Genomic Duplications Structural Variation and Disease Evan Eichler









- Slides: 53

Genomic Duplications, Structural Variation and Disease Evan Eichler Howard Hughes Medical Institute University of Washington April 3 rd, 2006, Frontiers in Genomics

Genomic Variation Mutational mechanisms underlying genetic variation? Sequence • • Single base-pair changes – point mutations Small insertions/deletions– frameshift, microsatellite, minisatellite Mobile elements—retroelement insertions (300 bp -10 kb in size) Large-scale genomic variation (>10 kb) – Large-scale Deletions – Segmental Duplications • Chromosomal variation—translocations, inversions, fusions. Cytogenetics

Global Analysis of Segmental Duplications Question: What is the organization, mechanism and impact of recent human segmental duplications? >90% and > 1 kb in length Intrachromosomal Interchromosomal Segmental Duplications Approaches: • Computational a) Whole genome assembly comparison b) Whole genome shotgun sequence detection strategies • Experimental Comparative sequence analysis, array comparative genomic hybridization, comparative FISH

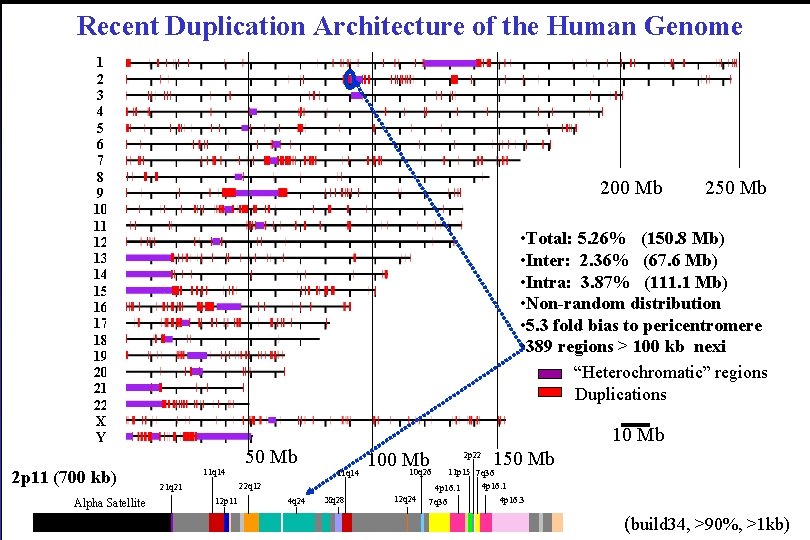
Recent Duplication Architecture of the Human Genome 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 X Y 2 p 11 (700 kb) Alpha Satellite 200 Mb 250 Mb • Total: 5. 26% (150. 8 Mb) • Inter: 2. 36% (67. 6 Mb) • Intra: 3. 87% (111. 1 Mb) • Non-random distribution • 5. 3 fold bias to pericentromere • 389 regions > 100 kb nexi “Heterochromatic” regions Duplications 10 Mb 50 Mb 11 q 14 100 Mb 10 q 26 22 q 12 21 q 21 12 p 11 4 q 24 Xq 28 12 q 24 2 p 22 150 Mb 11 p 15 7 q 36 4 p 16. 1 7 q 36 4 p 16. 3 (build 34, >90%, >1 kb)

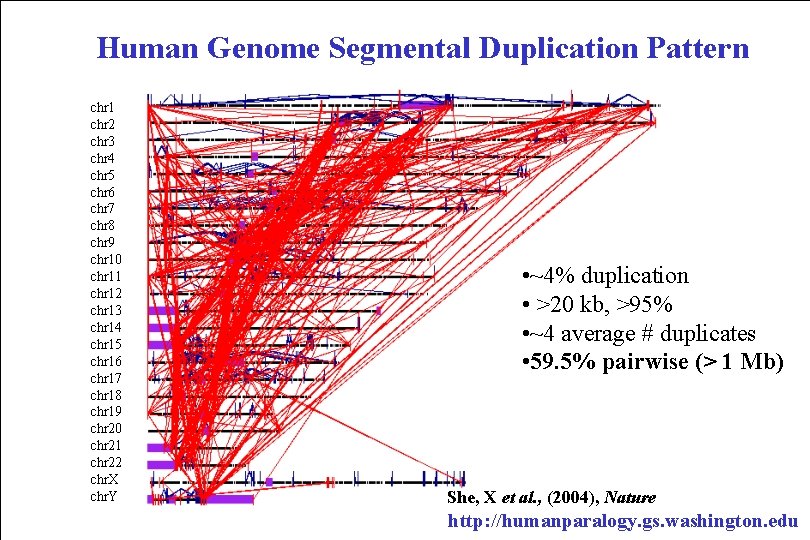
Human Genome Segmental Duplication Pattern chr 1 chr 2 chr 3 chr 4 chr 5 chr 6 chr 7 chr 8 chr 9 chr 10 chr 11 chr 12 chr 13 chr 14 chr 15 chr 16 chr 17 chr 18 chr 19 chr 20 chr 21 chr 22 chr. X chr. Y • ~4% duplication • >20 kb, >95% • ~4 average # duplicates • 59. 5% pairwise (> 1 Mb) She, X et al. , (2004), Nature http: //humanparalogy. gs. washington. edu

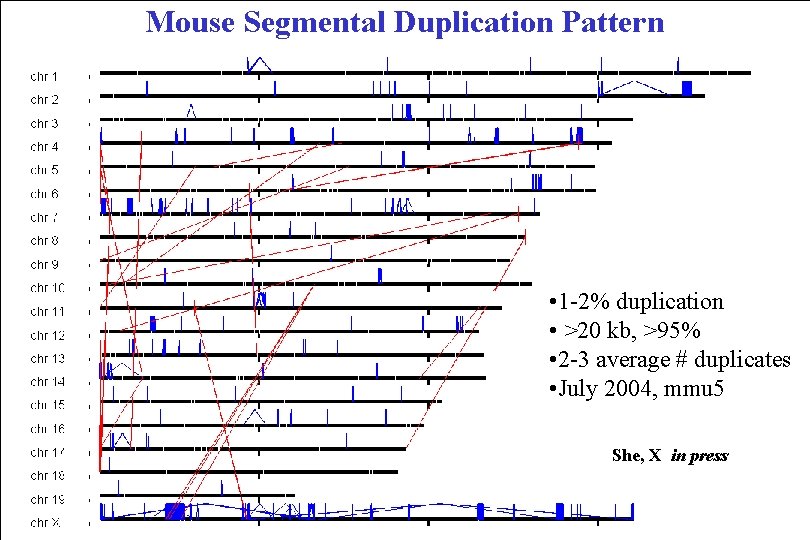
Mouse Segmental Duplication Pattern • 1 -2% duplication • >20 kb, >95% • 2 -3 average # duplicates • July 2004, mmu 5 She, X in press

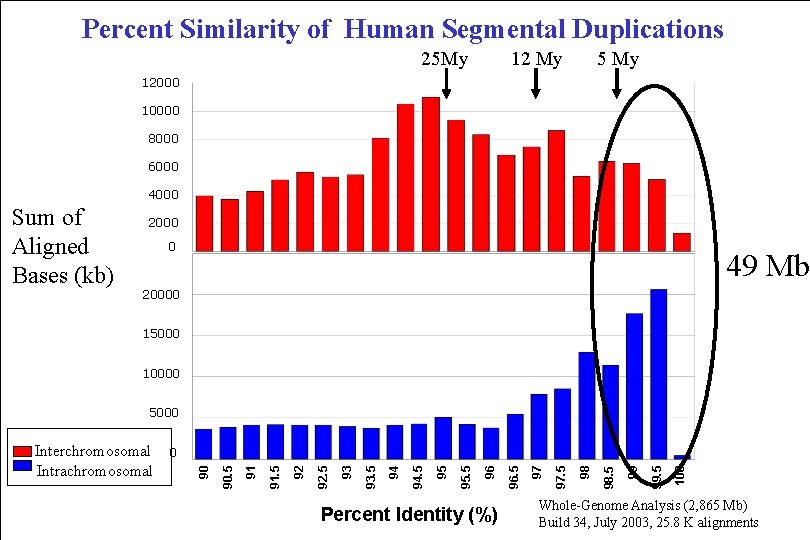
Percent Similarity of Human Segmental Duplications 25 My 12 My 5 My 12000 10000 8000 6000 4000 Sum of Aligned Bases (kb) 2000 0 49 Mb 20000 15000 10000 5000 Percent Identity (%) 100 99. 5 99 98. 5 98 97. 5 97 96. 5 96 95. 5 95 94 93. 5 93 92. 5 92 91. 5 91 90. 5 0 90 Interchromosomal Intrachromosomal Whole-Genome Analysis (2, 865 Mb) Build 34, July 2003, 25. 8 K alignments

