Genomics 101 DNA sequencing Alignment Gene identification Gene






- Slides: 28

Genomics 101 • DNA sequencing • Alignment • Gene identification • Gene expression • Genome evolution • …

Next Few Topics • Gene Recognition Finding genes in DNA with computational methods • Large-scale alignment & multiple alignment Comparing whole genomes, or large families of genes • Gene Expression and Regulation Measuring the expression of many genes at a time Finding elements in DNA that control the expression of genes

Gene Recognition Credits for slides: Marina Alexandersson Lior Pachter Serge Saxonov

Reading • GENSCAN • Easy. Gene • SLAM • Twinscan Optional: Chris Burge’s Thesis

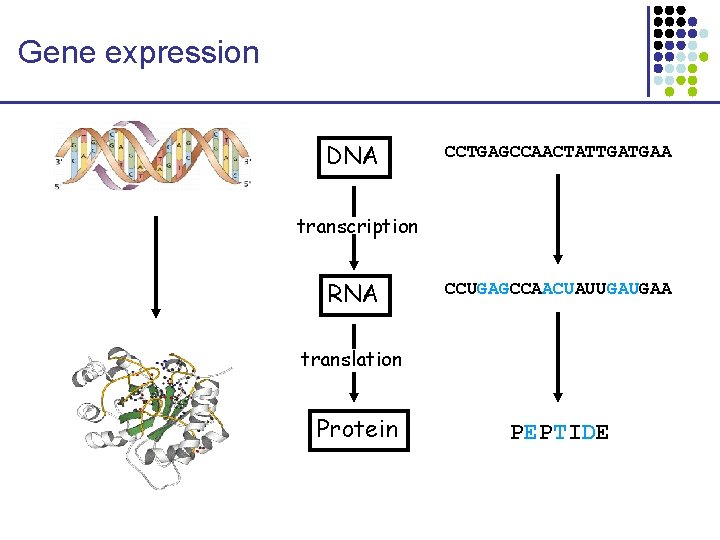
Gene expression DNA CCTGAGCCAACTATTGATGAA transcription RNA CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE

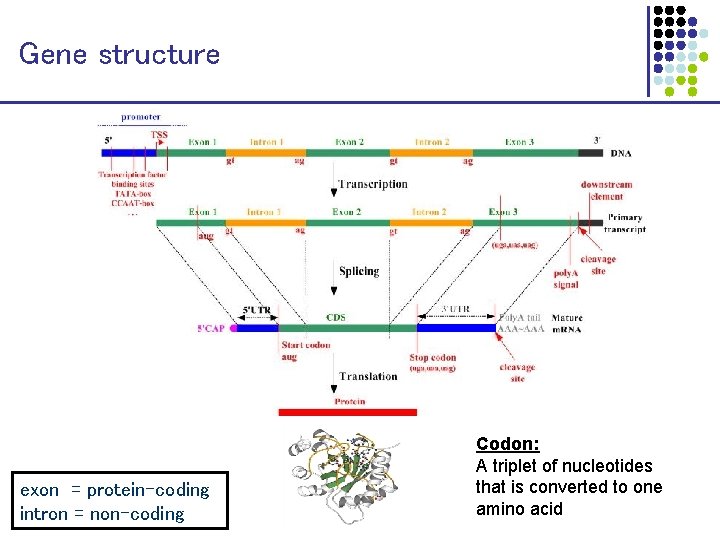
Gene structure intron 1 exon 1 intron 2 exon 3 transcription splicing translation exon = protein-coding intron = non-coding Codon: A triplet of nucleotides that is converted to one amino acid

Where are the genes?

