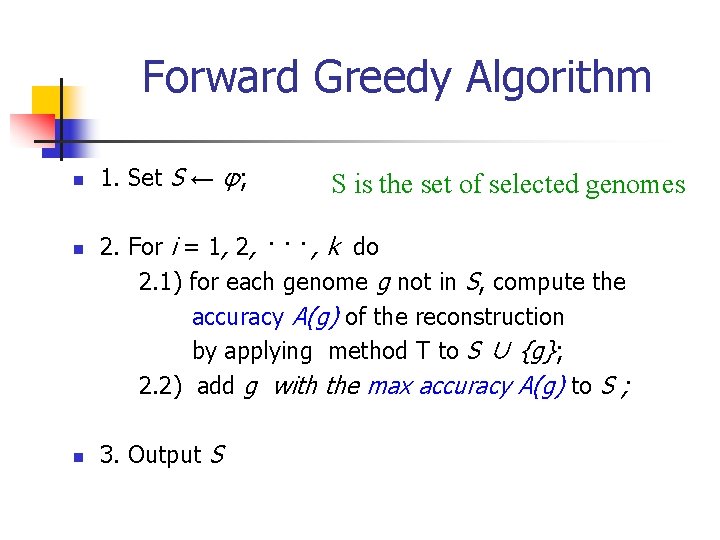
Forward Greedy Algorithm n n n 1. Set S ← φ; S is the set of selected genomes 2. For i = 1, 2, · · · , k do 2. 1) for each genome g not in S, compute the accuracy A(g) of the reconstruction by applying method T to S ∪ {g}; 2. 2) add g with the max accuracy A(g) to S ; 3. Output S

Backward Greedy Algorithm n n n 1. Let S contain all the given genomes; 2. For i = 1, 2, · · · , n − k do 2. 1) for each genome g in S, compute the accuracy A’(g) of the reconstruction by applying T to S − {g}; 2. 2) remove g from S if A’(g) is the max over all g’s; 3. Output S

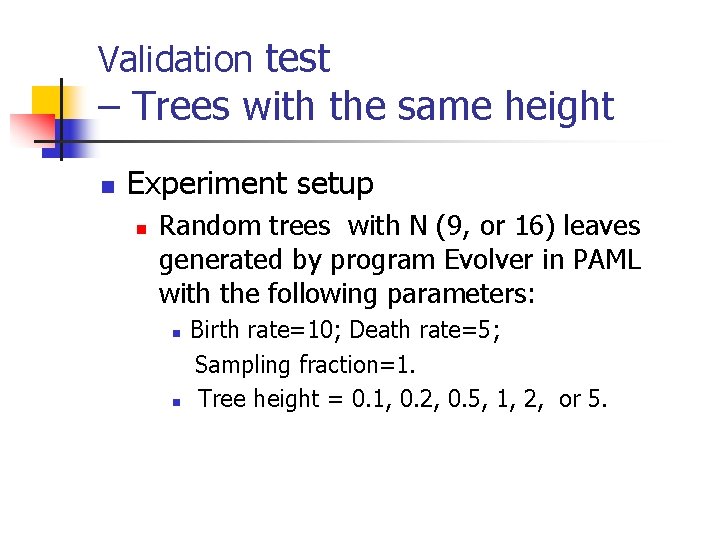
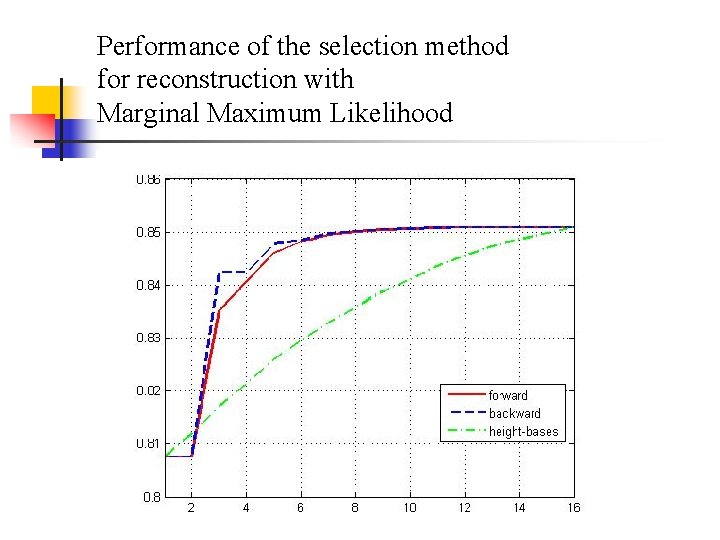
Validation test – Trees with the same height n Experiment setup n Random trees with N (9, or 16) leaves generated by program Evolver in PAML with the following parameters: n n Birth rate=10; Death rate=5; Sampling fraction=1. Tree height = 0. 1, 0. 2, 0. 5, 1, 2, or 5.

