Genome Evolution Amos Tanay 2010 Genome evolution Lecture

![Genome Evolution. Amos Tanay 2010 Dynamic programming to compute the total probability up[5] ? Genome Evolution. Amos Tanay 2010 Dynamic programming to compute the total probability up[5] ?](https://slidetodoc.com/presentation_image_h2/e96903420019daff133a24e27e8fce16/image-11.jpg)
![Genome Evolution. Amos Tanay 2010 Computing marginals and posteriors down 5] ? up[3] S Genome Evolution. Amos Tanay 2010 Computing marginals and posteriors down 5] ? up[3] S](https://slidetodoc.com/presentation_image_h2/e96903420019daff133a24e27e8fce16/image-12.jpg)

- Slides: 49

Genome Evolution. Amos Tanay 2010 Genome evolution Lecture 6: Inference through sampling. Basic phylogenetics

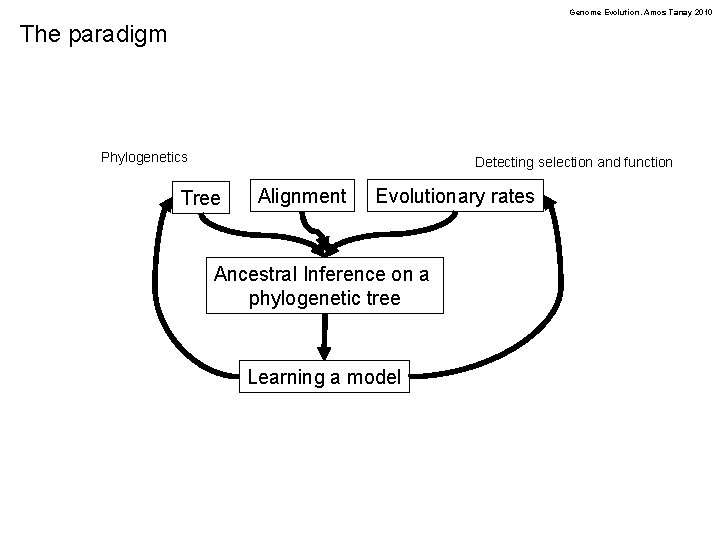
Genome Evolution. Amos Tanay 2010 The paradigm Phylogenetics Detecting selection and function Tree Alignment Evolutionary rates Ancestral Inference on a phylogenetic tree Learning a model

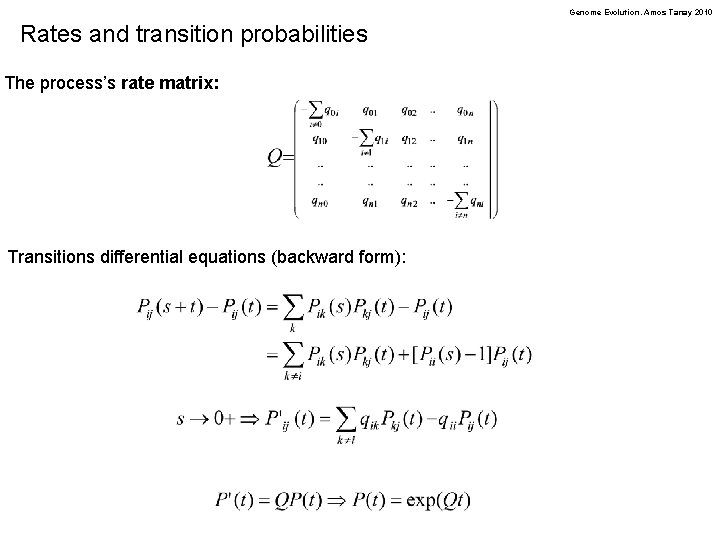
Genome Evolution. Amos Tanay 2010 Rates and transition probabilities The process’s rate matrix: Transitions differential equations (backward form):

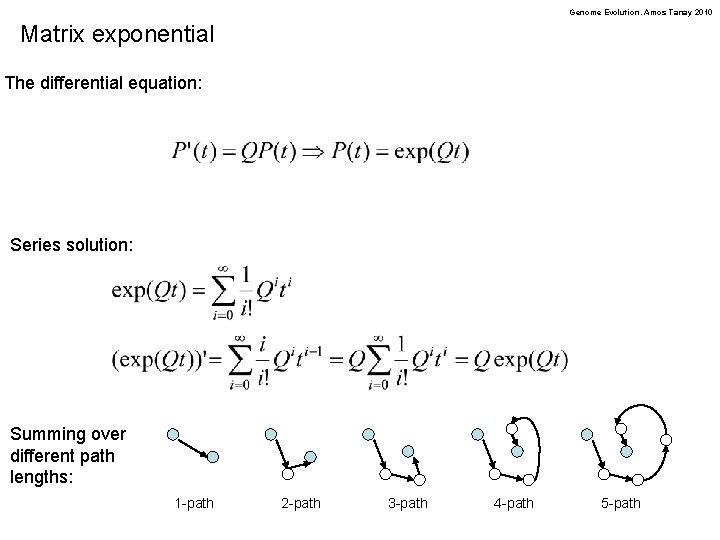
Genome Evolution. Amos Tanay 2010 Matrix exponential The differential equation: Series solution: Summing over different path lengths: 1 -path 2 -path 3 -path 4 -path 5 -path

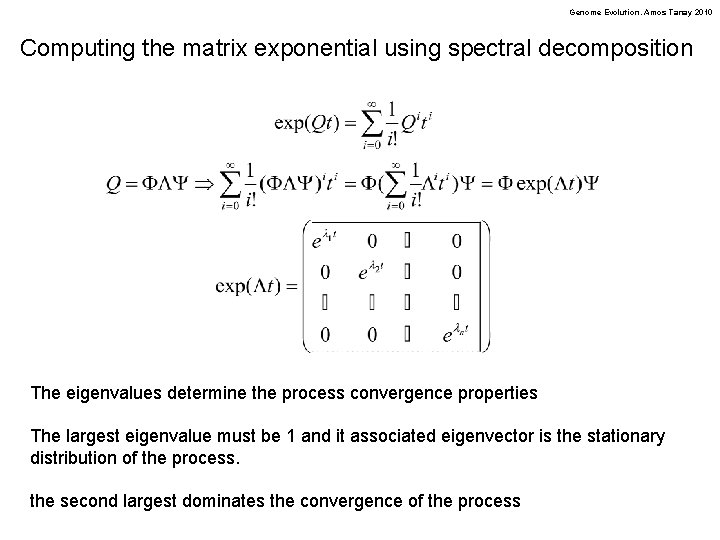
Genome Evolution. Amos Tanay 2010 Computing the matrix exponential using spectral decomposition The eigenvalues determine the process convergence properties The largest eigenvalue must be 1 and it associated eigenvector is the stationary distribution of the process. the second largest dominates the convergence of the process

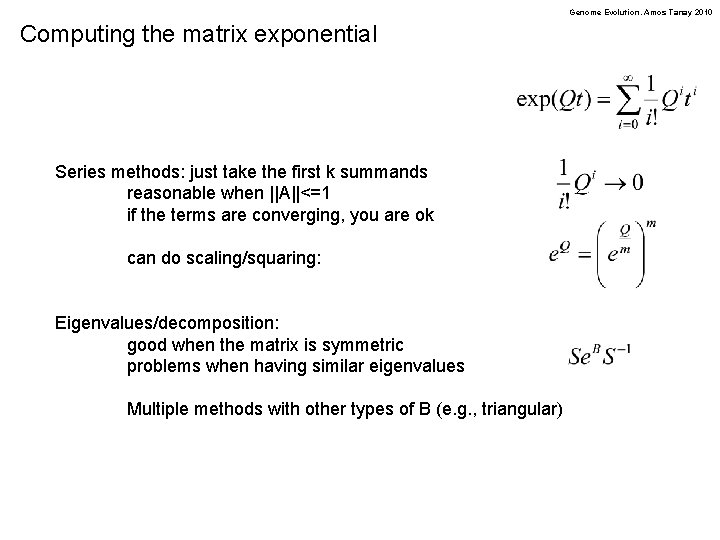
Genome Evolution. Amos Tanay 2010 Computing the matrix exponential Series methods: just take the first k summands reasonable when ||A||<=1 if the terms are converging, you are ok can do scaling/squaring: Eigenvalues/decomposition: good when the matrix is symmetric problems when having similar eigenvalues Multiple methods with other types of B (e. g. , triangular)