In humans: ~22, 000 genes ~1. 5% of human DNA

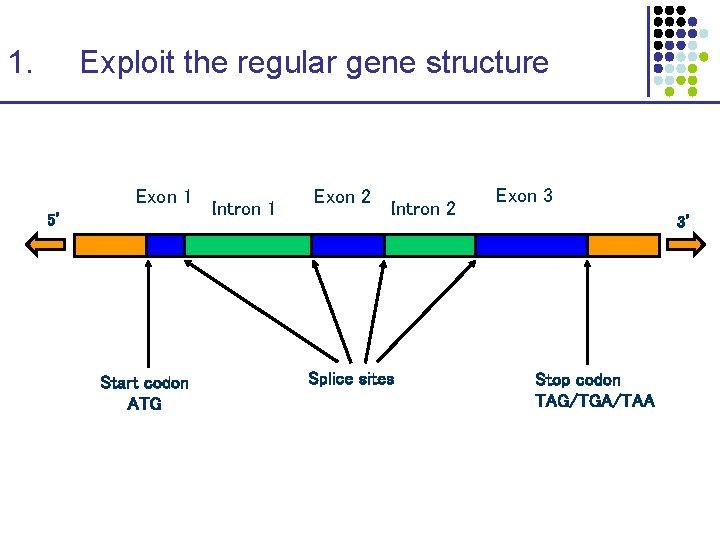
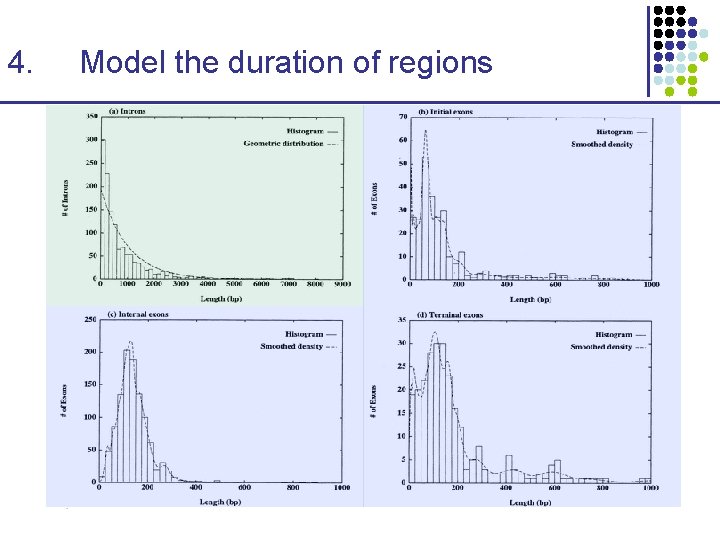
Finding Genes 1. Exploit the regular gene structure ATG—Exon 1—Intron 1—Exon 2—…—Exon. N—STOP 2. Recognize “coding bias” CAG-CGA-GAC-TAT-TTA-GAT-AAC-ACA-CAT-GAA-… 3. Recognize splice sites Intron—c. AGt—Exon—g. GTgag—Intron 4. Model the duration of regions Introns tend to be much longer than exons, in mammals Exons are biased to have a given minimum length 5. Use cross-species comparison Gene structure is conserved in mammals Exons are more similar (~85%) than introns

Approaches to gene finding • Homology § BLAST, Procrustes. • Ab initio § Genscan, Genie, Gene. ID. • Hybrids § Genome. Scan, Genie. EST, Twinscan, SGP, ROSETTA, CEM, TBLASTX, SLAM.

1. Exploit the regular gene structure Exon 1 5’ Start codon ATG Intron 1 Exon 2 Intron 2 Splice sites Exon 3 3’ Stop codon TAG/TGA/TAA