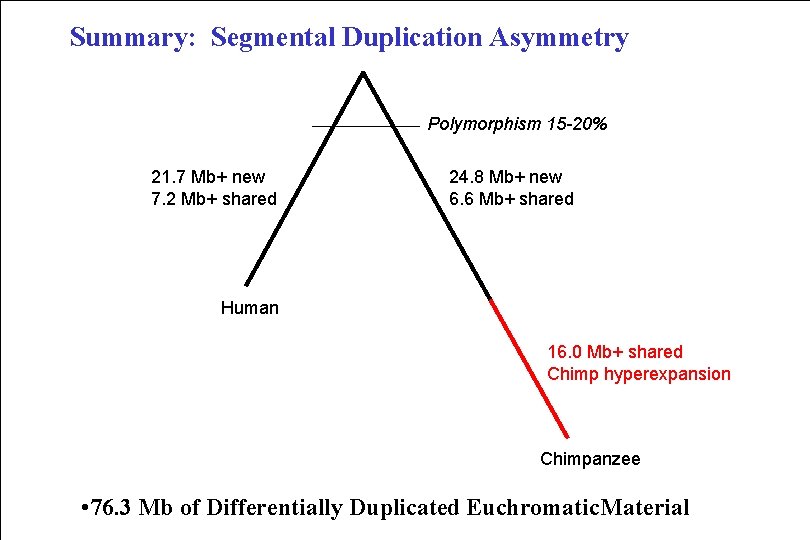
Summary: Segmental Duplication Asymmetry Polymorphism 15 -20% 21. 7 Mb+ new 7. 2 Mb+ shared 24. 8 Mb+ new 6. 6 Mb+ shared Human 16. 0 Mb+ shared Chimp hyperexpansion Chimpanzee • 76. 3 Mb of Differentially Duplicated Euchromatic. Material

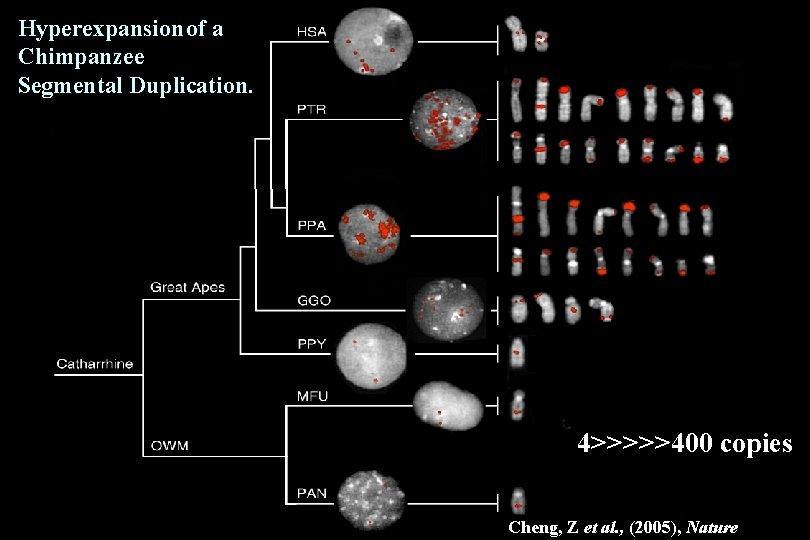
Hyperexpansion of a Chimpanzee Segmental Duplication. 4>>>>>400 copies Cheng, Z et al. , (2005), Nature

Human Segmental Duplications Properties • • Large (>10 kb) Recent (>95% identity) Interspersed (60% are separated by more than 1 Mb) Modular (duplicon architecture) ~389 acceptor regions • 2. 7% Genetic Difference, human vs. chimpanzee What impact in terms of human variation?

Models of Disease • • Rare Duplication-mediated Structural Variation Rare Duplication-Mediated Structural Variation • Common Fine-Scale Structural Variation

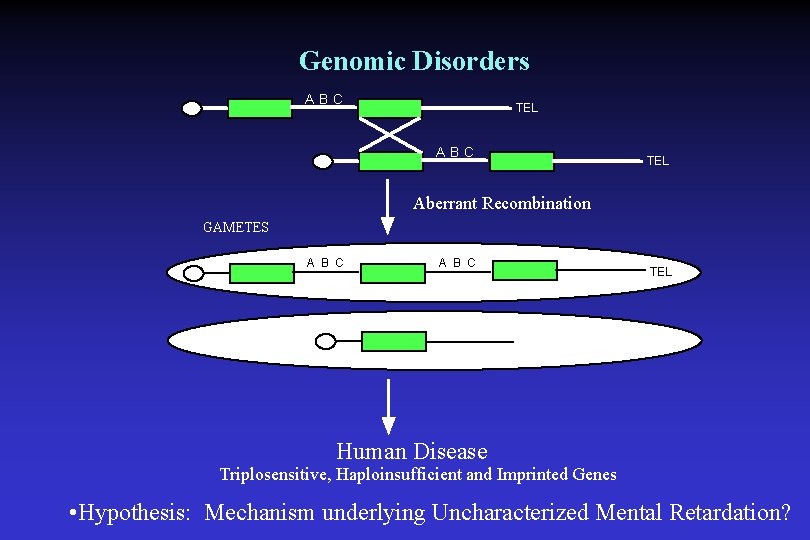
Genomic Disorders ABC TEL Aberrant Recombination GAMETES A B C TEL Human Disease Triplosensitive, Haploinsufficient and Imprinted Genes • Hypothesis: Mechanism underlying Uncharacterized Mental Retardation?

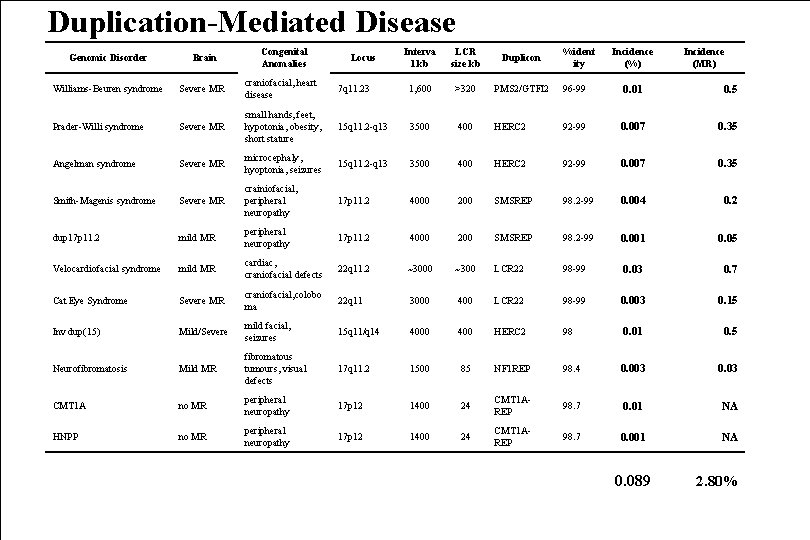
Duplication-Mediated Disease Genomic Disorder Brain Congenital Anomalies Locus Interva l kb LCR size kb Duplicon %ident ity Incidence (%) Incidence (MR) Williams-Beuren syndrome Severe MR craniofacial, heart disease 7 q 11. 23 1, 600 >320 PMS 2/GTFI 2 96 -99 0. 01 0. 5 Prader-Willi syndrome Severe MR small hands, feet, hypotonia, obesity, short stature 15 q 11. 2 -q 13 3500 400 HERC 2 92 -99 0. 007 0. 35 Angelman syndrome Severe MR microcephaly, hyoptonia, seizures 15 q 11. 2 -q 13 3500 400 HERC 2 92 -99 0. 007 0. 35 Smith-Magenis syndrome Severe MR crainiofacial, peripheral neuropathy 17 p 11. 2 4000 200 SMSREP 98. 2 -99 0. 004 0. 2 dup 17 p 11. 2 mild MR peripheral neuropathy 17 p 11. 2 4000 200 SMSREP 98. 2 -99 0. 001 0. 05 Velocardiofacial syndrome mild MR cardiac, craniofacial defects 22 q 11. 2 ~3000 ~300 LCR 22 98 -99 0. 03 0. 7 Cat Eye Syndrome Severe MR craniofacial, colobo ma 22 q 11 3000 400 LCR 22 98 -99 0. 003 0. 15 Inv dup(15) Mild/Severe mild facial, seizures 15 q 11/q 14 4000 400 HERC 2 98 0. 01 0. 5 Neurofibromatosis Mild MR fibromatous tumours, visual defects 17 q 11. 2 1500 85 NF 1 REP 98. 4 0. 003 0. 03 CMT 1 A no MR peripheral neuropathy 17 p 12 1400 24 CMT 1 AREP 98. 7 0. 01 NA HNPP no MR peripheral neuropathy 17 p 12 1400 24 CMT 1 AREP 98. 7 0. 001 NA 0. 089 2. 80%