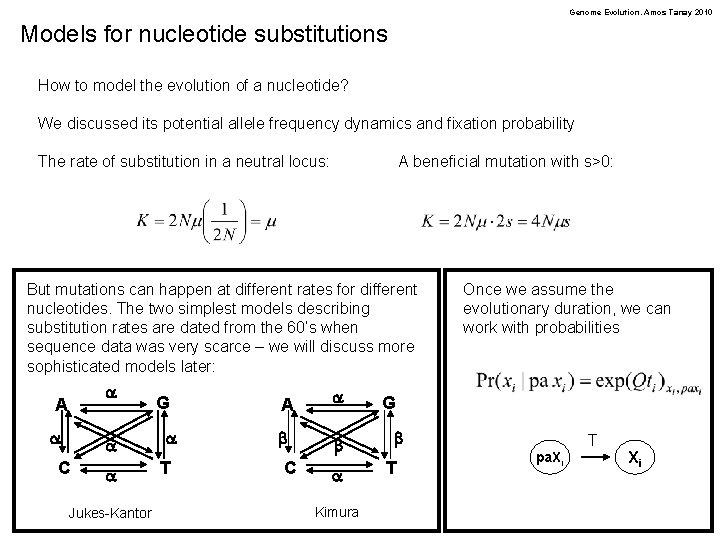
Genome Evolution. Amos Tanay 2010 Models for nucleotide substitutions How to model the evolution of a nucleotide? We discussed its potential allele frequency dynamics and fixation probability The rate of substitution in a neutral locus: A beneficial mutation with s>0: But mutations can happen at different rates for different nucleotides. The two simplest models describing substitution rates are dated from the 60’s when sequence data was very scarce – we will discuss more sophisticated models later: a A a a C a Jukes-Kantor G a T A a b b C a Kimura Once we assume the evolutionary duration, we can work with probabilities G b T T pa. Xi Xi

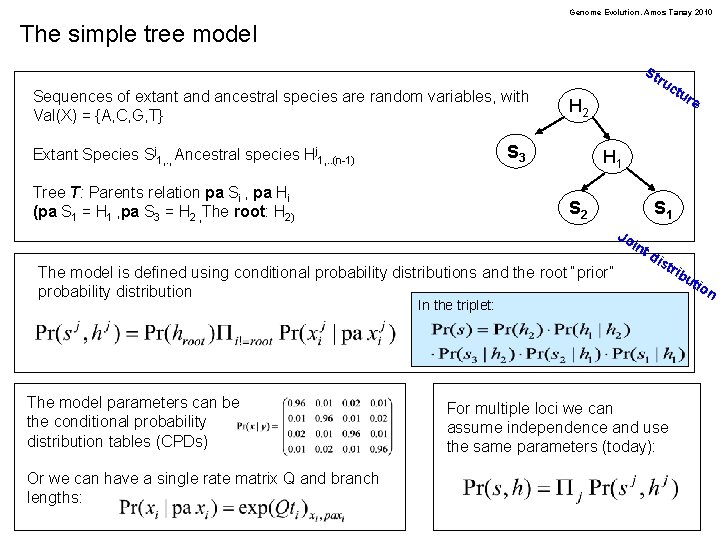
Genome Evolution. Amos Tanay 2010 The simple tree model St Sequences of extant and ancestral species are random variables, with Val(X) = {A, C, G, T} ru H 2 S 3 Extant Species Sj 1, . , Ancestral species Hj 1, . . (n-1) Tree T: Parents relation pa Si , pa Hi (pa S 1 = H 1 , pa S 3 = H 2 , The root: H 2) ctu re H 1 S 2 S 1 Jo int The model is defined using conditional probability distributions and the root “prior” probability distribution dis tri In the triplet: The model parameters can be the conditional probability distribution tables (CPDs) Or we can have a single rate matrix Q and branch lengths: For multiple loci we can assume independence and use the same parameters (today): bu tio n

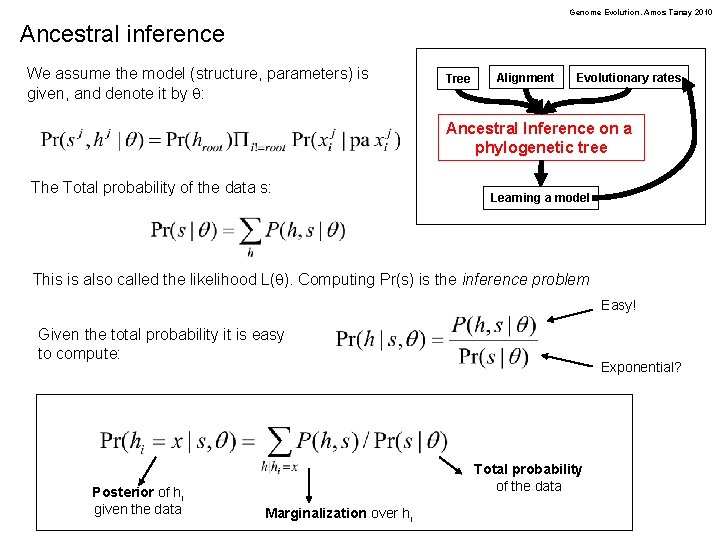
Genome Evolution. Amos Tanay 2010 Ancestral inference We assume the model (structure, parameters) is given, and denote it by q: Tree Alignment Evolutionary rates Ancestral Inference on a phylogenetic tree The Total probability of the data s: Learning a model This is also called the likelihood L(q). Computing Pr(s) is the inference problem Easy! Given the total probability it is easy to compute: Posterior of hi given the data Exponential? Total probability of the data Marginalization over hi

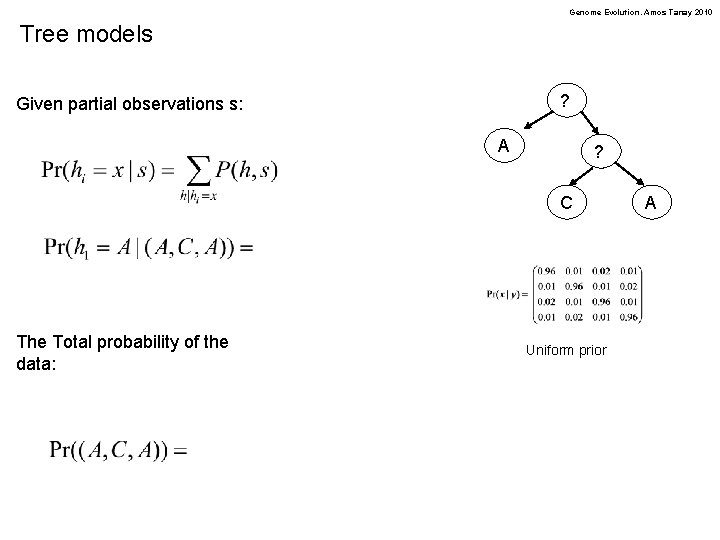
Genome Evolution. Amos Tanay 2010 Tree models ? Given partial observations s: A ? C The Total probability of the data: Uniform prior A
![Genome Evolution Amos Tanay 2010 Dynamic programming to compute the total probability up5 Genome Evolution. Amos Tanay 2010 Dynamic programming to compute the total probability up[5] ?](https://slidetodoc.com/presentation_image_h2/e96903420019daff133a24e27e8fce16/image-11.jpg)
Genome Evolution. Amos Tanay 2010 Dynamic programming to compute the total probability up[5] ? S 3 ? S 2 up[4] S 1 Algorithm (Following Felsenstein 1981): Up(i): if(extant) { up[i][a] = (a==Si ? 1: 0); return} up(r(i)), up(l(i)) iter on a up[i][a] = Down(i): down[i][a]= Sb, c Pr(Xl(i)=b|Xi=a) up[l(i)][b] Pr(Xr(i)=c|Xi=a) up[r(i)][c] Pr(Xsib(i)=b|Xpar(i)=c) up[sib(i)][b] Pr(Xi=a|Xpar(i)=c) down[par(i)][c] down(r(i)), down(l(i)) Algorithm: up(root); LL = 0; foreach a { L += log(Pr(root=a)up[root][a]) down[root][a]=Pr(root=a) } down(r(root)); down(l(root)); Felsentstein
![Genome Evolution Amos Tanay 2010 Computing marginals and posteriors down 5 up3 S Genome Evolution. Amos Tanay 2010 Computing marginals and posteriors down 5] ? up[3] S](https://slidetodoc.com/presentation_image_h2/e96903420019daff133a24e27e8fce16/image-12.jpg)
Genome Evolution. Amos Tanay 2010 Computing marginals and posteriors down 5] ? up[3] S 3 ? S 2 Felsentstein down[4] S 1 Algorithm (Following Felsenstein 1981): Up(i): if(extant) { up[i][a] = (a==Si ? 1: 0); return} up(r(i)), up(l(i)) iter on a up[i][a] = Down(i): down[i][a]= Sb, c Pr(Xl(i)=b|Xi=a) up[l(i)][b] Pr(Xr(i)=c|Xi=a) up[r(i)][c] Sb, c Pr(Xsib(i)=b|Xpar(i)=c) up[sib(i)][b] Pr(Xi=a|Xpar(i)=c) down[par(i)][c] down(r(i)), down(l(i)) Algorithm: up(root); LL = 0; foreach a { L += log(Pr(root=a)up[root][a]) down[root][a]=Pr(root=a) } P(hi|s) = up[i][c]*down[i][c]/ down(r(root)); down(l(root)); ( up[i][j]down[i][j]) Sj