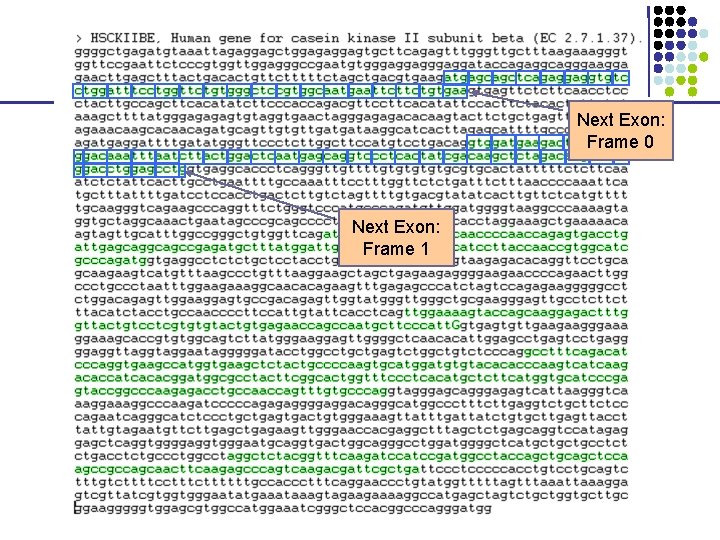
Next Exon: Frame 0 Next Exon: Frame 1

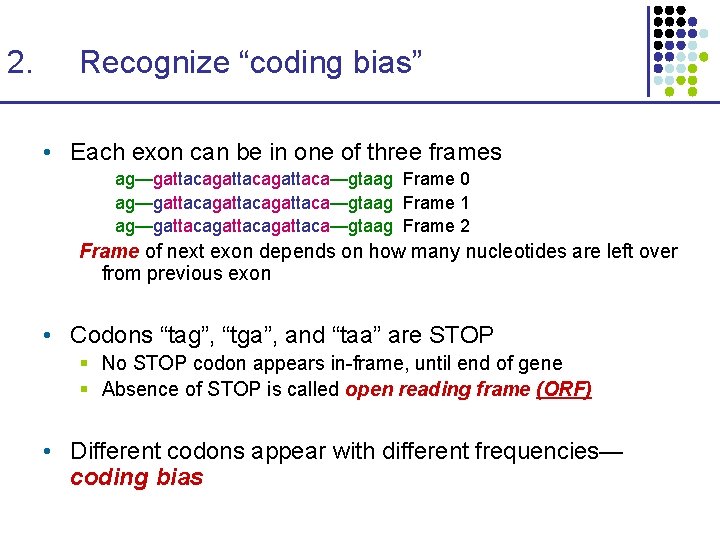
2. Recognize “coding bias” • Each exon can be in one of three frames ag—gattacagattaca—gtaag Frame 0 ag—gattacagattaca—gtaag Frame 1 ag—gattacagattaca—gtaag Frame 2 Frame of next exon depends on how many nucleotides are left over from previous exon • Codons “tag”, “tga”, and “taa” are STOP § No STOP codon appears in-frame, until end of gene § Absence of STOP is called open reading frame (ORF) • Different codons appear with different frequencies— coding bias

2. Recognize “coding bias” Amino Acid Isoleucine Leucine Valine Phenylalanine Methionine Cysteine Alanine Glycine Proline Threonine Serine Tyrosine Tryptophan Glutamine Asparagine Histidine Glutamic acid Aspartic acid Lysine Arginine Stop codons Stop SLC I L V F M C A G P T S Y W Q N H E D K R DNA codons ATT, ATC, ATA CTT, CTC, CTA, CTG, TTA, TTG GTT, GTC, GTA, GTG TTT, TTC ATG TGT, TGC GCT, GCC, GCA, GCG GGT, GGC, GGA, GGG CCT, CCC, CCA, CCG ACT, ACC, ACA, ACG TCT, TCC, TCA, TCG, AGT, AGC TAT, TAC TGG CAA, CAG AAT, AAC CAT, CAC GAA, GAG GAT, GAC AAA, AAG CGT, CGC, CGA, CGG, AGA, AGG TAA, TAG, TGA Can map 61 non-stop codons to frequencies & take log-odds ratios