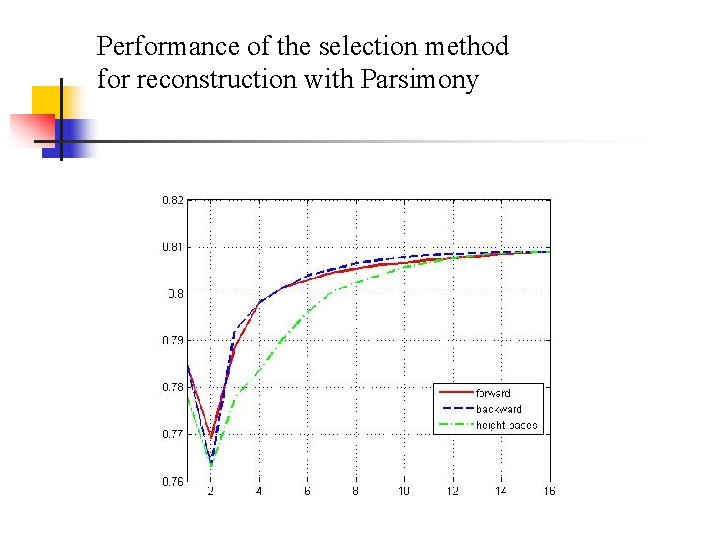
Performance of the selection method for reconstruction with Parsimony

Performance of the selection method for reconstruction with Marginal Maximum Likelihood

Performance of the selection method for reconstruction with Joint Maximum Likelihood

Marginal Maximum Likelihood

Parsimony Method

Concluding remarks n Reconstruction accuracy is not monotone increasing with the taxon sampling size in unbalanced trees for Parsimony method --- Another kind of “inconsistency” 1. One implication of this observation is that Parsimony, ML method might not explore the full power of incorporating fossil record into current data. Hence, modification might probably be needed. 2. Caution should be used in drawing conclusion on testing hypothesis with ancestral state reconstruction.

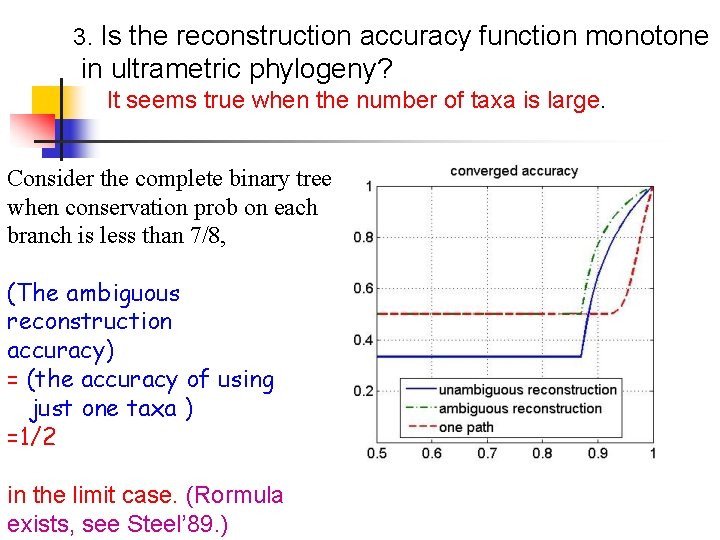
3. Is the reconstruction accuracy function monotone in ultrametric phylogeny? It seems true when the number of taxa is large. Consider the complete binary tree when conservation prob on each branch is less than 7/8, (The ambiguous reconstruction accuracy) = (the accuracy of using just one taxa ) =1/2 in the limit case. (Rormula exists, see Steel’ 89. )

Concluding remarks n n n Formulate the genome selection for reconstruction problem Two greedy algorithms proposed for the problem Validation test shows that the reconstruction accuracy of using the genomes selected by the greedy algorithms are comparable to the max reconstruction accuracy.