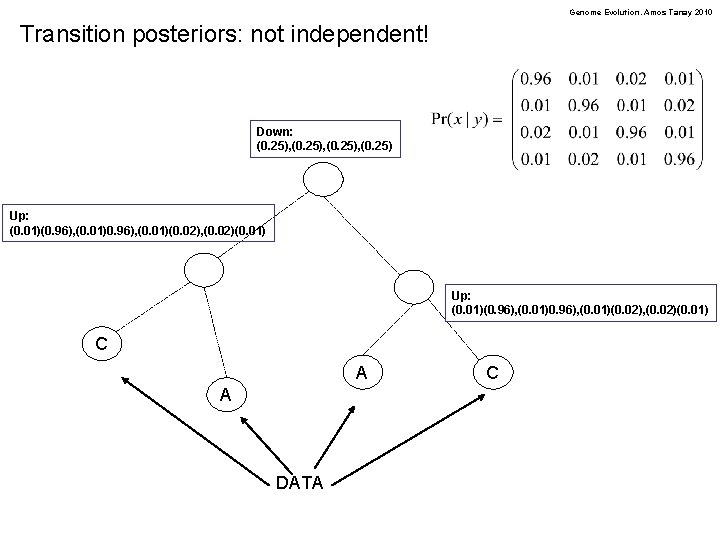
Genome Evolution. Amos Tanay 2010 Transition posteriors: not independent! Down: (0. 25), (0. 25) Up: (0. 01)(0. 96), (0. 01)0. 96), (0. 01)(0. 02), (0. 02)(0. 01) C A A DATA C

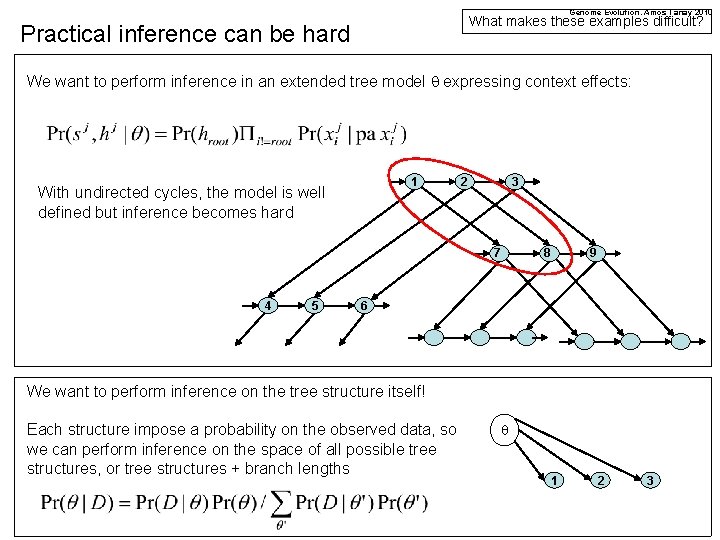
Genome Evolution. Amos Tanay 2010 What makes these examples difficult? Practical inference can be hard We want to perform inference in an extended tree model q expressing context effects: 1 With undirected cycles, the model is well defined but inference becomes hard 2 3 7 4 5 8 9 6 We want to perform inference on the tree structure itself! Each structure impose a probability on the observed data, so we can perform inference on the space of all possible tree structures, or tree structures + branch lengths q 1 2 3

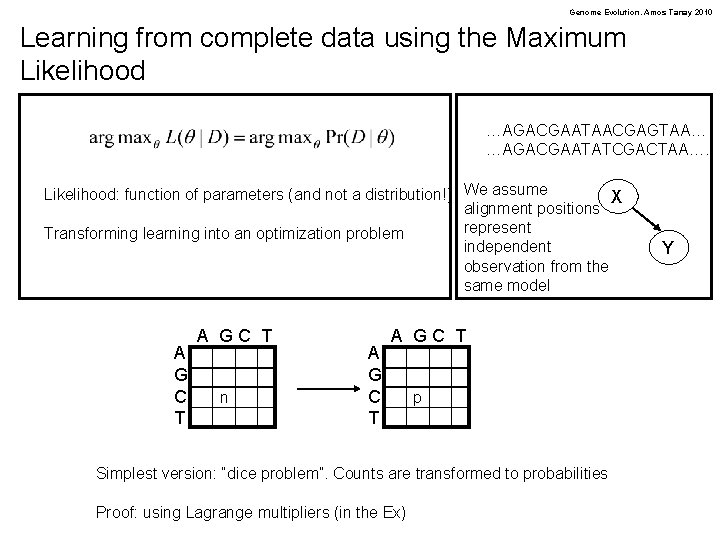
Genome Evolution. Amos Tanay 2010 Learning from complete data using the Maximum Likelihood …AGACGAATAACGAGTAA… …AGACGAATATCGACTAA…. Likelihood: function of parameters (and not a distribution!) We assume X alignment positions represent Transforming learning into an optimization problem independent observation from the same model A G C T A GC T n A G C T A GC T p Simplest version: “dice problem”. Counts are transformed to probabilities Proof: using Lagrange multipliers (in the Ex) Y

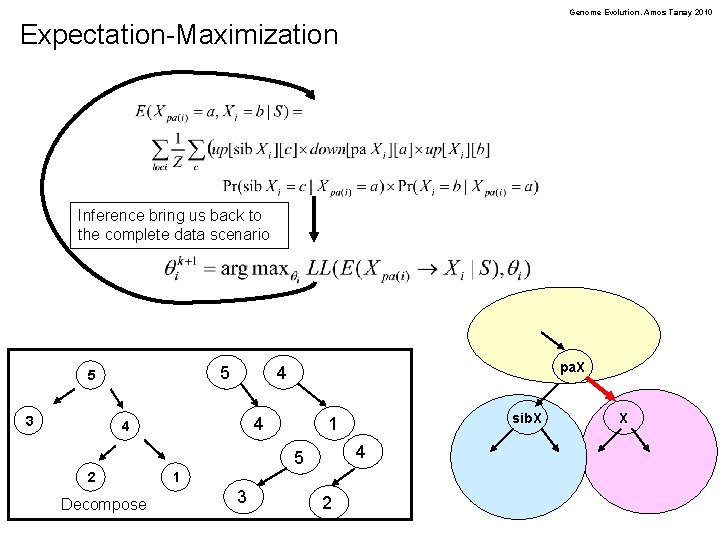
Genome Evolution. Amos Tanay 2010 Expectation-Maximization Inference bring us back to the complete data scenario 5 5 3 pa. X 4 4 5 2 Decompose 1 3 sib. X 1 2 X

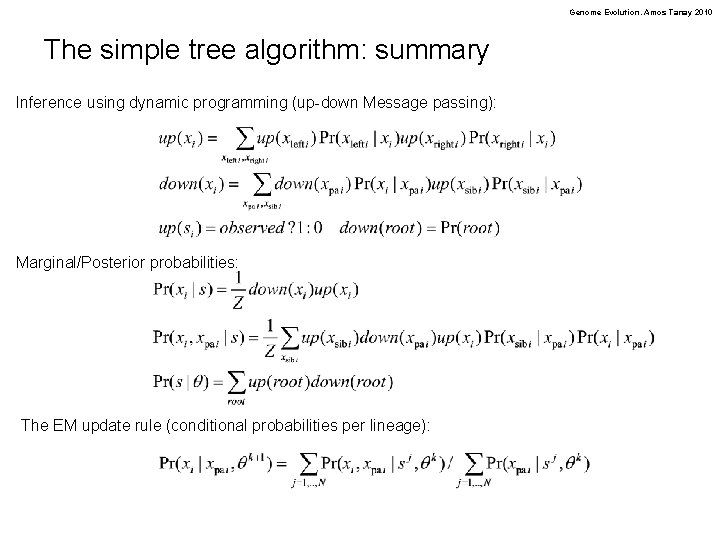
Genome Evolution. Amos Tanay 2010 The simple tree algorithm: summary Inference using dynamic programming (up-down Message passing): Marginal/Posterior probabilities: The EM update rule (conditional probabilities per lineage):

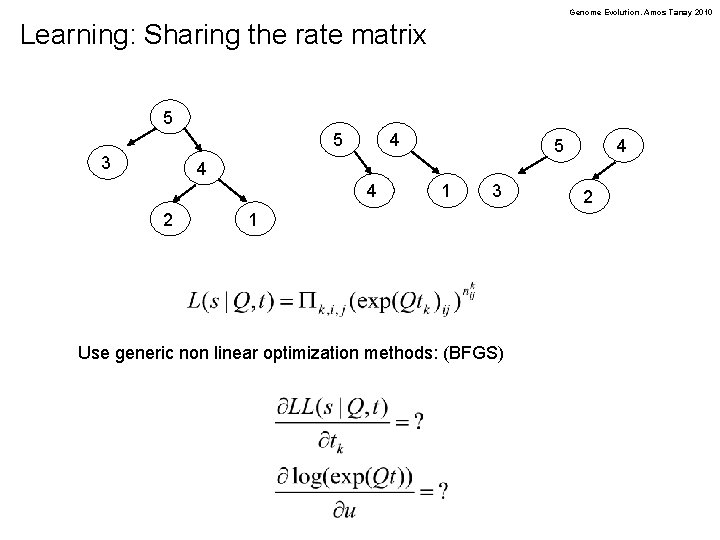
Genome Evolution. Amos Tanay 2010 Learning: Sharing the rate matrix 5 5 3 4 5 4 4 4 2 1 3 1 Use generic non linear optimization methods: (BFGS) 2