atg caggtgag cagatg ggtgag cagttg ggtgag caggcc ggtgag tga

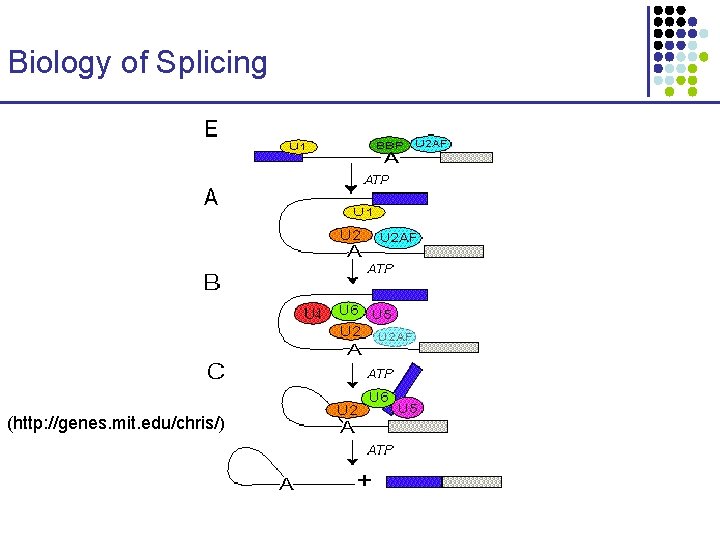
Biology of Splicing (http: //genes. mit. edu/chris/)

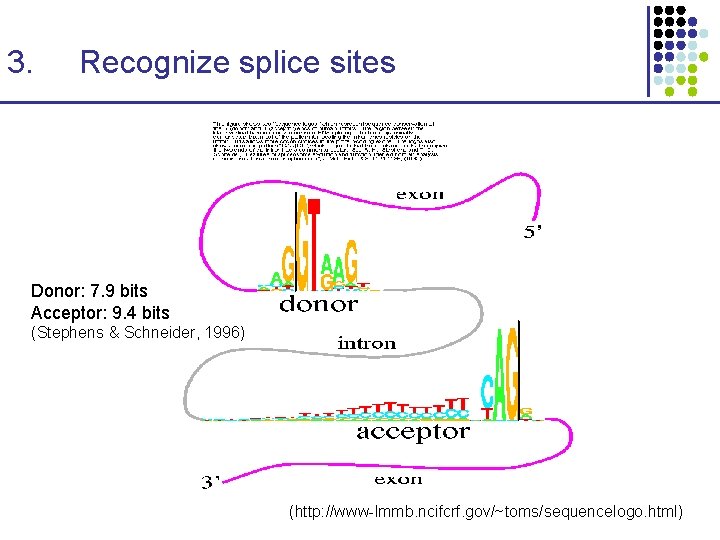
3. Recognize splice sites Donor: 7. 9 bits Acceptor: 9. 4 bits (Stephens & Schneider, 1996) (http: //www-lmmb. ncifcrf. gov/~toms/sequencelogo. html)

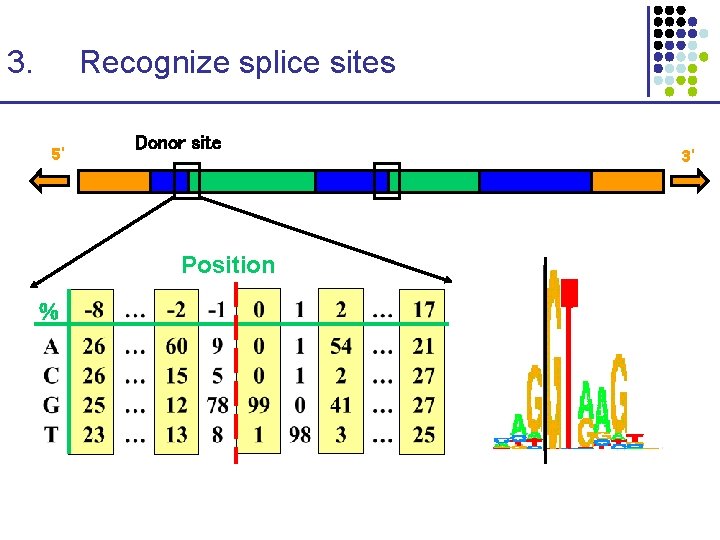
3. Recognize splice sites 5’ Donor site Position % 3’