Thanks You!

A Biological Example n Boreoeutherian ancestor n n n From Encode project 4 states at leaf nodes Expected accuracy at the root node

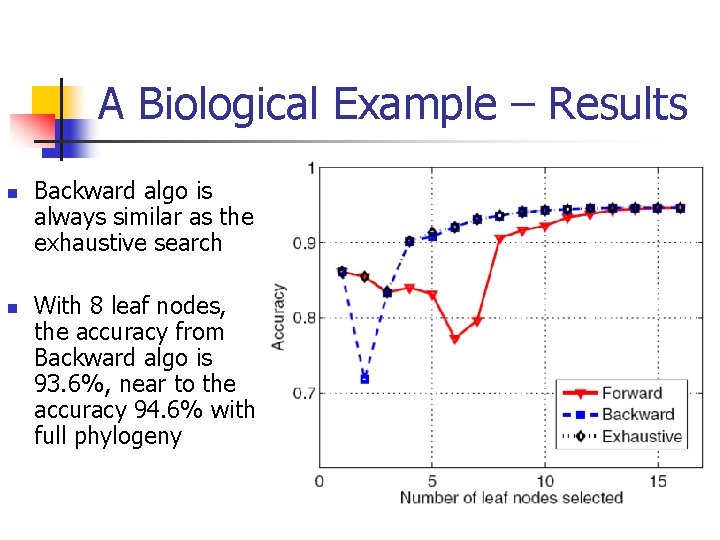
A Biological Example – Results n n Backward algo is always similar as the exhaustive search With 8 leaf nodes, the accuracy from Backward algo is 93. 6%, near to the accuracy 94. 6% with full phylogeny

Outline n Introduction to phylogeny reconstruction accuracy analysis n More genomes are not necessarily better for reconstruction accuracy n Greedy algorithms for genome selection problem n Validation test n Conclusion

Conclusion n Formulate the genome selection for reconstruction problem Two greedy algorithms proposed for the problem Validation test shows that the reconstruction accuracy of using the genomes selected by the greedy algorithms are comparable to the max reconstruction accuracy.

Fitch Parsimony method Given character states in the leave nodes the method reconstructs a subset of states at each internal nodes by the following rule: {0} {0, 1} 0 1 0

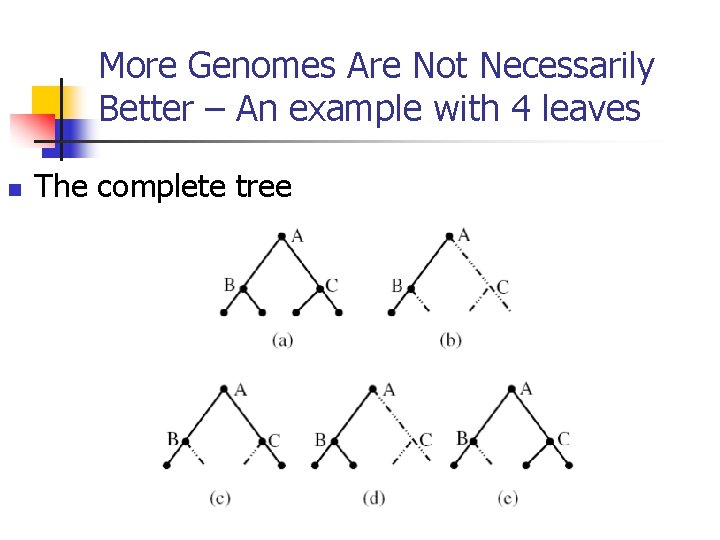
More Genomes Are Not Necessarily Better – An example with 4 leaves n The complete tree

More Genomes Are Not Necessarily Better – An example with 4 leaves n n The unambiguous reconstruction accuracy of using one genome is Ppath= p 2+(1 -p)2; The unambiguous reconstruction accuracy of using all the 4 genomes is Pwhole= Ppath – 3 p 2(1 -p)2; More genomes give more noise!