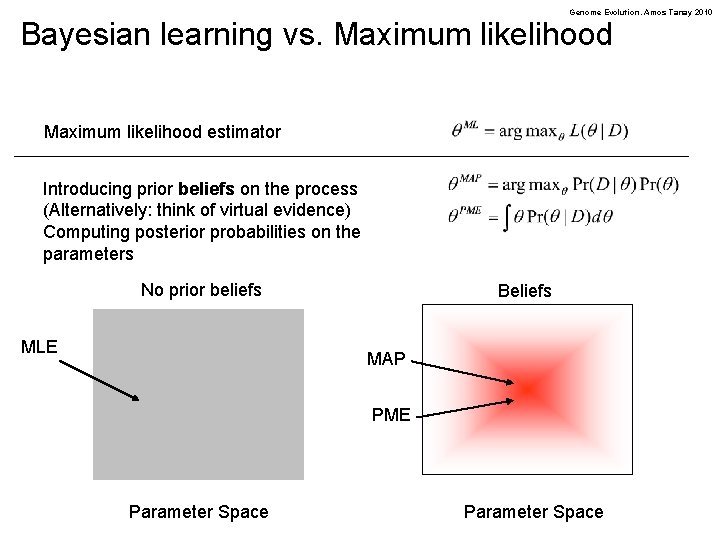
Genome Evolution. Amos Tanay 2010 Bayesian learning vs. Maximum likelihood estimator Introducing prior beliefs on the process (Alternatively: think of virtual evidence) Computing posterior probabilities on the parameters No prior beliefs MLE Beliefs MAP PME Parameter Space

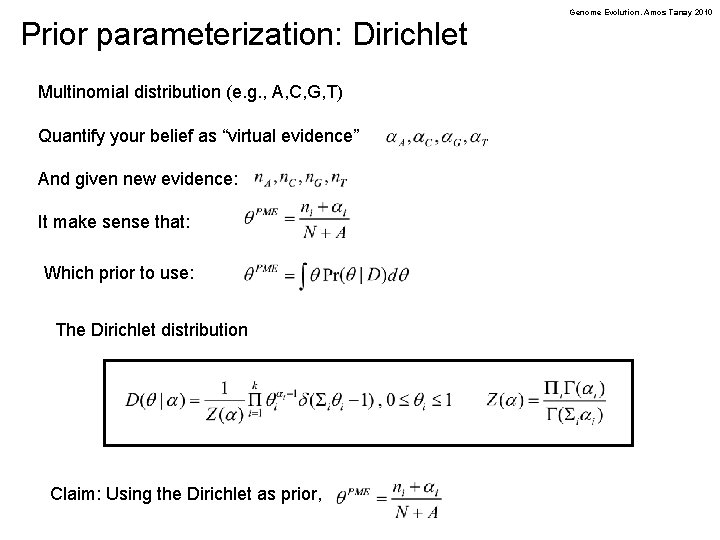
Prior parameterization: Dirichlet Multinomial distribution (e. g. , A, C, G, T) Quantify your belief as “virtual evidence” And given new evidence: It make sense that: Which prior to use: The Dirichlet distribution Claim: Using the Dirichlet as prior, Genome Evolution. Amos Tanay 2010

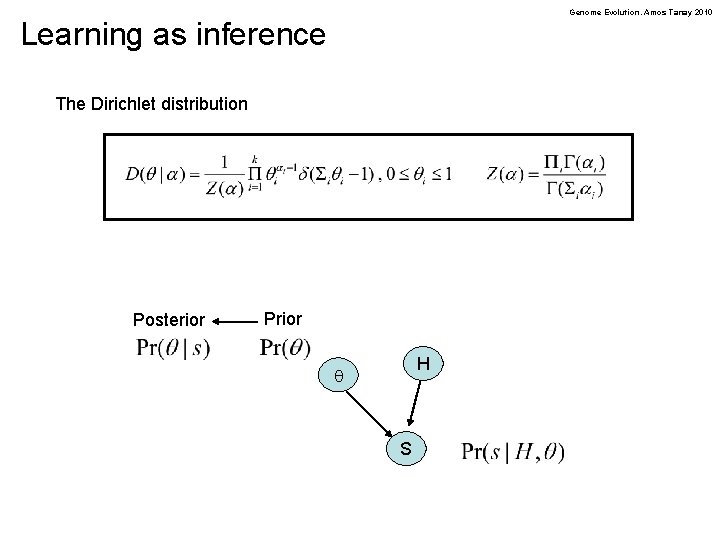
Genome Evolution. Amos Tanay 2010 Learning as inference The Dirichlet distribution Posterior Prior H q S

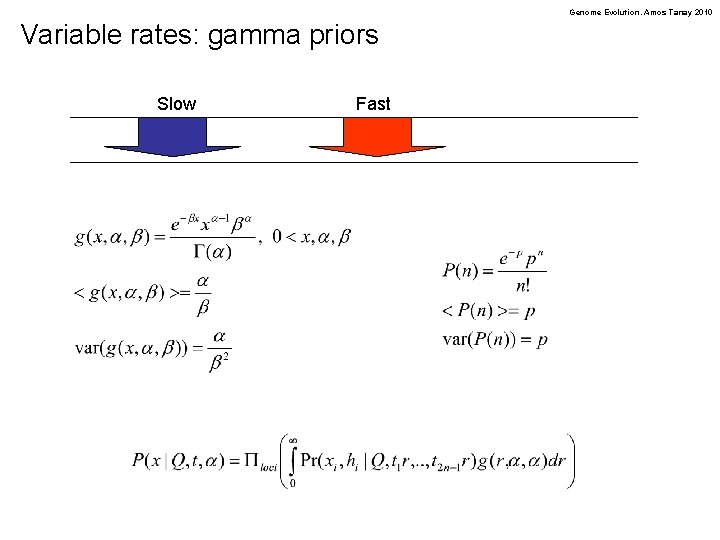
Genome Evolution. Amos Tanay 2010 Variable rates: gamma priors Slow Fast

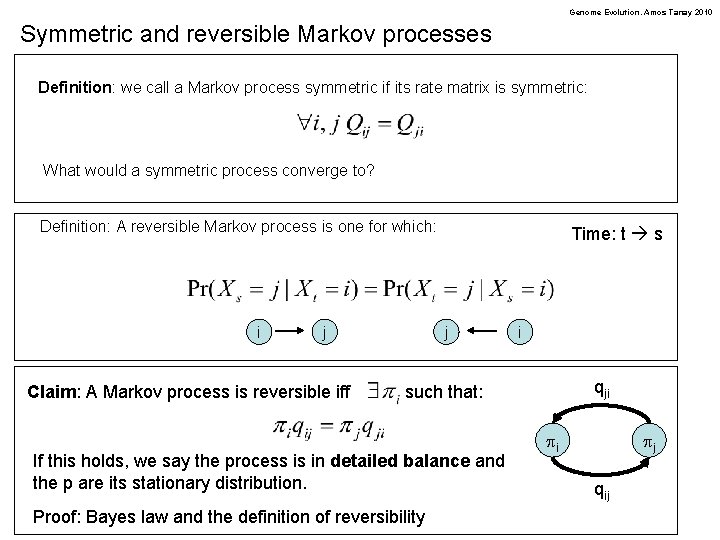
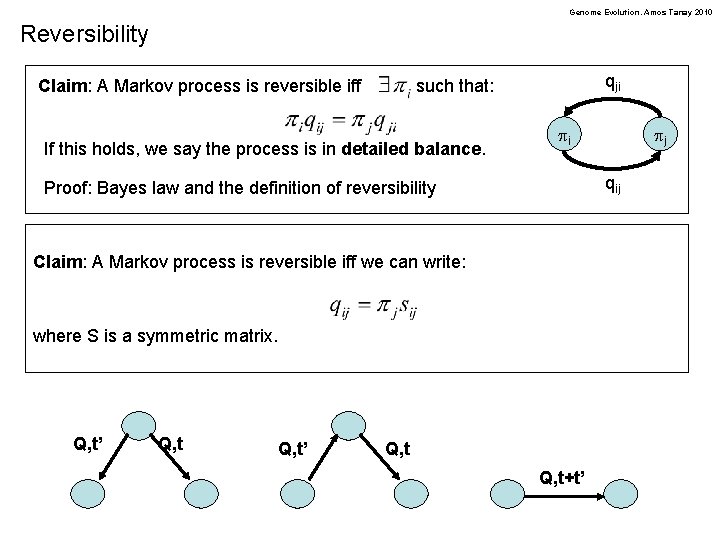
Genome Evolution. Amos Tanay 2010 Symmetric and reversible Markov processes Definition: we call a Markov process symmetric if its rate matrix is symmetric: What would a symmetric process converge to? Definition: A reversible Markov process is one for which: i j Claim: A Markov process is reversible iff Time: t s j i qji such that: If this holds, we say the process is in detailed balance and the p are its stationary distribution. Proof: Bayes law and the definition of reversibility pi pj qij

Genome Evolution. Amos Tanay 2010 Reversibility Claim: A Markov process is reversible iff qji such that: If this holds, we say the process is in detailed balance. pi qij Proof: Bayes law and the definition of reversibility Claim: A Markov process is reversible iff we can write: where S is a symmetric matrix. Q, t’ pj Q, t+t’