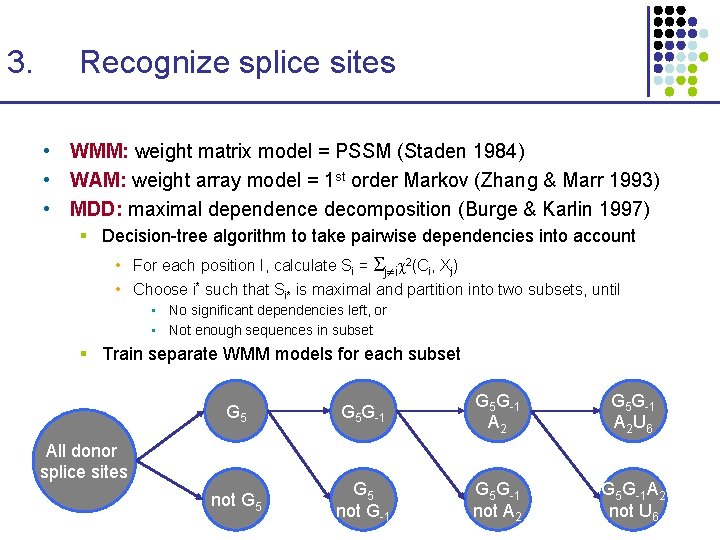
3. Recognize splice sites • WMM: weight matrix model = PSSM (Staden 1984) • WAM: weight array model = 1 st order Markov (Zhang & Marr 1993) • MDD: maximal dependence decomposition (Burge & Karlin 1997) § Decision-tree algorithm to take pairwise dependencies into account • For each position I, calculate Si = j i 2(Ci, Xj) • Choose i* such that Si* is maximal and partition into two subsets, until • No significant dependencies left, or • Not enough sequences in subset § Train separate WMM models for each subset G 5 G-1 A 2 U 6 not G 5 not G-1 G 5 G-1 not A 2 G 5 G-1 A 2 not U 6 All donor splice sites

4. Model the duration of regions

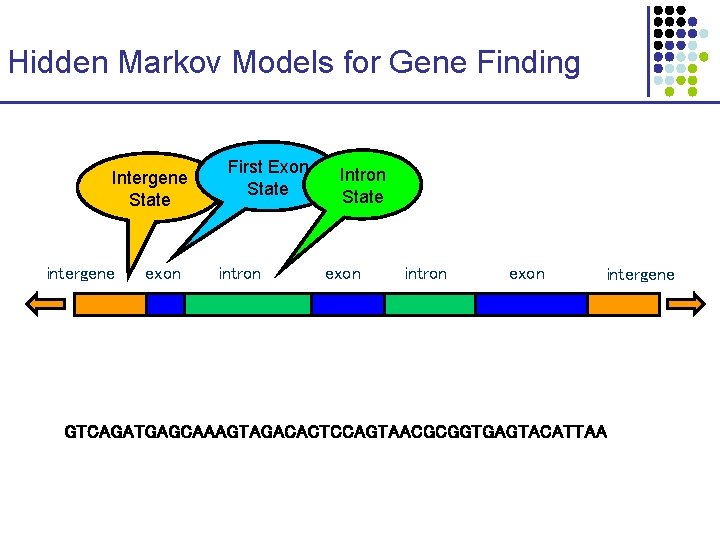
Hidden Markov Models for Gene Finding Intergene State intergene exon First Exon State intron Intron State exon intron exon intergene GTCAGATGAGCAAAGTAGACACTCCAGTAACGCGGTGAGTACATTAA