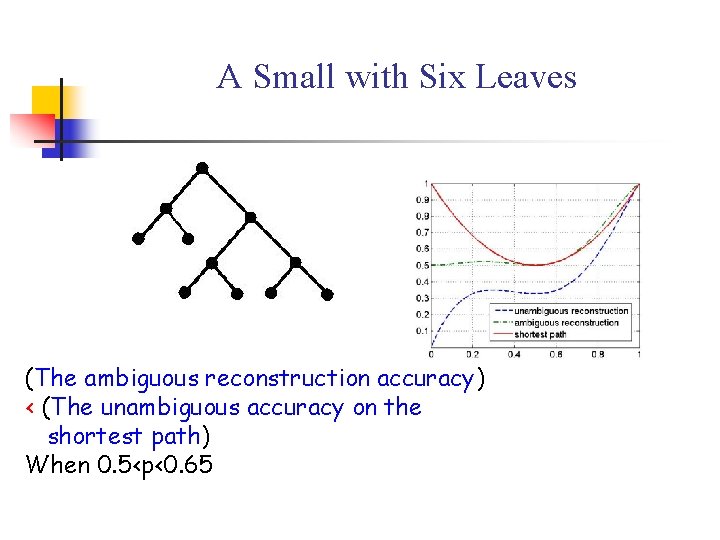
A Small with Six Leaves (The ambiguous reconstruction accuracy) < (The unambiguous accuracy on the shortest path) When 0. 5<p<0. 65

Reconstruction accuracy on complete phylogeny in limit case When conservation rate on each branch is less than 7/8, (The ambiguous reconstruction accuracy) = (the accuracy of using just one genome ) =1/2
 Louxin
Louxin Selecting text means selecting
Selecting text means selecting Chapter 18 genomes and their evolution
Chapter 18 genomes and their evolution Computational biology: genomes, networks, evolution
Computational biology: genomes, networks, evolution Mesquite ancestral state reconstruction
Mesquite ancestral state reconstruction Läkarutlåtande för livränta
Läkarutlåtande för livränta En lathund för arbete med kontinuitetshantering
En lathund för arbete med kontinuitetshantering Tack för att ni lyssnade bild
Tack för att ni lyssnade bild Redogör för vad psykologi är
Redogör för vad psykologi är Centrum för kunskap och säkerhet
Centrum för kunskap och säkerhet Formuö
Formuö Mat för idrottare
Mat för idrottare Inköpsprocessen steg för steg
Inköpsprocessen steg för steg Selecting the promotional mix
Selecting the promotional mix Urban torhamn
Urban torhamn Elektronik för barn
Elektronik för barn Informationskartläggning
Informationskartläggning Acquiring information systems and applications
Acquiring information systems and applications Borra hål för knoppar
Borra hål för knoppar Tack för att ni har lyssnat
Tack för att ni har lyssnat Vishnuismen
Vishnuismen Write the ( 5 ) rules in the selection of delta
Write the ( 5 ) rules in the selection of delta The logical view of data is:
The logical view of data is: Ministerstyre för och nackdelar
Ministerstyre för och nackdelar Guidelines for selecting proper device based controls
Guidelines for selecting proper device based controls Myndigheten för delaktighet
Myndigheten för delaktighet Tack för att ni lyssnade
Tack för att ni lyssnade Treserva lathund
Treserva lathund Handledning reflektionsmodellen
Handledning reflektionsmodellen Formel för lufttryck
Formel för lufttryck The process of selecting training and evaluating employees
The process of selecting training and evaluating employees Perception
Perception Project identification and selection process
Project identification and selection process Dissect the broad area into sub areas
Dissect the broad area into sub areas Sten karttecken
Sten karttecken Särskild löneskatt för pensionskostnader
Särskild löneskatt för pensionskostnader Ekologiskt fotavtryck
Ekologiskt fotavtryck Selecting organizing and interpreting information
Selecting organizing and interpreting information Rbk mätning
Rbk mätning Likability brand elements examples
Likability brand elements examples Selecting an hris
Selecting an hris Verktyg för automatisering av utbetalningar
Verktyg för automatisering av utbetalningar Kung som dog 1611
Kung som dog 1611 Presentera för publik crossboss
Presentera för publik crossboss Boverket ka
Boverket ka Delegerande ledarstil
Delegerande ledarstil Tobinskatten för och nackdelar
Tobinskatten för och nackdelar Meios steg för steg
Meios steg för steg