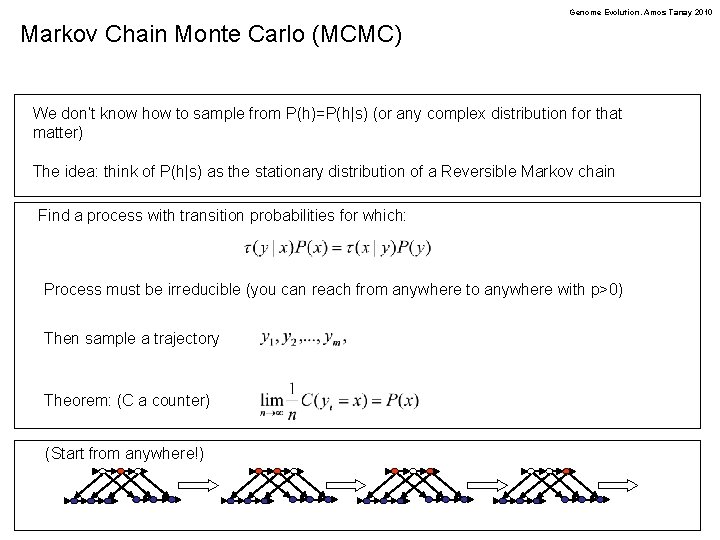
Genome Evolution. Amos Tanay 2010 Markov Chain Monte Carlo (MCMC) We don’t know how to sample from P(h)=P(h|s) (or any complex distribution for that matter) The idea: think of P(h|s) as the stationary distribution of a Reversible Markov chain Find a process with transition probabilities for which: Process must be irreducible (you can reach from anywhere to anywhere with p>0) Then sample a trajectory Theorem: (C a counter) (Start from anywhere!)

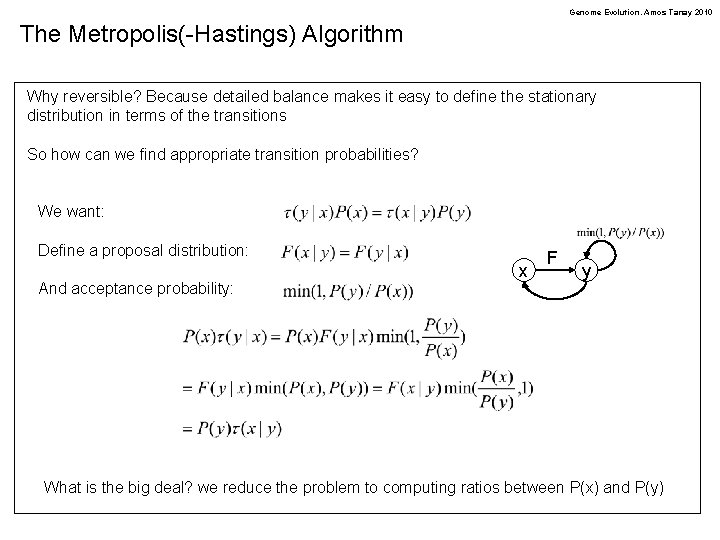
Genome Evolution. Amos Tanay 2010 The Metropolis(-Hastings) Algorithm Why reversible? Because detailed balance makes it easy to define the stationary distribution in terms of the transitions So how can we find appropriate transition probabilities? We want: Define a proposal distribution: And acceptance probability: x F y What is the big deal? we reduce the problem to computing ratios between P(x) and P(y)

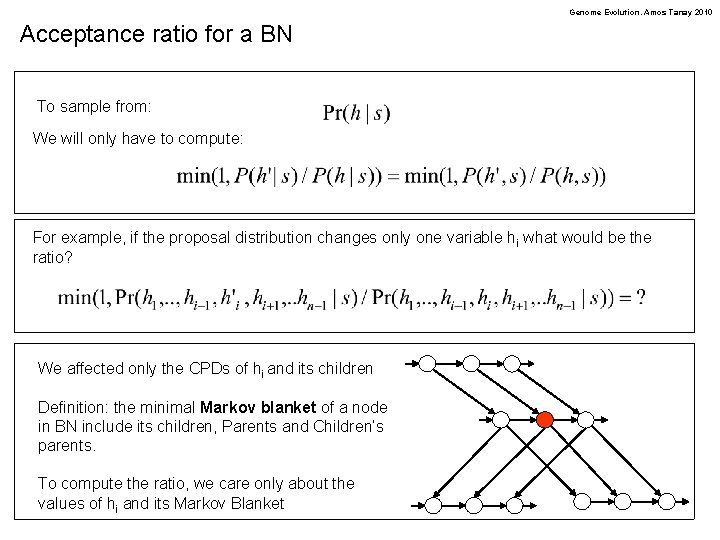
Genome Evolution. Amos Tanay 2010 Acceptance ratio for a BN To sample from: We will only have to compute: For example, if the proposal distribution changes only one variable hi what would be the ratio? We affected only the CPDs of hi and its children Definition: the minimal Markov blanket of a node in BN include its children, Parents and Children’s parents. To compute the ratio, we care only about the values of hi and its Markov Blanket

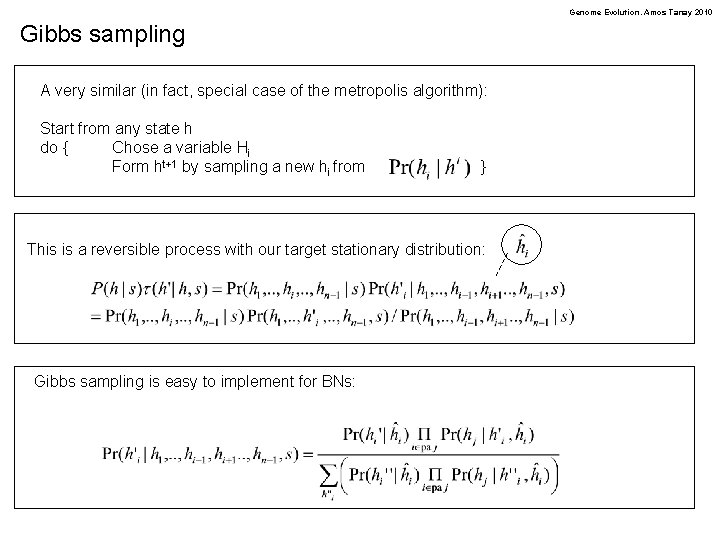
Genome Evolution. Amos Tanay 2010 Gibbs sampling A very similar (in fact, special case of the metropolis algorithm): Start from any state h do { Chose a variable Hi Form ht+1 by sampling a new hi from } This is a reversible process with our target stationary distribution: Gibbs sampling is easy to implement for BNs:

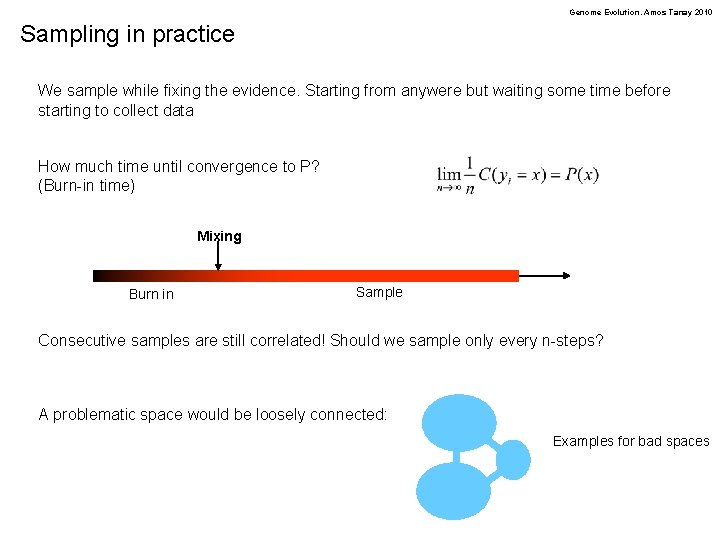
Genome Evolution. Amos Tanay 2010 Sampling in practice We sample while fixing the evidence. Starting from anywere but waiting some time before starting to collect data How much time until convergence to P? (Burn-in time) Mixing Burn in Sample Consecutive samples are still correlated! Should we sample only every n-steps? A problematic space would be loosely connected: Examples for bad spaces

Genome Evolution. Amos Tanay 2010 Inferring/learning phylogenetic trees Distance based methods: computing pairwise distances and building a tree based only on those (how would you implement this? ) More elaborated methods use a scoring scheme that take the whole tree into account, using Parsimony or likelihood Likelihood methods: universal rate matrices (BLOSSOM 62, PAM) Searching for the optimal tree (min parsimony or max likelihood) is NP hard Many search heuristics were developed, finding a high quality solution and repeating computation using partial dataset to test for the robustness of particular features (Bootstrap) Bayesian inference methods – assuming some prior on trees (e. g. uniform) and trying to sample trees from the probability space P(q|D). Using MCMC, we only need a proposal distribution that span all possible trees and a way to compute the likelihood ratio between two trees (polynomial for the simple tree model) From sampling we can extract any desirable parameter on the tree (number of time X, Y and Z are in the same clade)