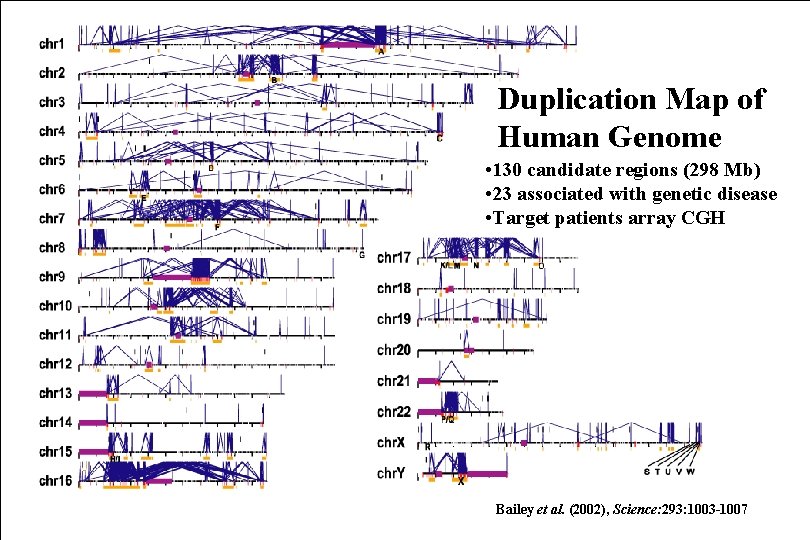
Duplication Map of Human Genome • 130 candidate regions (298 Mb) • 23 associated with genetic disease • Target patients array CGH Bailey et al. (2002), Science: 293: 1003 -1007

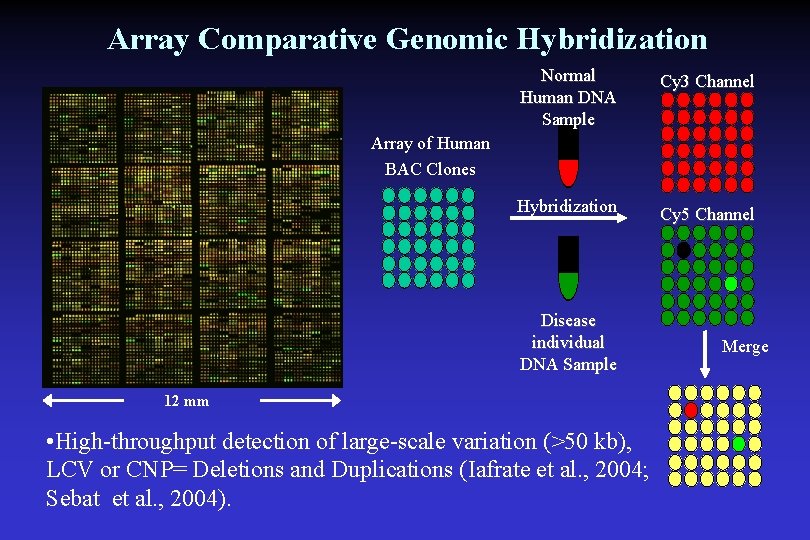
Array Comparative Genomic Hybridization Normal Human DNA Sample Cy 3 Channel Hybridization Cy 5 Channel Array of Human BAC Clones Disease individual DNA Sample 12 mm • High-throughput detection of large-scale variation (>50 kb), LCV or CNP= Deletions and Duplications (Iafrate et al. , 2004; Sebat et al. , 2004). Merge

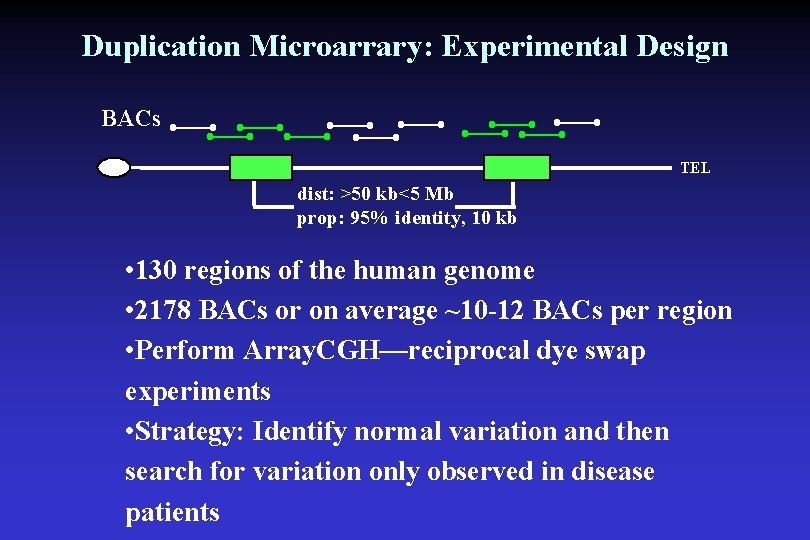
Duplication Microarrary: Experimental Design BACs TEL dist: >50 kb<5 Mb prop: 95% identity, 10 kb • 130 regions of the human genome • 2178 BACs or on average ~10 -12 BACs per region • Perform Array. CGH—reciprocal dye swap experiments • Strategy: Identify normal variation and then search for variation only observed in disease patients

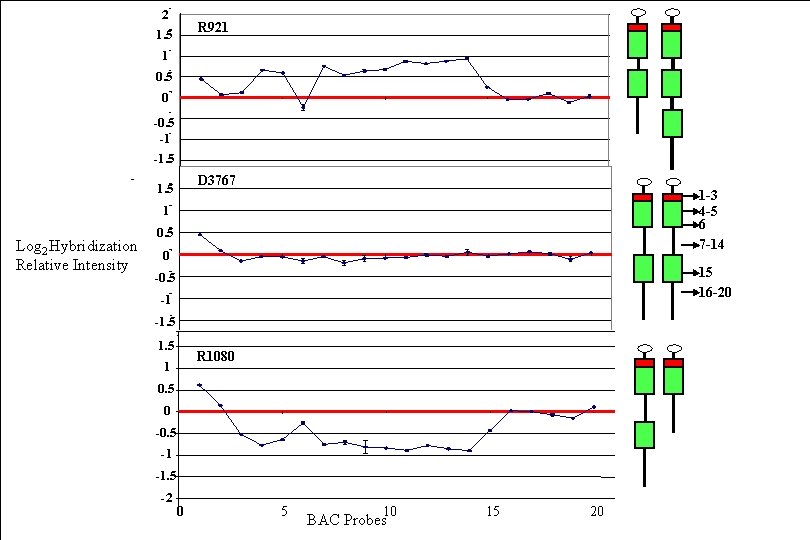
2 R 921 1. 5 1 0. 5 0 -0. 5 -1 -1. 5 D 3767 1. 5 5 10 15 20 1 -3 4 -5 6 7 -14 1 Log 2 Hybridization Relative Intensity 0. 5 0 Hybridization -0. 5 15 16 -20 -1 -1. 5 0 R 1080 1 5 10 15 20 0. 5 0 -0. 5 -1 -1. 5 -2 0 5 10 BAC Probes 15 20

Study Populations • Normal unaffected (diversity panel and Hap. Map Samples). Target= 800 samples, Completed: 75 + 269 samples=344 total—Identified additional 257 CNPs. • Idiopathic Mental Retardation: Target =900 samples; (400 samples Flint, 500 CWRU samples); 291 complete

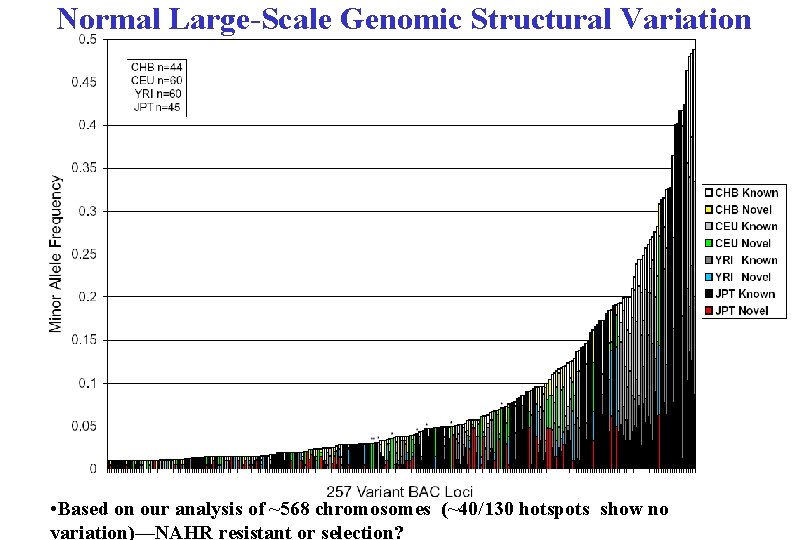
Normal Large-Scale Genomic Structural Variation • Based on our analysis of ~568 chromosomes (~40/130 hotspots show no variation)—NAHR resistant or selection?