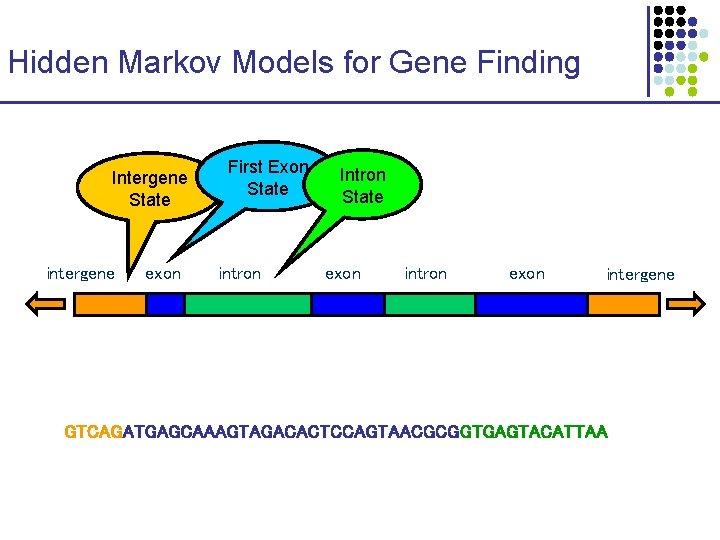
Hidden Markov Models for Gene Finding Intergene State intergene exon First Exon State intron Intron State exon intron exon intergene GTCAGATGAGCAAAGTAGACACTCCAGTAACGCGGTGAGTACATTAA

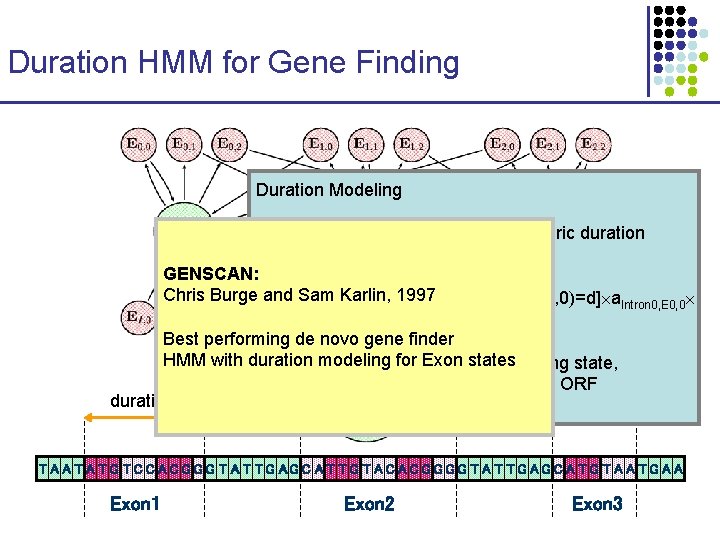
Duration HMM for Gene Finding Duration Modeling Introns: regular HMM states—geometric duration Exons: special duration model GENSCAN: Chris Burge. Vand Sam Karlin, 1997 E 0, 0(i) = maxd=1…D { Prob[duration(E 0, 0)=d] a. Intron 0, E 0, 0 j=i-d+1…ie. E 0, 0(xj) } Best performing de novo gene finder HMM with duration for Exon states where modeling i is an admissible exon-ending state, D is restricted by the longest ORF duration T A A T G T C C A C G G G T A T TG AG C A T T G T A C G G T A T T G A G C A T G T A A T G A A Exon 1 Exon 2 Exon 3