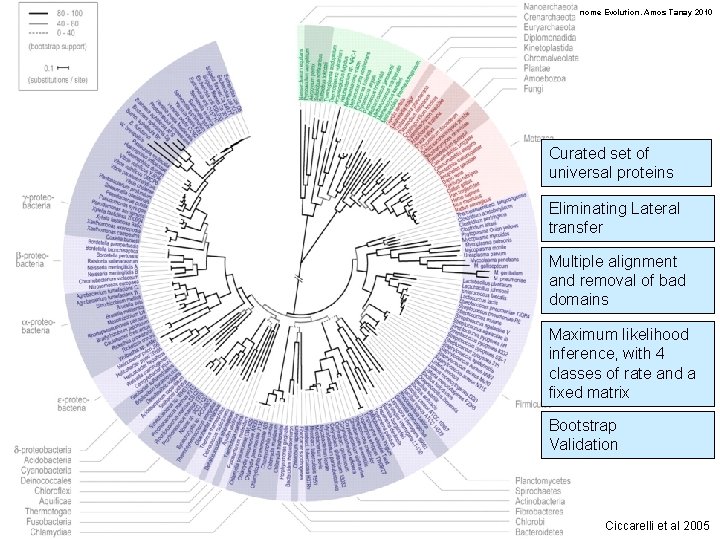
Genome Evolution. Amos Tanay 2010 Curated set of universal proteins Eliminating Lateral transfer Multiple alignment and removal of bad domains Maximum likelihood inference, with 4 classes of rate and a fixed matrix Bootstrap Validation Ciccarelli et al 2005

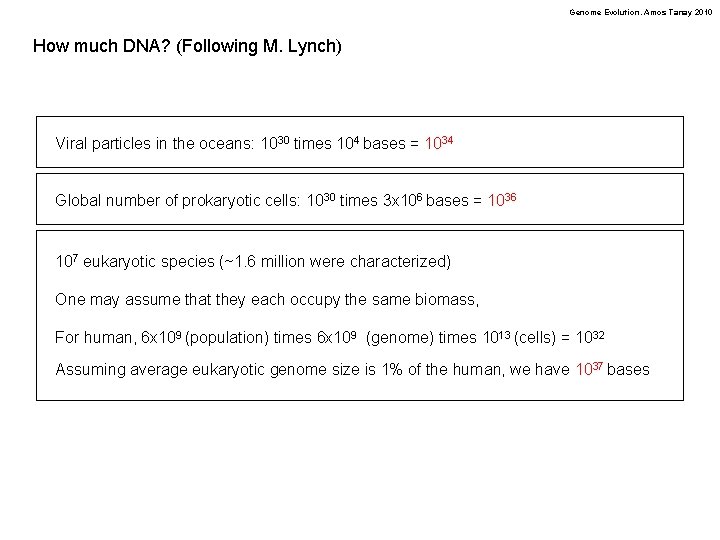
Genome Evolution. Amos Tanay 2010 How much DNA? (Following M. Lynch) Viral particles in the oceans: 1030 times 104 bases = 1034 Global number of prokaryotic cells: 1030 times 3 x 106 bases = 1036 107 eukaryotic species (~1. 6 million were characterized) One may assume that they each occupy the same biomass, For human, 6 x 109 (population) times 6 x 109 (genome) times 1013 (cells) = 1032 Assuming average eukaryotic genome size is 1% of the human, we have 1037 bases

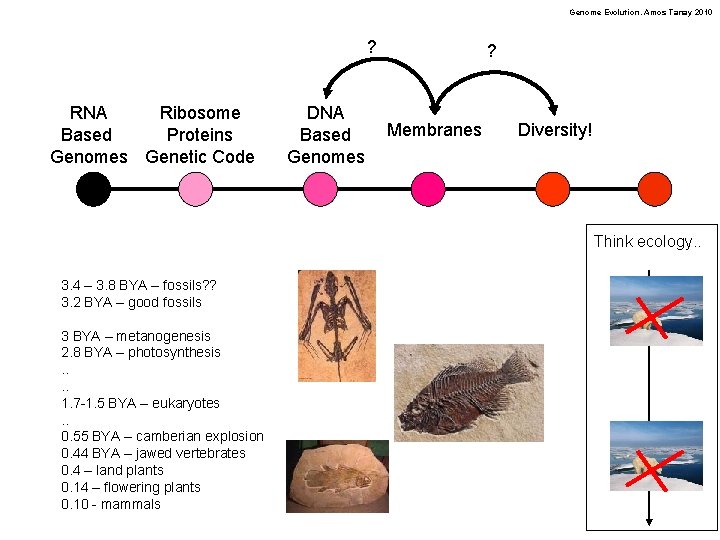
Genome Evolution. Amos Tanay 2010 ? RNA Ribosome Based Proteins Genomes Genetic Code DNA Based Genomes ? Membranes Diversity! Think ecology. . 3. 4 – 3. 8 BYA – fossils? ? 3. 2 BYA – good fossils 3 BYA – metanogenesis 2. 8 BYA – photosynthesis. . 1. 7 -1. 5 BYA – eukaryotes. . 0. 55 BYA – camberian explosion 0. 44 BYA – jawed vertebrates 0. 4 – land plants 0. 14 – flowering plants 0. 10 - mammals

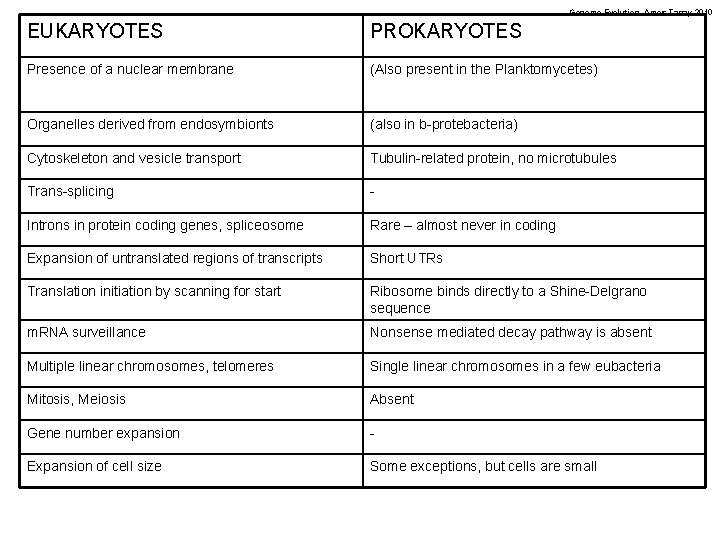
Genome Evolution. Amos Tanay 2010 EUKARYOTES PROKARYOTES Presence of a nuclear membrane (Also present in the Planktomycetes) Organelles derived from endosymbionts (also in b-protebacteria) Cytoskeleton and vesicle transport Tubulin-related protein, no microtubules Trans-splicing - Introns in protein coding genes, spliceosome Rare – almost never in coding Expansion of untranslated regions of transcripts Short UTRs Translation initiation by scanning for start Ribosome binds directly to a Shine-Delgrano sequence m. RNA surveillance Nonsense mediated decay pathway is absent Multiple linear chromosomes, telomeres Single linear chromosomes in a few eubacteria Mitosis, Meiosis Absent Gene number expansion - Expansion of cell size Some exceptions, but cells are small

Genome Evolution. Amos Tanay 2010

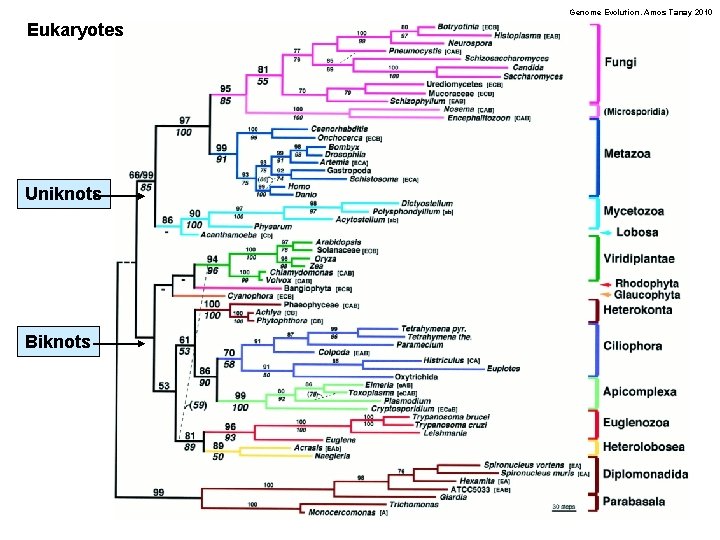
Genome Evolution. Amos Tanay 2010 Eukaryotes Uniknots Biknots