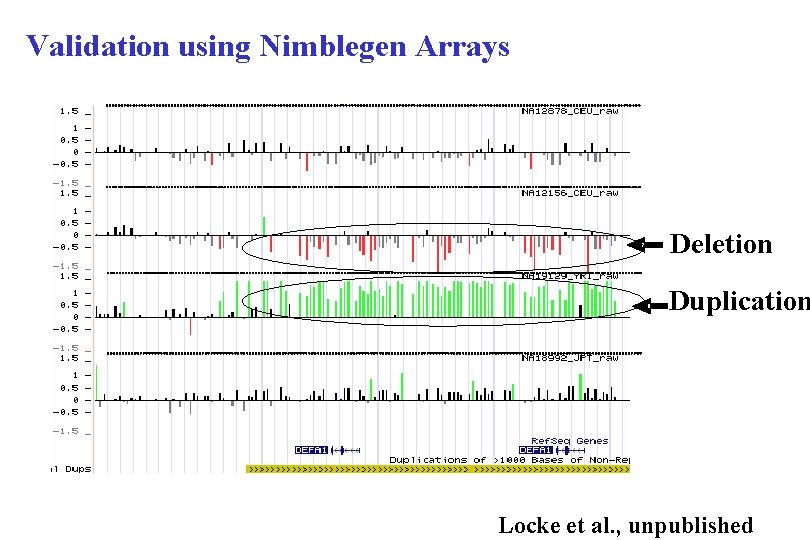
Validation using Nimblegen Arrays Deletion Duplication Locke et al. , unpublished

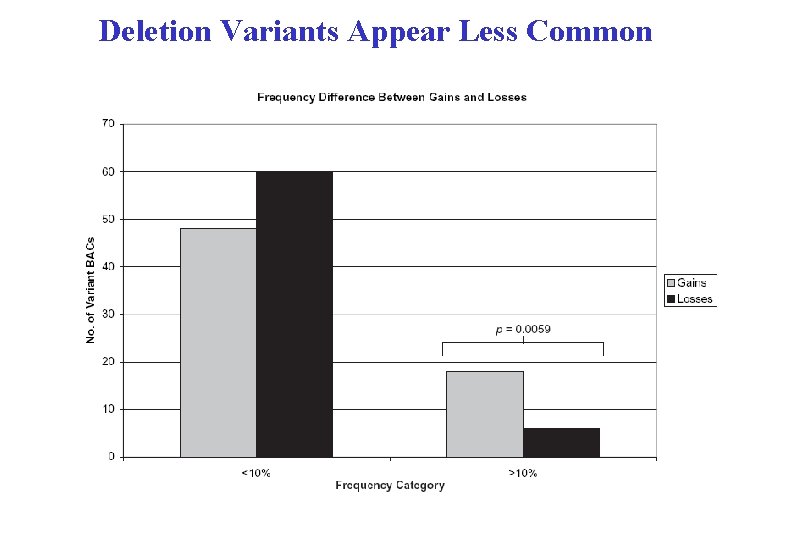
Deletion Variants Appear Less Common

Study Populations • Normal unaffected (diversity panel and Hap. Map Samples). Target= 800 samples, Completed: 75 + 269 samples=344 total—Identified additional 257 CNPs. • Idiopathic Mental Retardation: Target =900 samples; (400 samples Flint, 500 CWRU samples); 291 complete

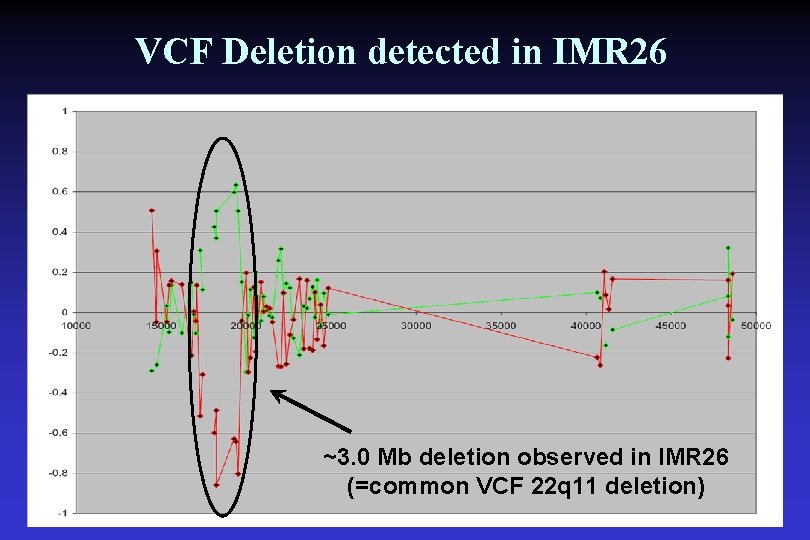
VCF Deletion detected in IMR 26 ~3. 0 Mb deletion observed in IMR 26 (=common VCF 22 q 11 deletion)

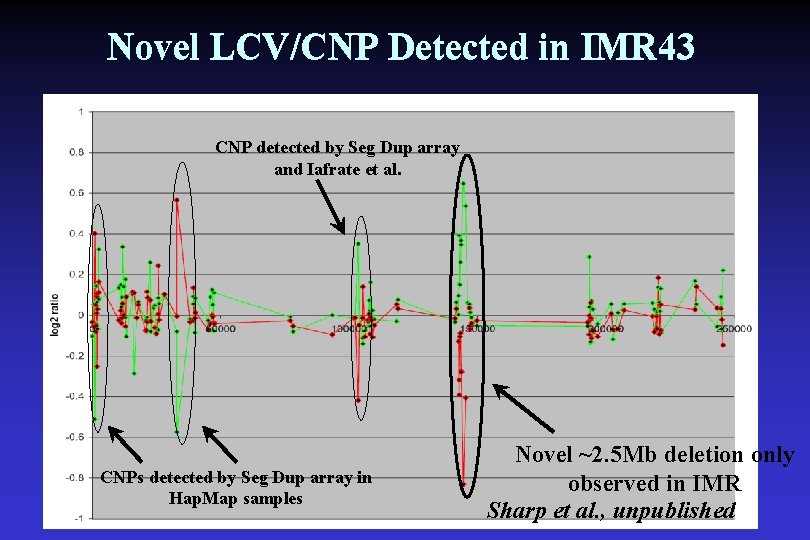
Novel LCV/CNP Detected in IMR 43 CNP detected by Seg Dup array and Iafrate et al. CNPs detected by Seg Dup array in Hap. Map samples Novel ~2. 5 Mb deletion only observed in IMR Sharp et al. , unpublished

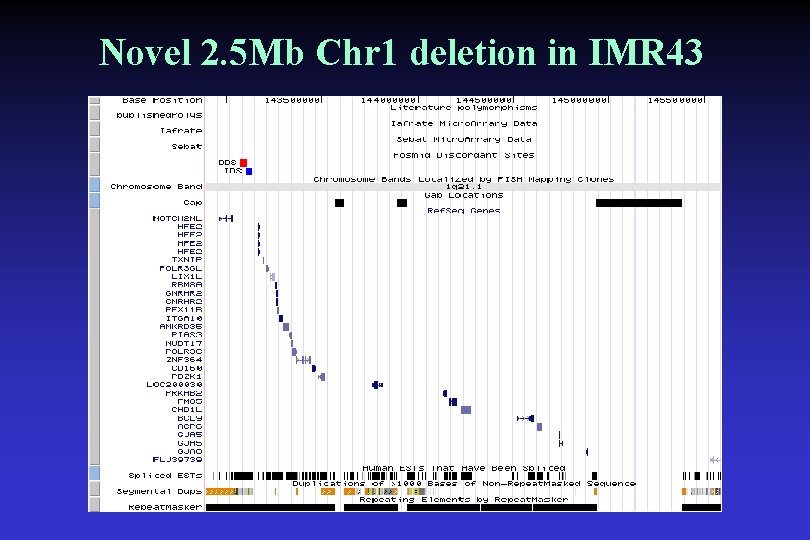
Novel 2. 5 Mb Chr 1 deletion in IMR 43

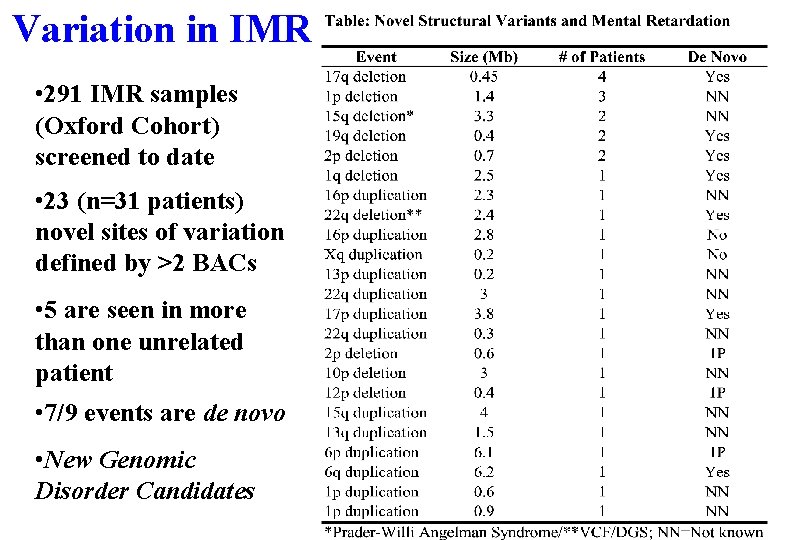
Variation in IMR • 291 IMR samples (Oxford Cohort) screened to date • 23 (n=31 patients) novel sites of variation defined by >2 BACs • 5 are seen in more than one unrelated patient • 7/9 events are de novo • New Genomic Disorder Candidates

Problems: • Array CGH has a lower limit to detect deletions (~30 kb) • Oligo-based approaches effectively sample a small fraction of the genome and extrapolate size indirectly 1. 2. 3. 4. Precise location of the rearrangement is unknown. Neither can identify subtle (5 -30 kb) variation Neither approach can detect inversions. Location and structure of the change unknown

Models of Disease • Rare Duplication-mediated Structural Variation • Common Fine-Scale Structural Variation

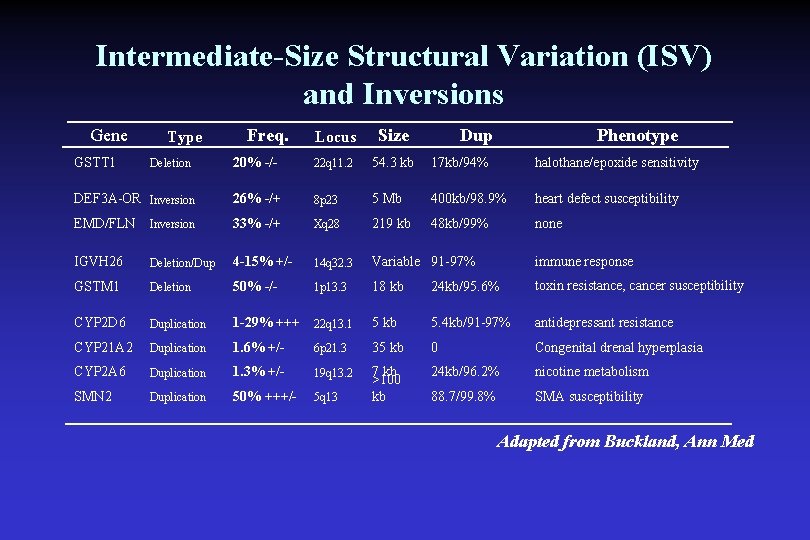
Intermediate-Size Structural Variation (ISV) and Inversions Gene Locus Size 20% -/- 22 q 11. 2 54. 3 kb 17 kb/94% halothane/epoxide sensitivity DEF 3 A-OR Inversion 26% -/+ 8 p 23 5 Mb 400 kb/98. 9% heart defect susceptibility EMD/FLN Inversion 33% -/+ Xq 28 219 kb 48 kb/99% none IGVH 26 Deletion/Dup 4 -15% +/- 14 q 32. 3 Variable 91 -97% GSTM 1 Deletion 50% -/- 1 p 13. 3 18 kb 24 kb/95. 6% toxin resistance, cancer susceptibility CYP 2 D 6 Duplication 1 -29% +++ 22 q 13. 1 5 kb 5. 4 kb/91 -97% antidepressant resistance CYP 21 A 2 Duplication 1. 6% +/- 6 p 21. 3 35 kb 0 Congenital drenal hyperplasia CYP 2 A 6 Duplication 1. 3% +/- 19 q 13. 2 24 kb/96. 2% nicotine metabolism SMN 2 Duplication 50% +++/- 5 q 13 7 kb >100 kb 88. 7/99. 8% SMA susceptibility GSTT 1 Type Deletion Freq. Dup Phenotype immune response Adapted from Buckland, Ann Med

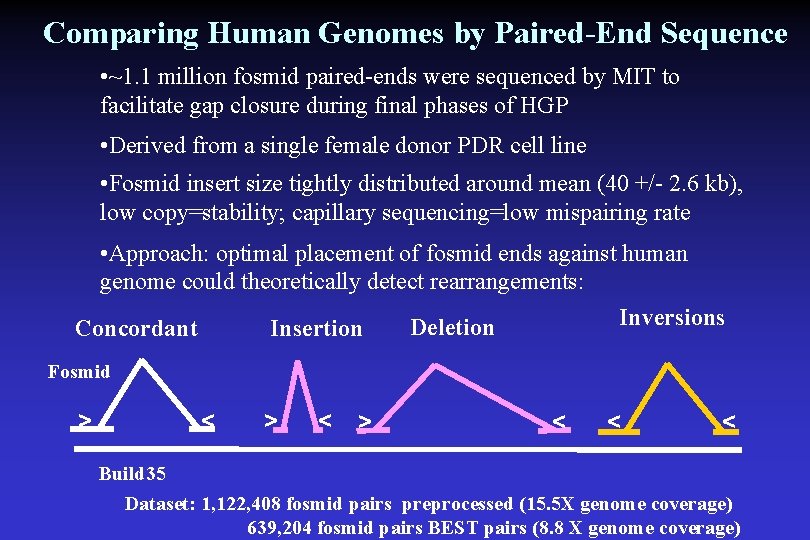
Comparing Human Genomes by Paired-End Sequence • ~1. 1 million fosmid paired-ends were sequenced by MIT to facilitate gap closure during final phases of HGP • Derived from a single female donor PDR cell line • Fosmid insert size tightly distributed around mean (40 +/- 2. 6 kb), low copy=stability; capillary sequencing=low mispairing rate • Approach: optimal placement of fosmid ends against human genome could theoretically detect rearrangements: Inversions Deletion Insertion Concordant Fosmid > < > < < < Build 35 Dataset: 1, 122, 408 fosmid pairs preprocessed (15. 5 X genome coverage) 639, 204 fosmid pairs BEST pairs (8. 8 X genome coverage)

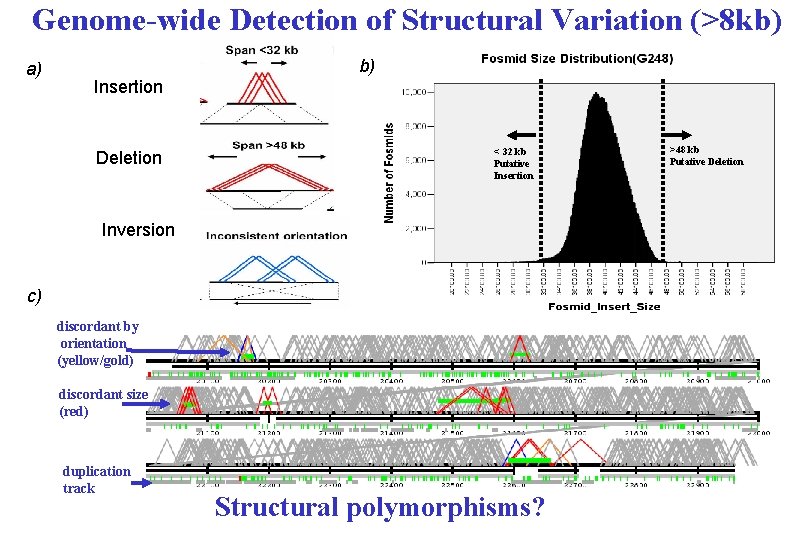
Genome-wide Detection of Structural Variation (>8 kb) a) b) Insertion Deletion < 32 kb Putative Insertion Inversion c) discordant by orientation (yellow/gold) discordant size (red) duplication track Structural polymorphisms? >48 kb Putative Deletion