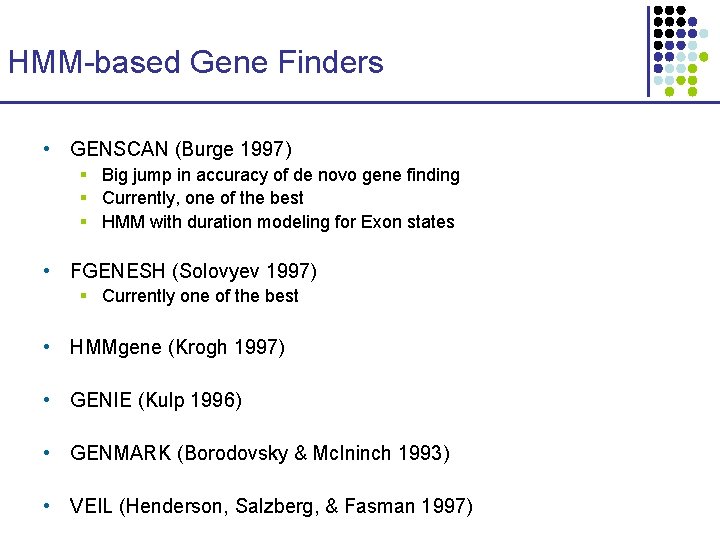
HMM-based Gene Finders • GENSCAN (Burge 1997) § Big jump in accuracy of de novo gene finding § Currently, one of the best § HMM with duration modeling for Exon states • FGENESH (Solovyev 1997) § Currently one of the best • HMMgene (Krogh 1997) • GENIE (Kulp 1996) • GENMARK (Borodovsky & Mc. Ininch 1993) • VEIL (Henderson, Salzberg, & Fasman 1997)

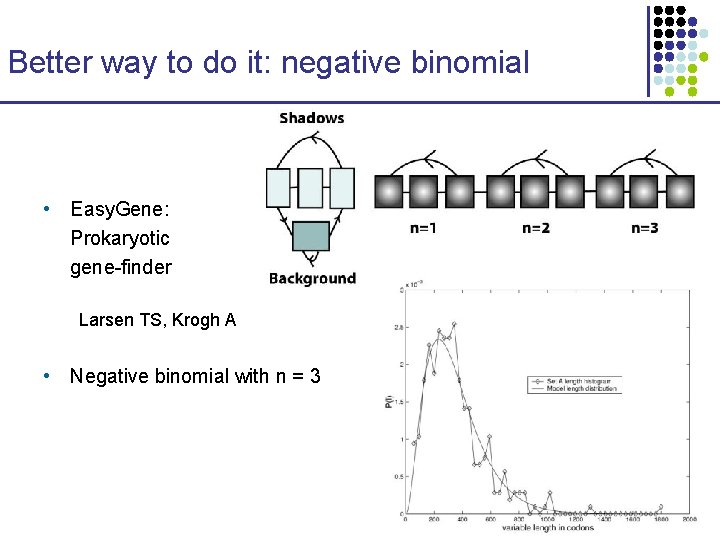
Better way to do it: negative binomial • Easy. Gene: Prokaryotic gene-finder Larsen TS, Krogh A • Negative binomial with n = 3

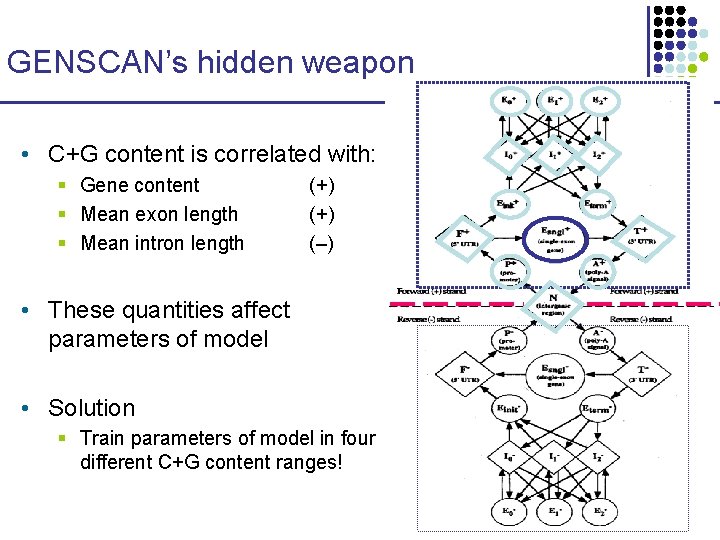
GENSCAN’s hidden weapon • C+G content is correlated with: § Gene content § Mean exon length § Mean intron length (+) (–) • These quantities affect parameters of model • Solution § Train parameters of model in four different C+G content ranges!