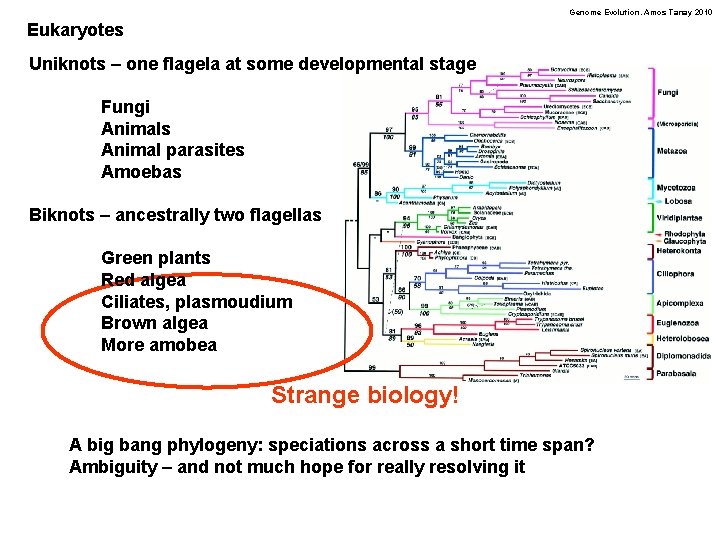
Genome Evolution. Amos Tanay 2010 Eukaryotes Uniknots – one flagela at some developmental stage Fungi Animals Animal parasites Amoebas Biknots – ancestrally two flagellas Green plants Red algea Ciliates, plasmoudium Brown algea More amobea Strange biology! A big bang phylogeny: speciations across a short time span? Ambiguity – and not much hope for really resolving it

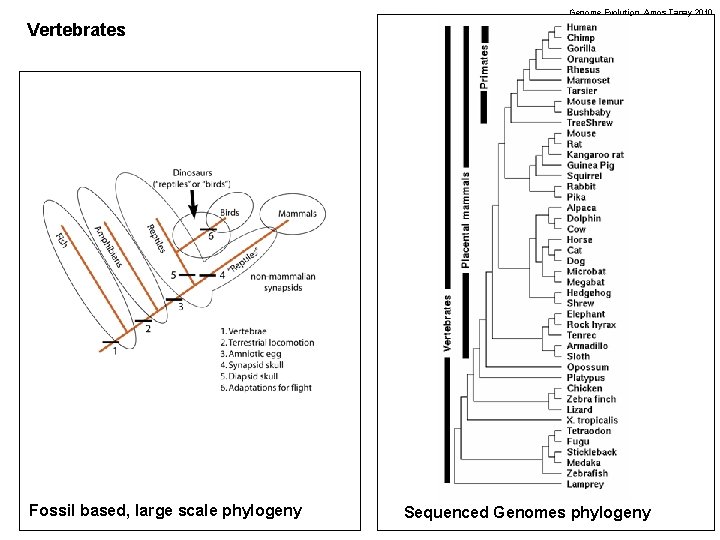
Genome Evolution. Amos Tanay 2010 Vertebrates Fossil based, large scale phylogeny Sequenced Genomes phylogeny

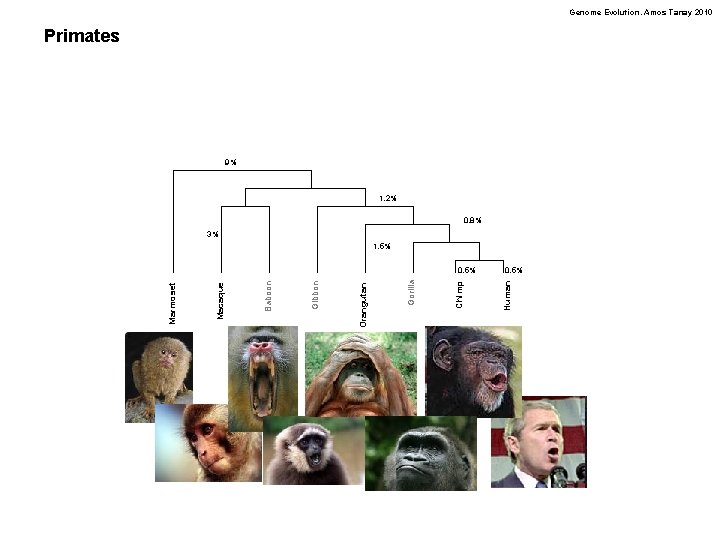
Genome Evolution. Amos Tanay 2010 Primates 9% 1. 2% 0. 8% 3% 1. 5% 0. 5% Human Chimp Gorilla Orangutan Gibbon Baboon Macaque Marmoset 0. 5%

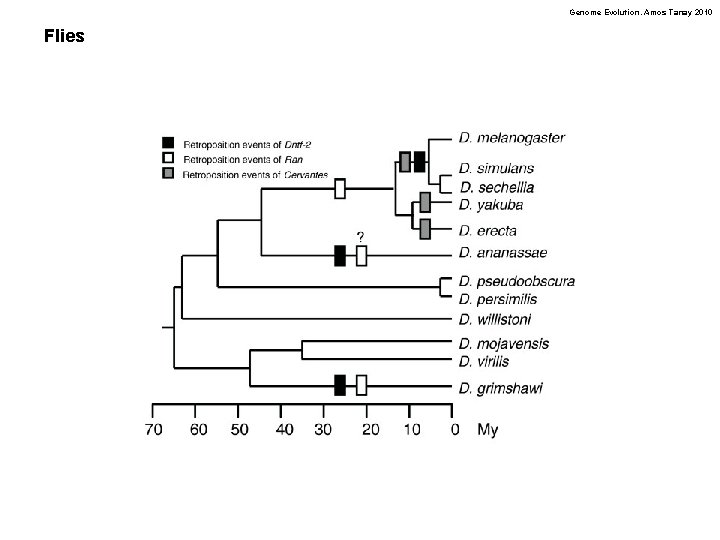
Genome Evolution. Amos Tanay 2010 Flies

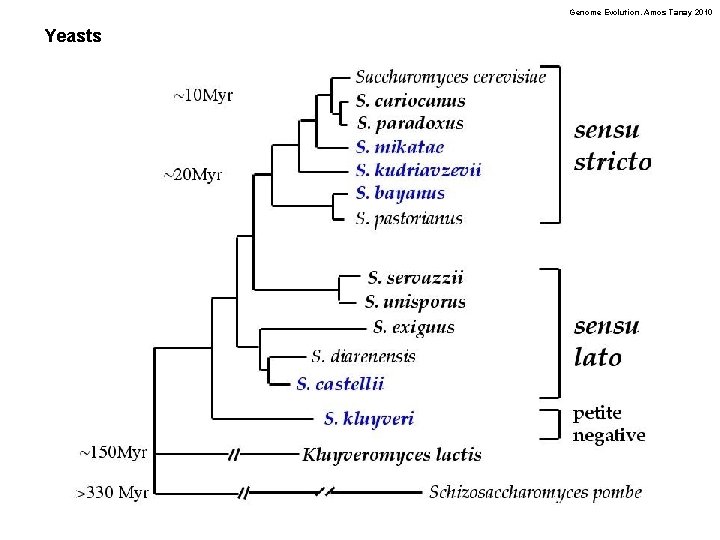
Genome Evolution. Amos Tanay 2010 Yeasts

Genome Evolution. Amos Tanay 2010 Inference by forward sampling • We did not cover these slides this year

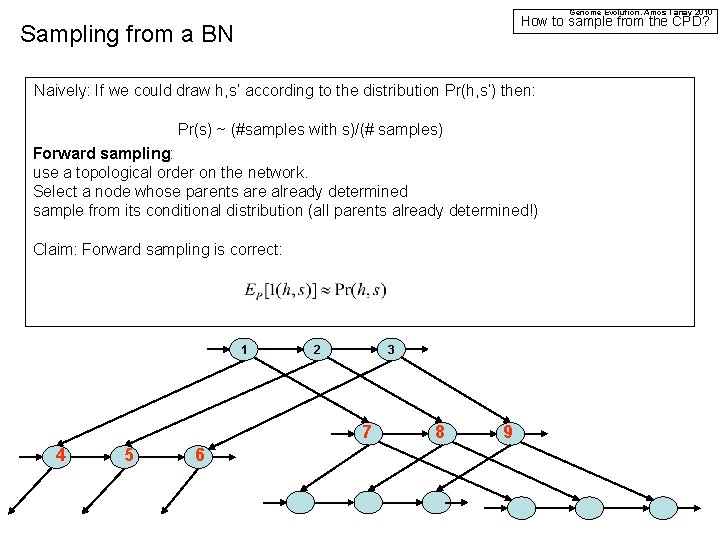
Genome Evolution. Amos Tanay 2010 How to sample from the CPD? Sampling from a BN Naively: If we could draw h, s’ according to the distribution Pr(h, s’) then: Pr(s) ~ (#samples with s)/(# samples) Forward sampling: use a topological order on the network. Select a node whose parents are already determined sample from its conditional distribution (all parents already determined!) Claim: Forward sampling is correct: 1 2 3 7 4 5 6 8 9