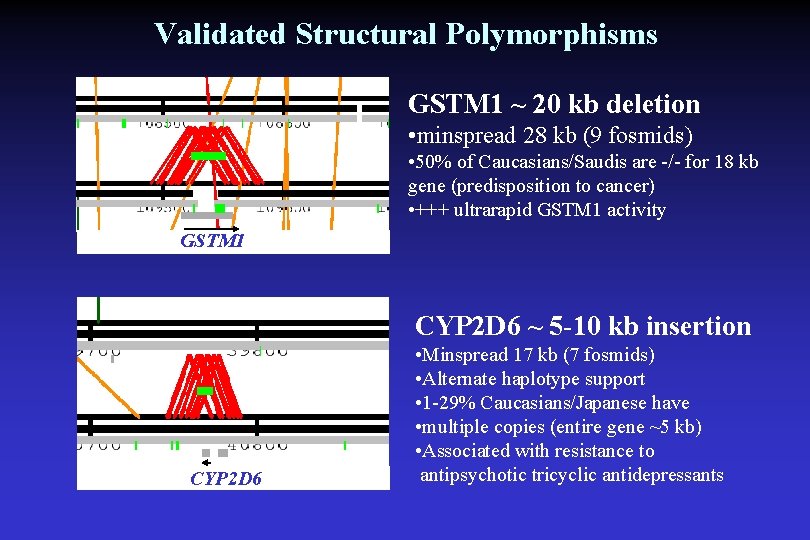
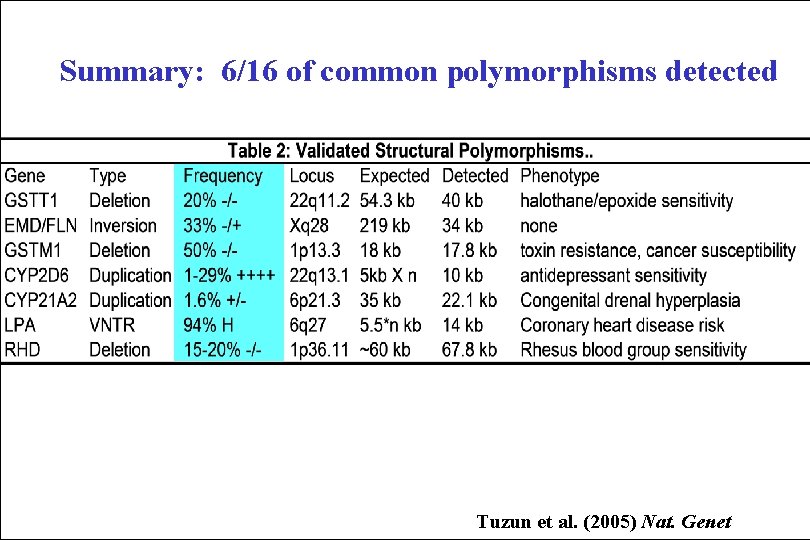
Validated Structural Polymorphisms GSTM 1 ~ 20 kb deletion • minspread 28 kb (9 fosmids) • 50% of Caucasians/Saudis are -/- for 18 kb gene (predisposition to cancer) • +++ ultrarapid GSTM 1 activity GSTM 1 CYP 2 D 6 ~ 5 -10 kb insertion CYP 2 D 6 • Minspread 17 kb (7 fosmids) • Alternate haplotype support • 1 -29% Caucasians/Japanese have • multiple copies (entire gene ~5 kb) • Associated with resistance to antipsychotic tricyclic antidepressants

Summary: 6/16 of common polymorphisms detected Tuzun et al. (2005) Nat. Genet

……Sequence the Structural Variation

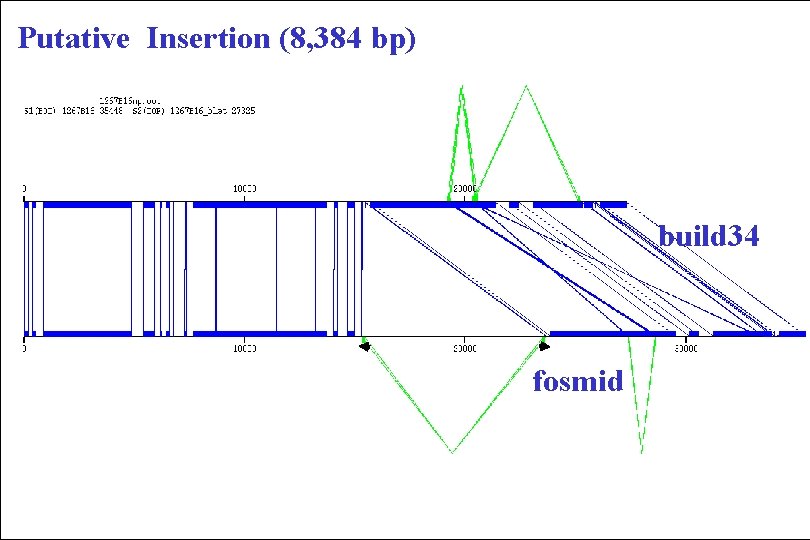
Putative Insertion (8, 384 bp) build 34 fosmid

Putative Deletion (14, 055 bp) build 34 fosmid

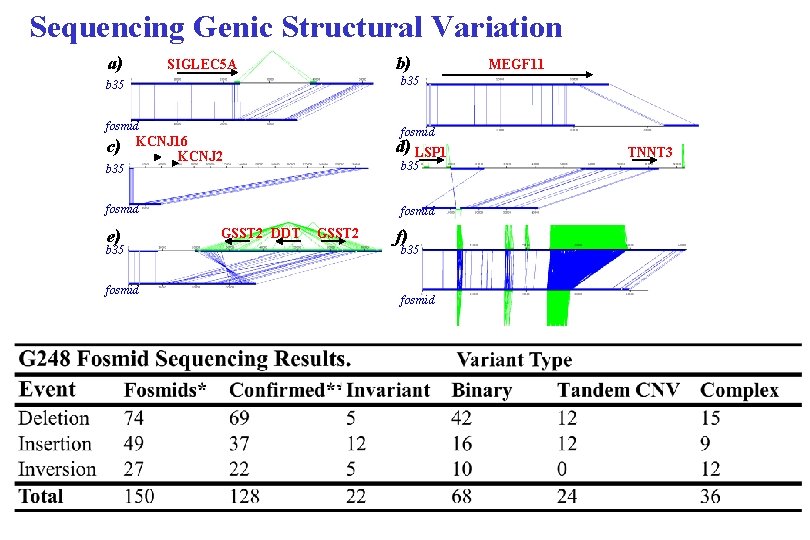
Sequencing Genic Structural Variation a) b) SIGLEC 5 A b 35 fosmid c) b 35 MEGF 11 fosmid KCNJ 16 KCNJ 2 d) LSP 1 b 35 fosmid e) b 35 fosmid GSST 2 DDT GSST 2 f) b 35 fosmid TNNT 3

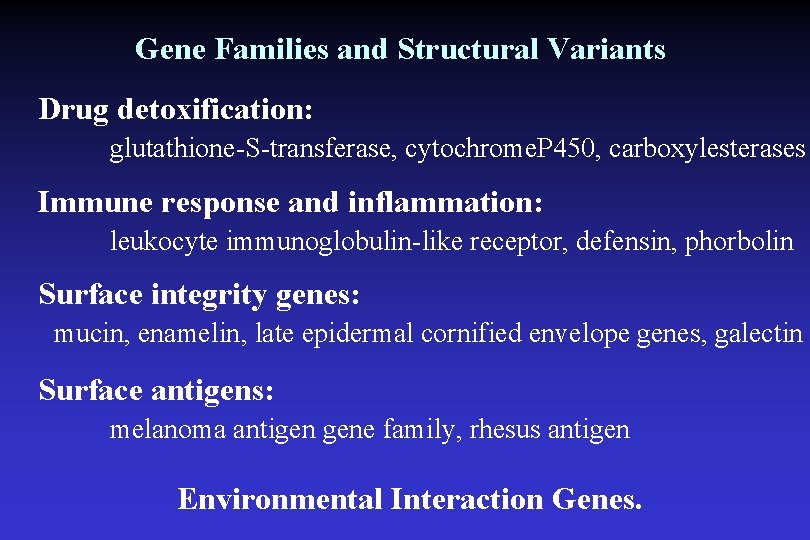
Gene Families and Structural Variants Drug detoxification: glutathione-S-transferase, cytochrome. P 450, carboxylesterases Immune response and inflammation: leukocyte immunoglobulin-like receptor, defensin, phorbolin Surface integrity genes: mucin, enamelin, late epidermal cornified envelope genes, galectin Surface antigens: melanoma antigen gene family, rhesus antigen Environmental Interaction Genes.

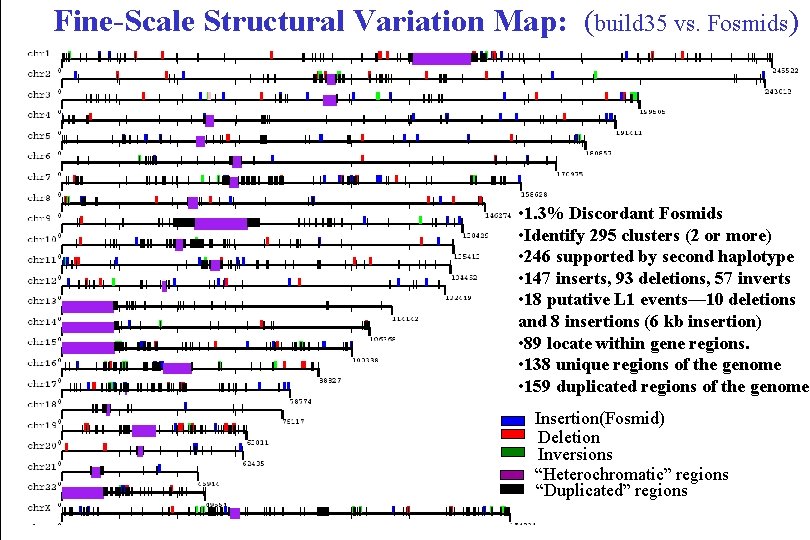
Fine-Scale Structural Variation Map: (build 35 vs. Fosmids) • 1. 3% Discordant Fosmids • Identify 295 clusters (2 or more) • 246 supported by second haplotype • 147 inserts, 93 deletions, 57 inverts • 18 putative L 1 events— 10 deletions and 8 insertions (6 kb insertion) • 89 locate within gene regions. • 138 unique regions of the genome • 159 duplicated regions of the genome Insertion(Fosmid) Deletion Inversions “Heterochromatic” regions “Duplicated” regions

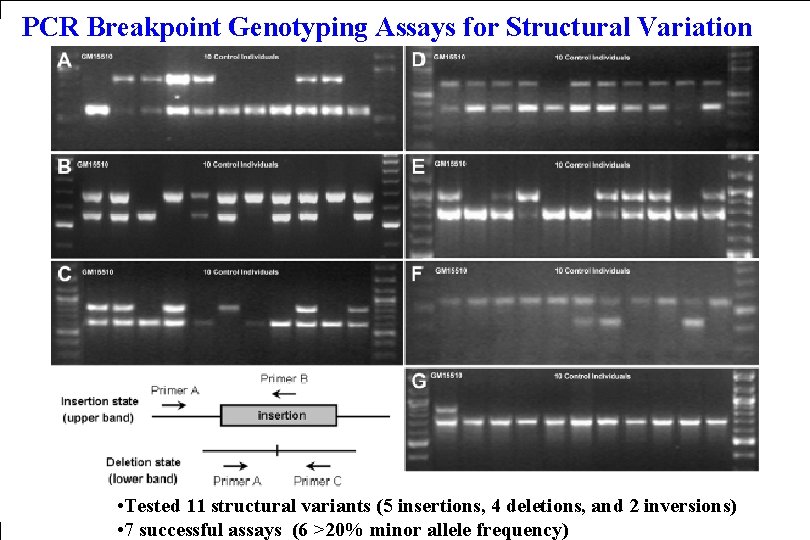
PCR Breakpoint Genotyping Assays for Structural Variation • Tested 11 structural variants (5 insertions, 4 deletions, and 2 inversions) • 7 successful assays (6 >20% minor allele frequency)

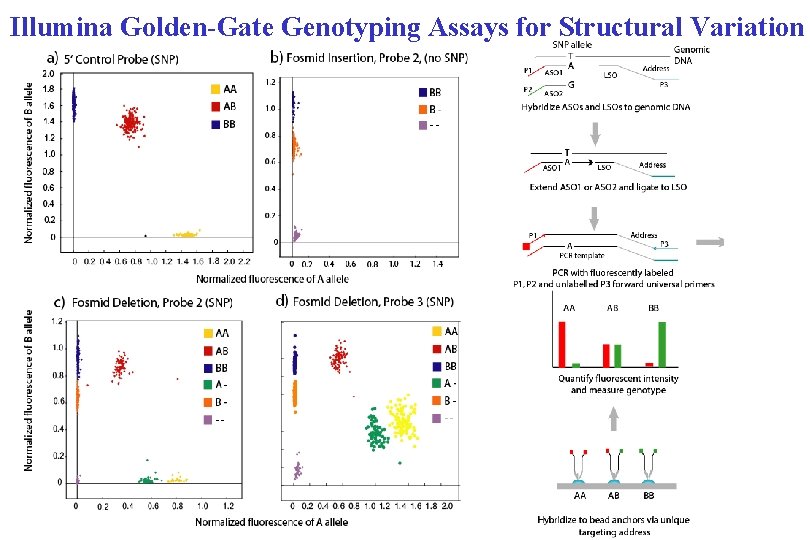
Illumina Golden-Gate Genotyping Assays for Structural Variation

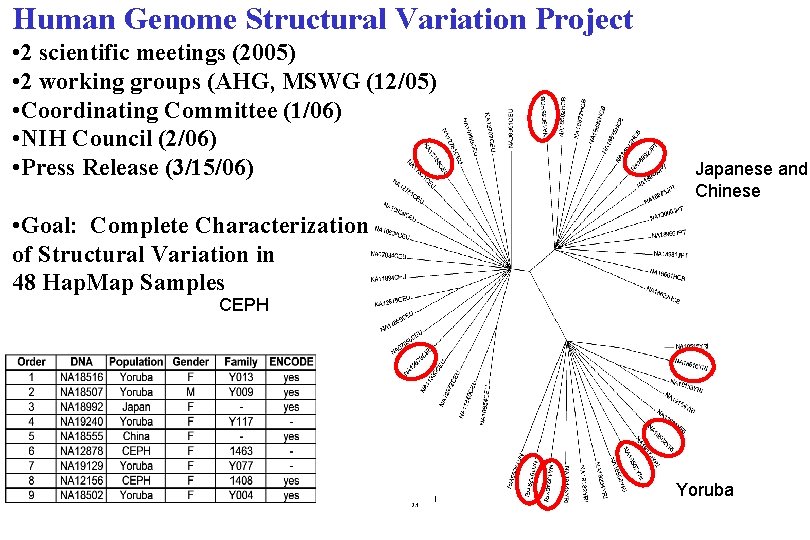
Human Genome Structural Variation Project • 2 scientific meetings (2005) • 2 working groups (AHG, MSWG (12/05) • Coordinating Committee (1/06) • NIH Council (2/06) • Press Release (3/15/06) Japanese and Chinese • Goal: Complete Characterization of Structural Variation in 48 Hap. Map Samples CEPH Yoruba

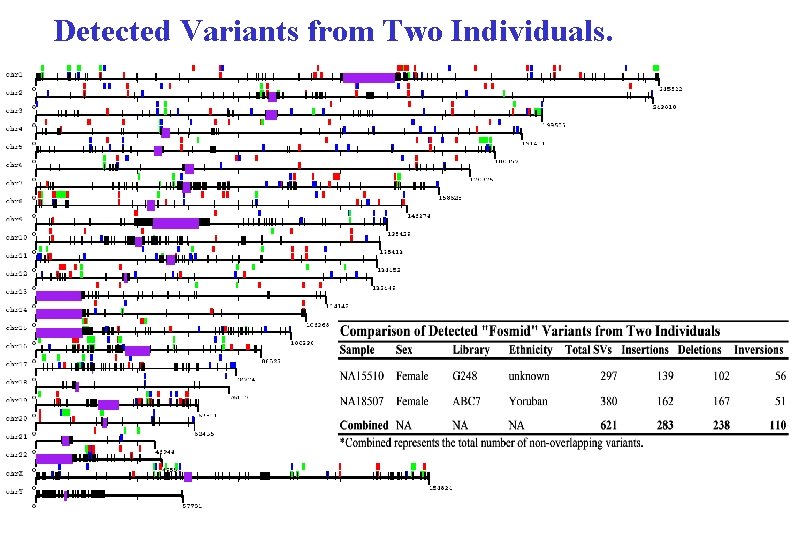
Detected Variants from Two Individuals.

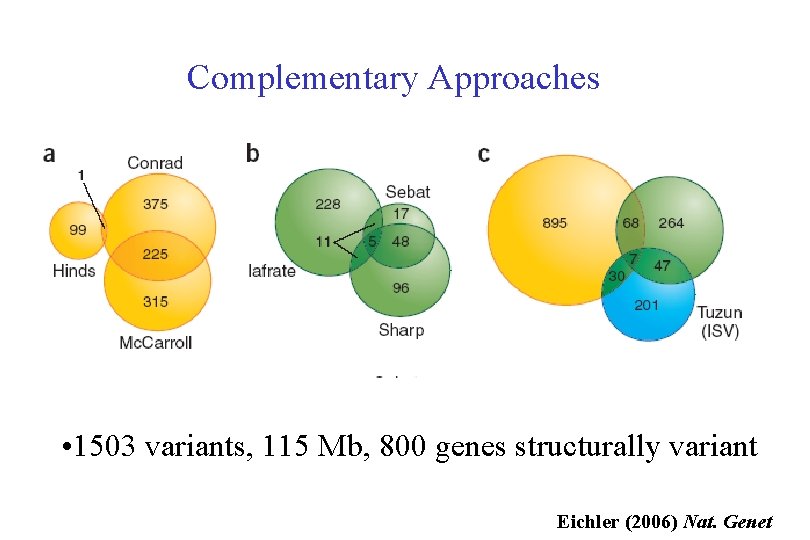
Complementary Approaches • 1503 variants, 115 Mb, 800 genes structurally variant Eichler (2006) Nat. Genet

Summary • Humans relatively unique in size, proportion and architecture of interspersed segmental duplications • Large-Scale Variation • Normals: Identified 257 CNPs using a targeted microarray to duplicated regions • IMR: Identified 23 sites (>2 BACs) unique to patients (n=291 probands) (5 are recurrent and 7 are confirmed de novo) Novel Genomic Disorders • Fine-Scale Variation: Developed an approach to map and sequence common fine-scale variation within the human Population, estimate ~200 -300 differences > 8 kb between 2 individuals.


Models of Human “Genetic” Disease 1) Simple Mendelian --one gene-one disease, familial, highly penetrant, small fraction of pop. Eg. cystic fibrosis 2) Chromosome Disease –large chromosomal regions, non-familial, sporadic, relatively high frequency Eg. Turner Syndrome 3) Genomic Disease –familial and/or recurrent, deletion or duplication of large # of genes, dosage effects. Eg. Prader-Willi Syndrome. 4) Complex Traits--multiple genes plus environment, familial, variably penetrant, large fraction of population, susceptibility genes eg. hypertension.

Acknowledgements Eichler Lab Eray Tuzun Andy Sharp Devin Locke Matthew Johnson Zhaoshi Jiang Jon Bleyhl Sean Mc. Grath Tera Newman Jeff Bailey Anne Morrison Lisa Pertz Ze Cheng Xinwei She James Sprague UCSF Dan Pinkel Donna Albertson CWRU/UChicago Stuart Schwartz Laurie Christ Oxford Jonathan Flint Samantha Knight UW Debbie Nickerson Mark Rieder Chris Carlson Josh Smith UWGSC Maynard Olson Rajinder Kaul Hillary Hayden Eric Haugen Agencourt Doug Smith NHGRI Jim Mullikin

……Finding Novel Human Sequence

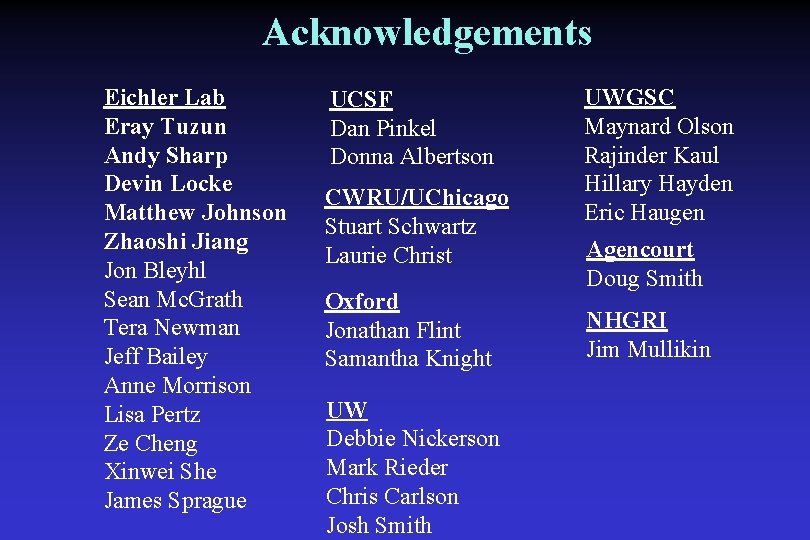
Sequence of Traversing Fosmid Fills Gaps Kaul et al, unpublished

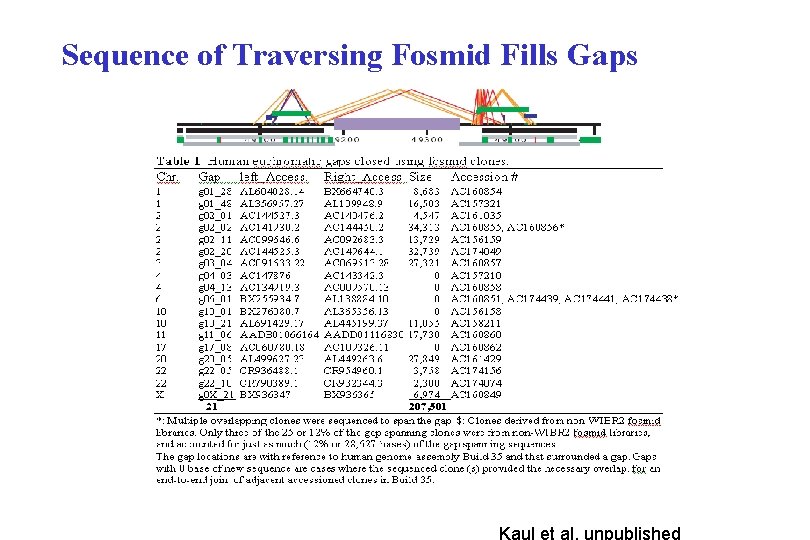
Singleton Fosmids Extend into Gaps Kaul et al, unpublished

Fosmid Pairs that fail to Map to build 35 • 4773 fosmid paired-end sequences fail to map to build 35. – 1613 have 150 bp >Q 30 at either end and have >100 bp unique seq • 1416 of these have no hit to HTGS BAC sequence • 1503 BLAST hit chimpanzee WGS but only 403 within chimp assembly • Estimate that represents ~10 -20 Mb. • 1503 of these selected for fingerprinting (4 enzymes). • Four independent restriction enzymes E ( co. R I, Hind III, Bgl II and Nsi I) • Contigs constructed from 1376 clones (95% success rate) using Composite Mutual Overlap Statistic (CMOS)

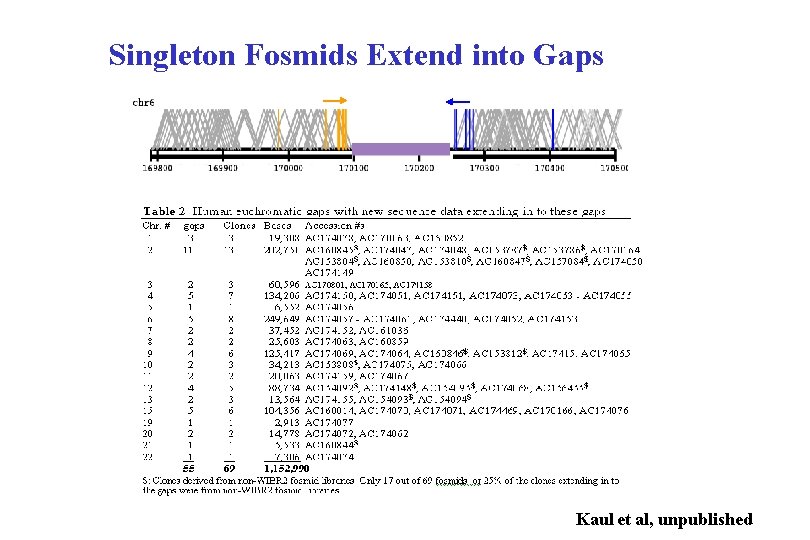
FISH Summary of Orphan. Fosmids • 52 contigs tested by FISH • 15 subtelomeric, 5 acrocentric and 5 pericentromeric • 22 interstitial euchromatin (9 corresponding to known gaps) • 10 contigs =no signals observed against 2 individuals (6/10 largest)