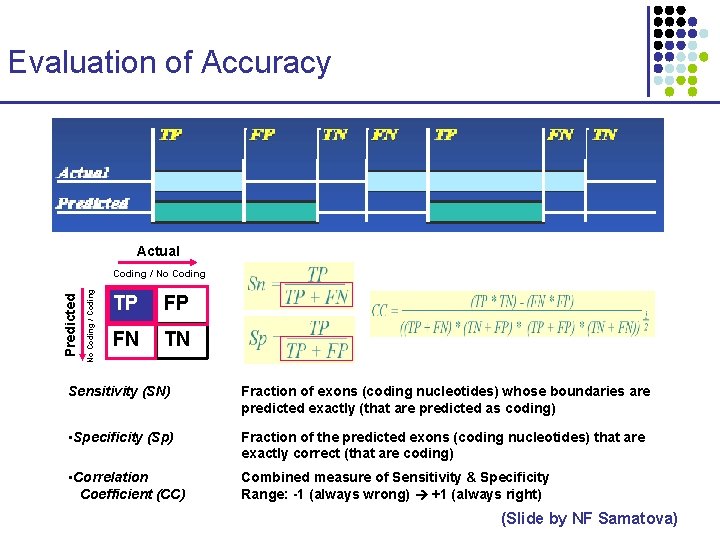
Evaluation of Accuracy TP FP TN FN TP FN TN Actual Predicted Actual No Coding / Coding Predicted Coding / No Coding TP FP FN TN Sensitivity (SN) Fraction of exons (coding nucleotides) whose boundaries are predicted exactly (that are predicted as coding) • Specificity (Sp) Fraction of the predicted exons (coding nucleotides) that are exactly correct (that are coding) • Correlation Coefficient (CC) Combined measure of Sensitivity & Specificity Range: -1 (always wrong) +1 (always right) (Slide by NF Samatova)

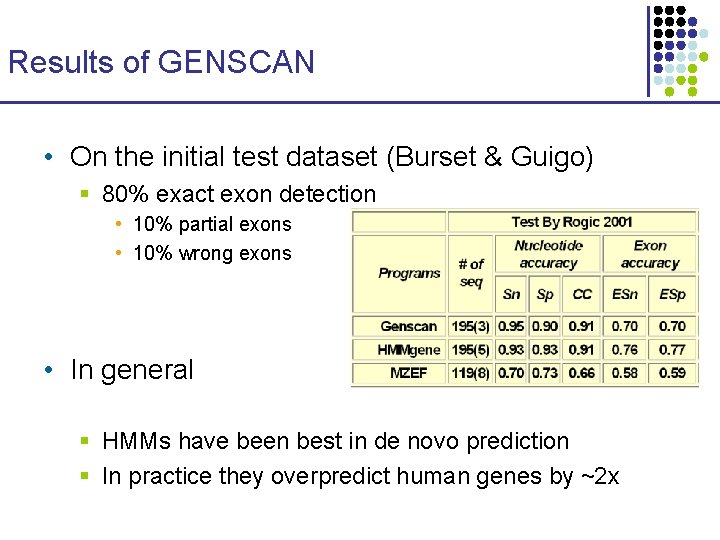
Results of GENSCAN • On the initial test dataset (Burset & Guigo) § 80% exact exon detection • 10% partial exons • 10% wrong exons • In general § HMMs have been best in de novo prediction § In practice they overpredict human genes by ~2 x