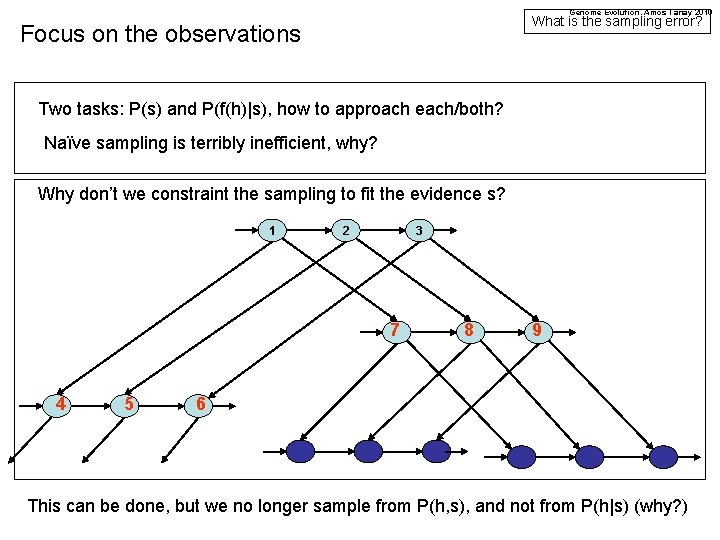
Genome Evolution. Amos Tanay 2010 What is the sampling error? Focus on the observations Two tasks: P(s) and P(f(h)|s), how to approach each/both? Naïve sampling is terribly inefficient, why? Why don’t we constraint the sampling to fit the evidence s? 1 2 3 7 4 5 8 9 6 This can be done, but we no longer sample from P(h, s), and not from P(h|s) (why? )

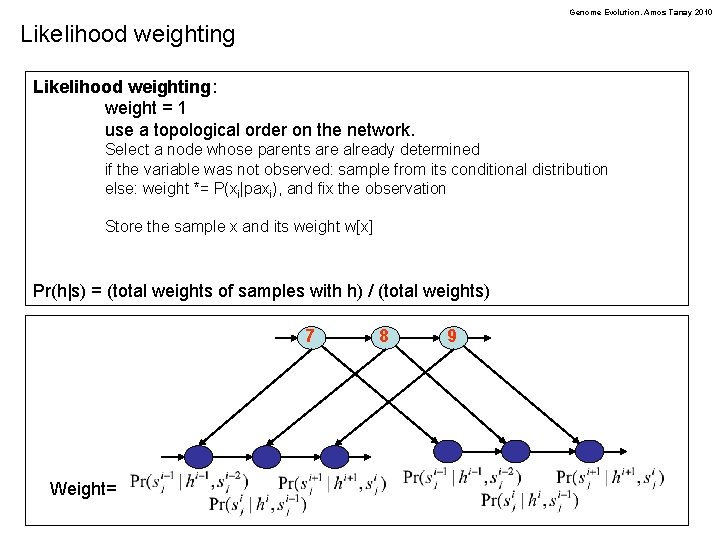
Genome Evolution. Amos Tanay 2010 Likelihood weighting: weight = 1 use a topological order on the network. Select a node whose parents are already determined if the variable was not observed: sample from its conditional distribution else: weight *= P(xi|paxi), and fix the observation Store the sample x and its weight w[x] Pr(h|s) = (total weights of samples with h) / (total weights) 7 Weight= 8 9

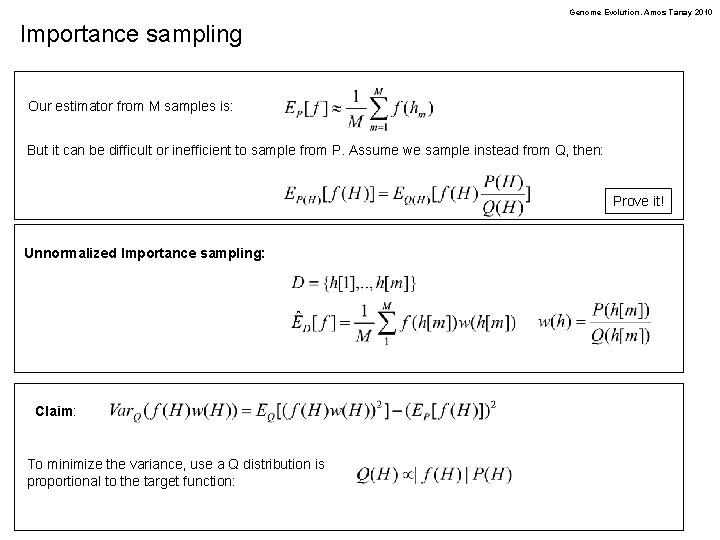
Genome Evolution. Amos Tanay 2010 Importance sampling Our estimator from M samples is: But it can be difficult or inefficient to sample from P. Assume we sample instead from Q, then: Prove it! Unnormalized Importance sampling: Claim: To minimize the variance, use a Q distribution is proportional to the target function:

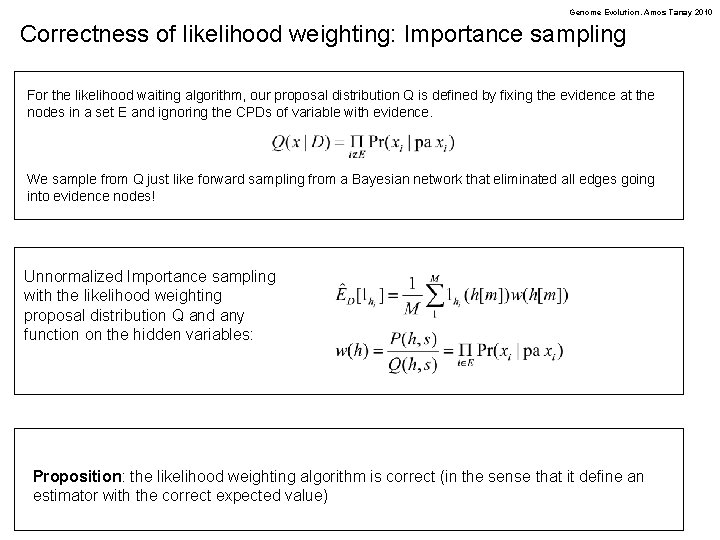
Genome Evolution. Amos Tanay 2010 Correctness of likelihood weighting: Importance sampling For the likelihood waiting algorithm, our proposal distribution Q is defined by fixing the evidence at the nodes in a set E and ignoring the CPDs of variable with evidence. We sample from Q just like forward sampling from a Bayesian network that eliminated all edges going into evidence nodes! Unnormalized Importance sampling with the likelihood weighting proposal distribution Q and any function on the hidden variables: Proposition: the likelihood weighting algorithm is correct (in the sense that it define an estimator with the correct expected value)

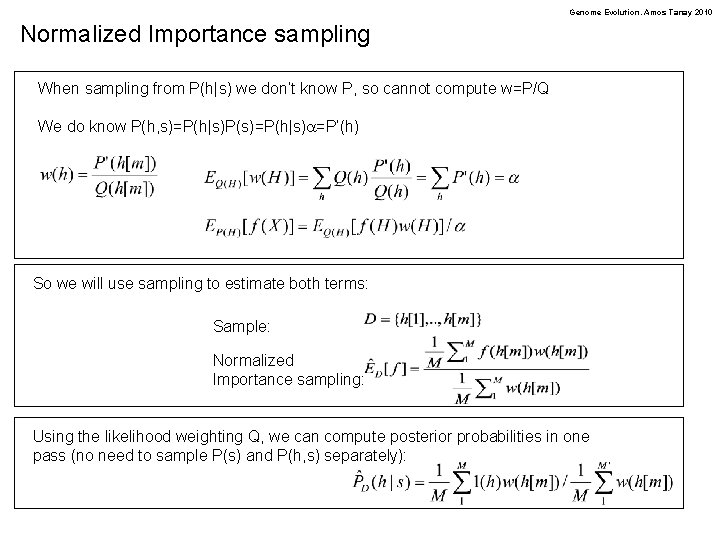
Genome Evolution. Amos Tanay 2010 Normalized Importance sampling When sampling from P(h|s) we don’t know P, so cannot compute w=P/Q We do know P(h, s)=P(h|s)P(s)=P(h|s)a=P’(h) So we will use sampling to estimate both terms: Sample: Normalized Importance sampling: Using the likelihood weighting Q, we can compute posterior probabilities in one pass (no need to sample P(s) and P(h, s) separately):

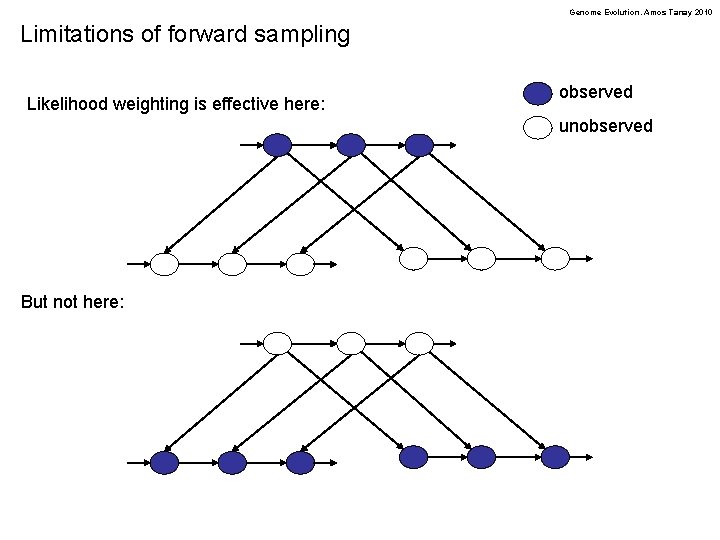
Genome Evolution. Amos Tanay 2010 Limitations of forward sampling Likelihood weighting is effective here: observed unobserved But not here: