Genome Evolution Amos Tanay 2009 Genome evolution Lecture





- Slides: 26

Genome Evolution. Amos Tanay 2009 Genome evolution Lecture 10: Comparative genomics, non coding sequences

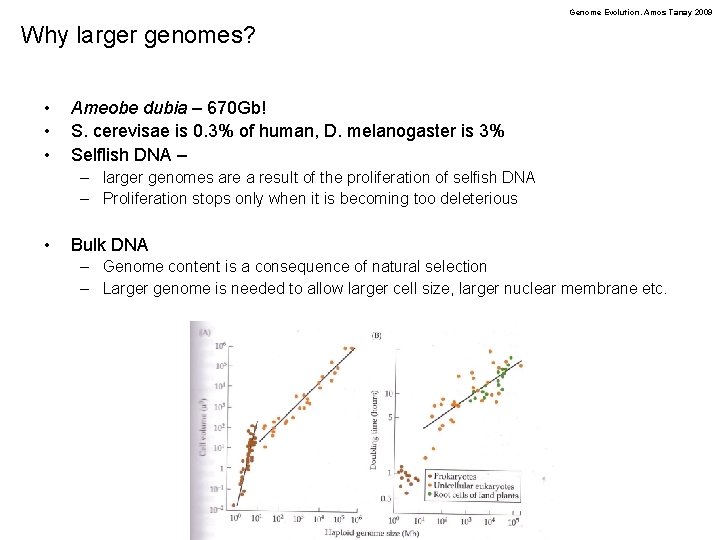
Genome Evolution. Amos Tanay 2009 Why larger genomes? • • • Ameobe dubia – 670 Gb! S. cerevisae is 0. 3% of human, D. melanogaster is 3% Selflish DNA – – larger genomes are a result of the proliferation of selfish DNA – Proliferation stops only when it is becoming too deleterious • Bulk DNA – Genome content is a consequence of natural selection – Larger genome is needed to allow larger cell size, larger nuclear membrane etc.

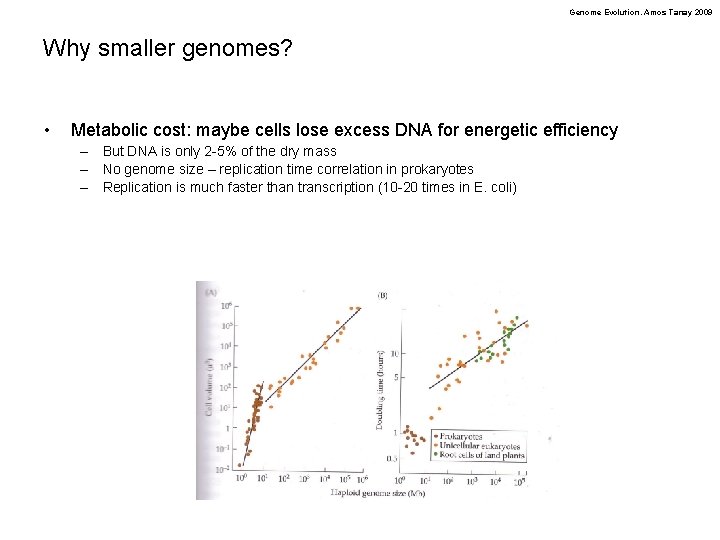
Genome Evolution. Amos Tanay 2009 Why smaller genomes? • Metabolic cost: maybe cells lose excess DNA for energetic efficiency – But DNA is only 2 -5% of the dry mass – No genome size – replication time correlation in prokaryotes – Replication is much faster than transcription (10 -20 times in E. coli)

Genome Evolution. Amos Tanay 2009 Mutational balance • Balance between deletions and insertions – May be different between species – Different balances may have been evolved • In flies, yeast laboratory evolution – 4 -fold more 4 kb spontaneous insertions • In mammals – More small deletions than insertions Mutational hazard • No loss of function for inert DNA – But is it truly not functional? • Gain of function mutations are still possible: – Transcription – Regulation Differences in population size may make DNA purging more effective for prokaryotes, small eukaryotes Differences in regulatory sophistication may make DNA mutational hazard less of a problem for metazoan

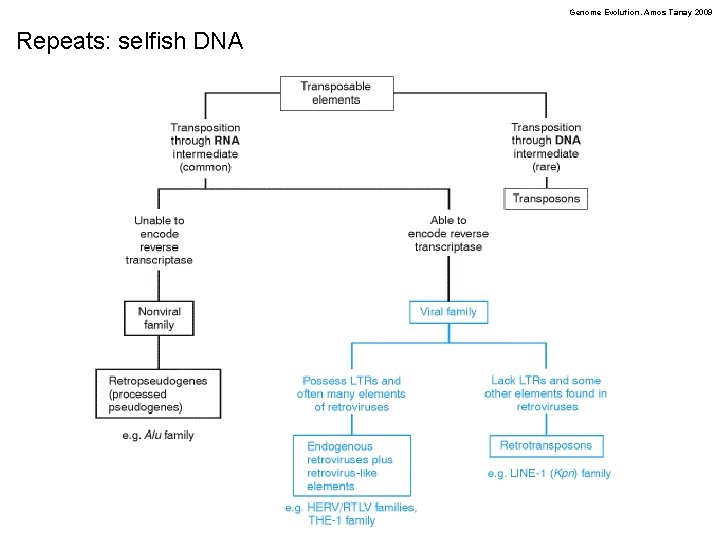
Genome Evolution. Amos Tanay 2009 Repeats: selfish DNA

Genome Evolution. Amos Tanay 2009 Retrotransposition via RNA Repetitive elements in the human genome Class Copies Genome Fraction LINEs 868, 000 20. 4% (only ~100 active!!) SINEs 1, 558, 000 (70% Alu) 13. 1% LTR elements 443, 000 8. 3% Transposons 294, 000 2. 8%

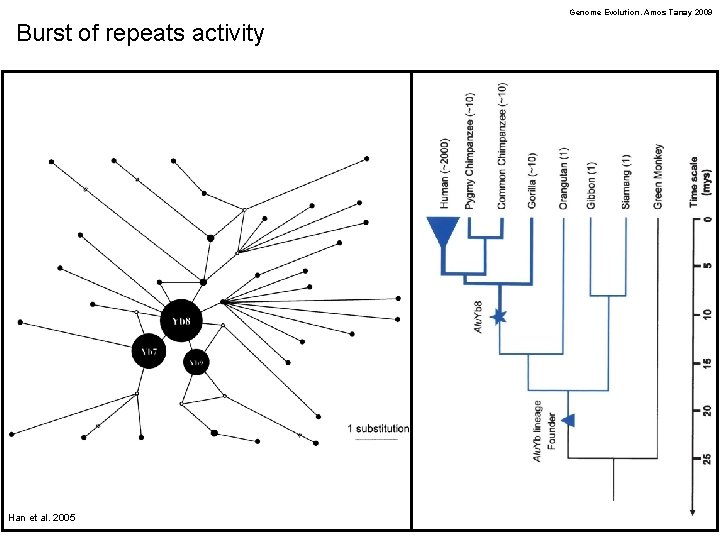
Genome Evolution. Amos Tanay 2009 Burst of repeats activity Han et al. 2005

Genome Evolution. Amos Tanay 2009 Age of repeats in the human genome

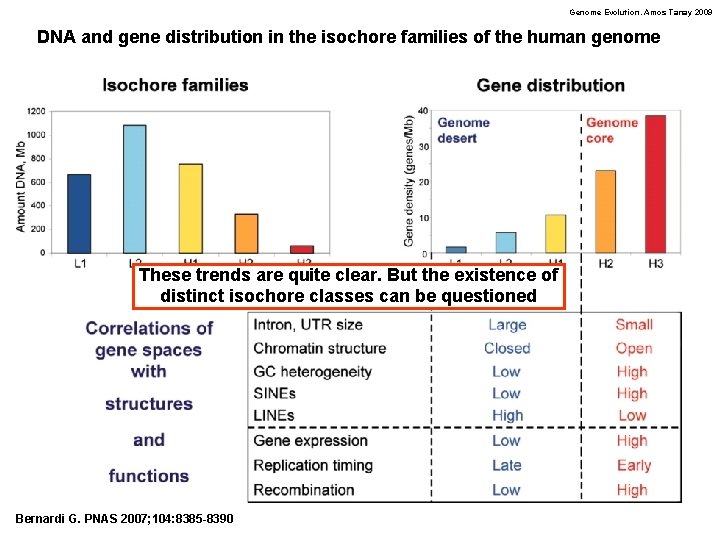
Genome Evolution. Amos Tanay 2009 DNA and gene distribution in the isochore families of the human genome These trends are quite clear. But the existence of distinct isochore classes can be questioned Bernardi G. PNAS 2007; 104: 8385 -8390

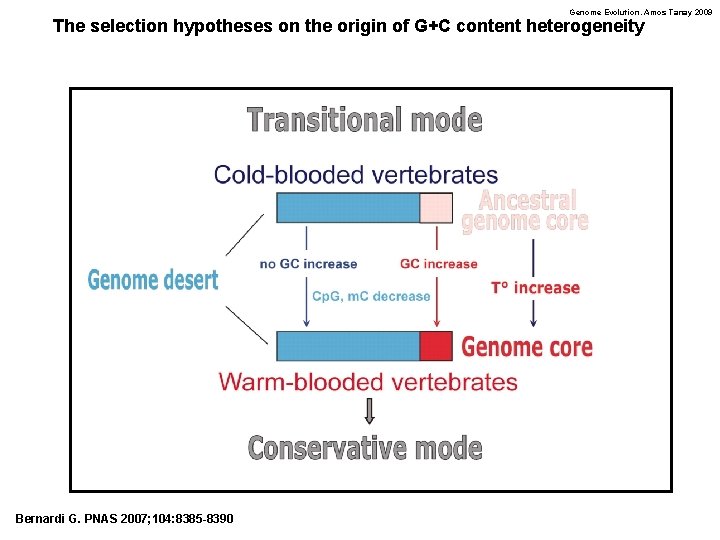
Genome Evolution. Amos Tanay 2009 The selection hypotheses on the origin of G+C content heterogeneity Bernardi G. PNAS 2007; 104: 8385 -8390

Genome Evolution. Amos Tanay 2009 Genomic information: Protein coding genes

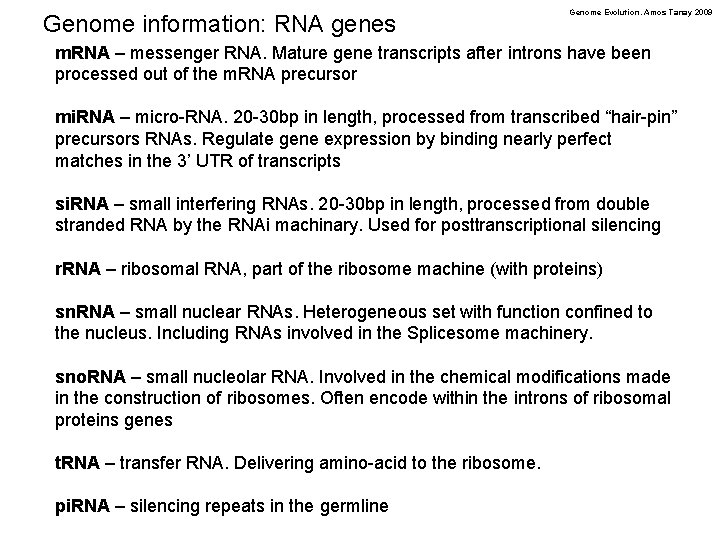
Genome information: RNA genes Genome Evolution. Amos Tanay 2009 m. RNA – messenger RNA. Mature gene transcripts after introns have been processed out of the m. RNA precursor mi. RNA – micro-RNA. 20 -30 bp in length, processed from transcribed “hair-pin” precursors RNAs. Regulate gene expression by binding nearly perfect matches in the 3’ UTR of transcripts si. RNA – small interfering RNAs. 20 -30 bp in length, processed from double stranded RNA by the RNAi machinary. Used for posttranscriptional silencing r. RNA – ribosomal RNA, part of the ribosome machine (with proteins) sn. RNA – small nuclear RNAs. Heterogeneous set with function confined to the nucleus. Including RNAs involved in the Splicesome machinery. sno. RNA – small nucleolar RNA. Involved in the chemical modifications made in the construction of ribosomes. Often encode within the introns of ribosomal proteins genes t. RNA – transfer RNA. Delivering amino-acid to the ribosome. pi. RNA – silencing repeats in the germline

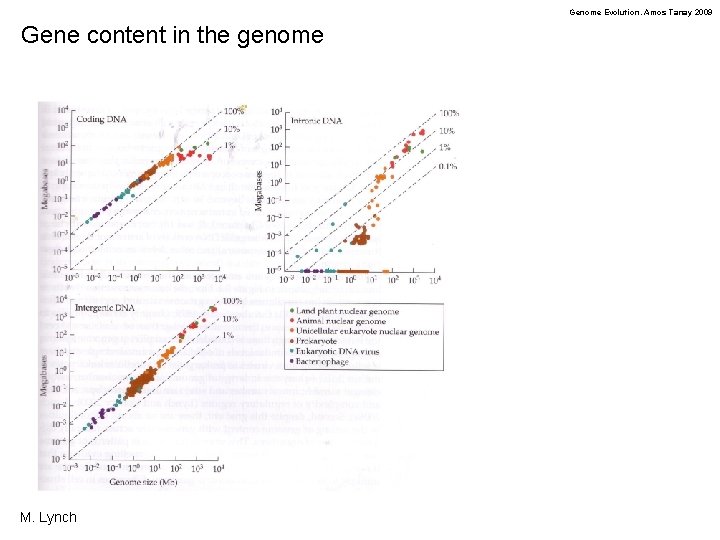
Genome Evolution. Amos Tanay 2009 Gene content in the genome M. Lynch

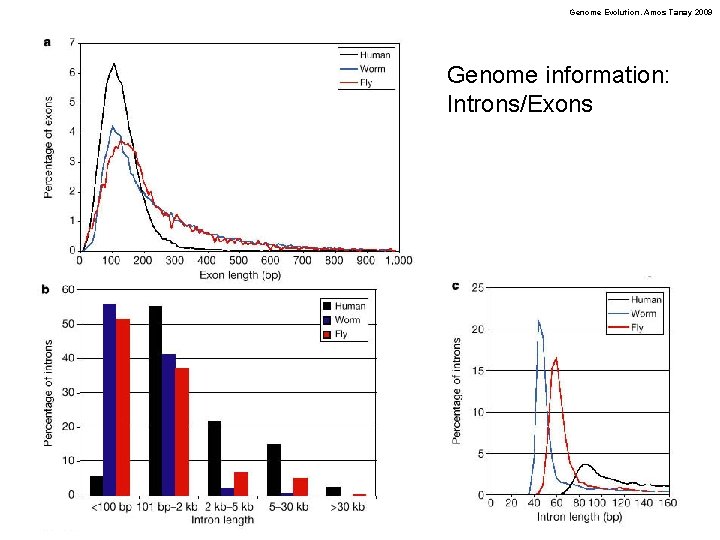
Genome Evolution. Amos Tanay 2009 Genome information: Introns/Exons

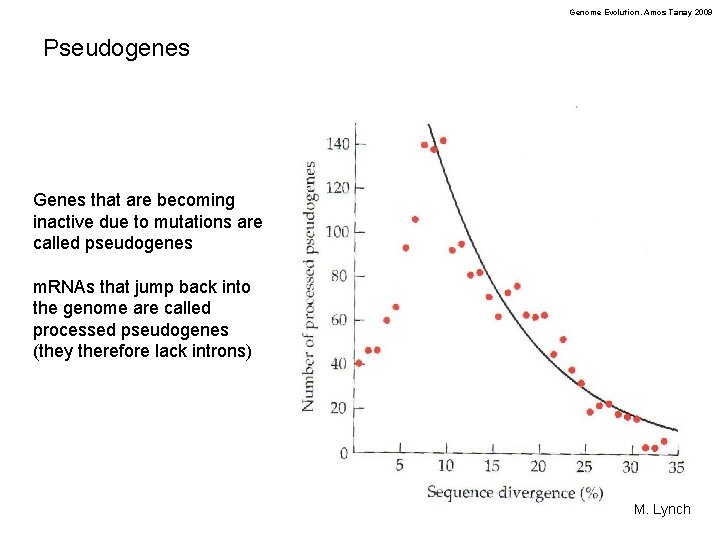
Genome Evolution. Amos Tanay 2009 Pseudogenes Genes that are becoming inactive due to mutations are called pseudogenes m. RNAs that jump back into the genome are called processed pseudogenes (they therefore lack introns) M. Lynch

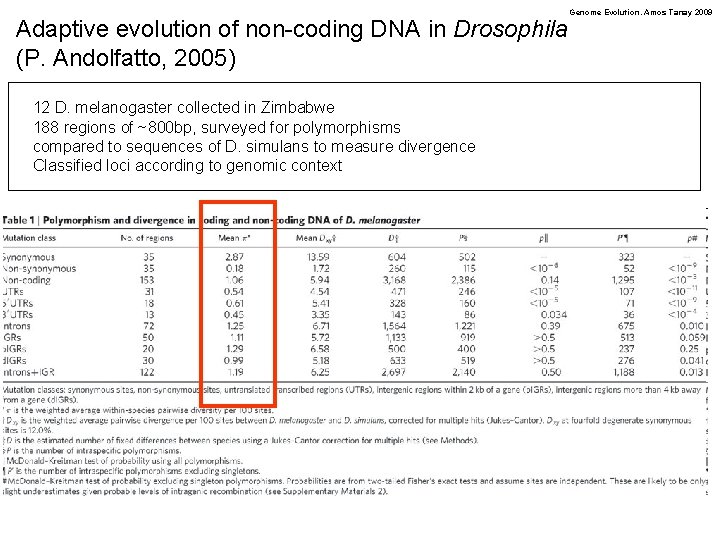
Adaptive evolution of non-coding DNA in Drosophila (P. Andolfatto, 2005) 12 D. melanogaster collected in Zimbabwe 188 regions of ~800 bp, surveyed for polymorphisms compared to sequences of D. simulans to measure divergence Classified loci according to genomic context Genome Evolution. Amos Tanay 2009

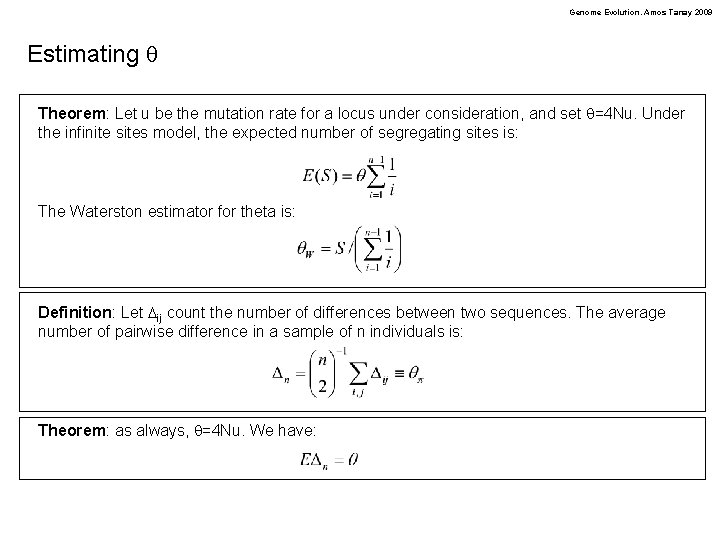
Genome Evolution. Amos Tanay 2009 Estimating q Theorem: Let u be the mutation rate for a locus under consideration, and set q=4 Nu. Under the infinite sites model, the expected number of segregating sites is: The Waterston estimator for theta is: Definition: Let Dij count the number of differences between two sequences. The average number of pairwise difference in a sample of n individuals is: Theorem: as always, q=4 Nu. We have:

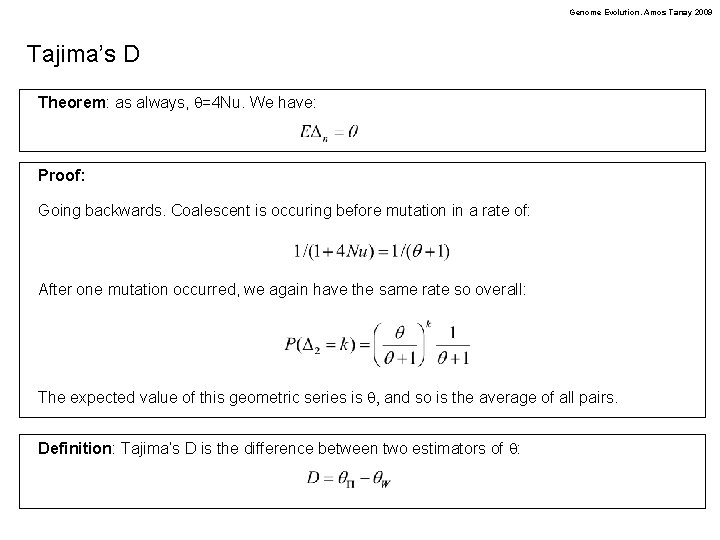
Genome Evolution. Amos Tanay 2009 Tajima’s D Theorem: as always, q=4 Nu. We have: Proof: Going backwards. Coalescent is occuring before mutation in a rate of: After one mutation occurred, we again have the same rate so overall: The expected value of this geometric series is q, and so is the average of all pairs. Definition: Tajima’s D is the difference between two estimators of q:

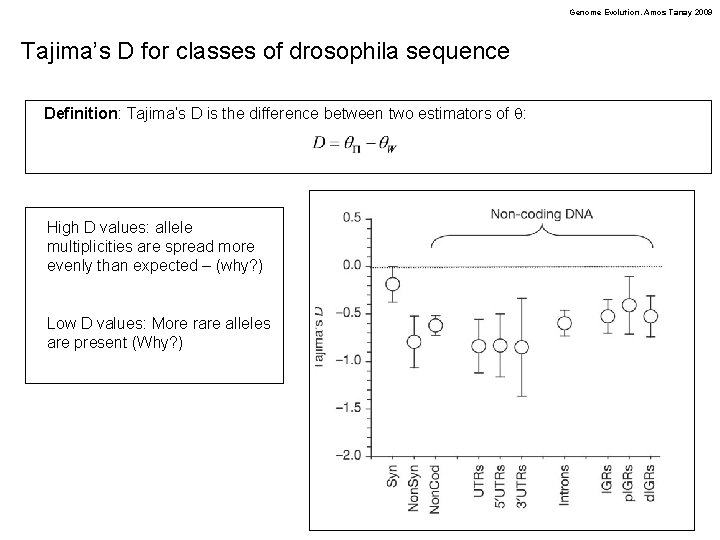
Genome Evolution. Amos Tanay 2009 Tajima’s D for classes of drosophila sequence Definition: Tajima’s D is the difference between two estimators of q: High D values: allele multiplicities are spread more evenly than expected – (why? ) Low D values: More rare alleles are present (Why? )

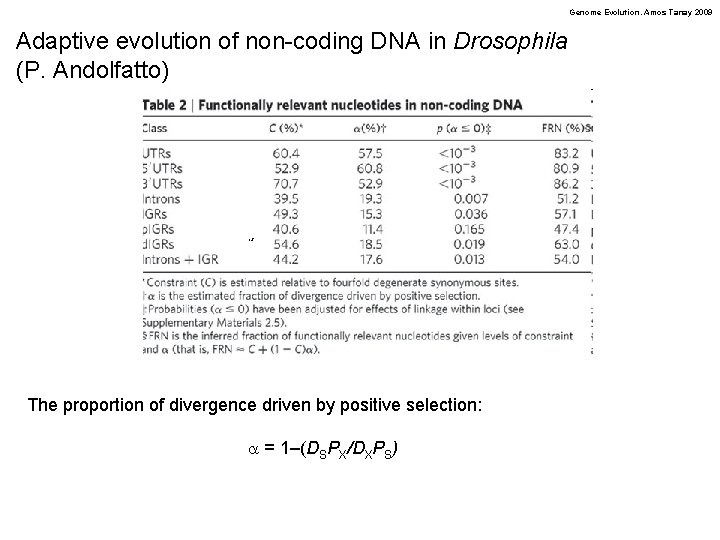
Genome Evolution. Amos Tanay 2009 Adaptive evolution of non-coding DNA in Drosophila (P. Andolfatto) The proportion of divergence driven by positive selection: a = 1–(DSPX/DXPS)

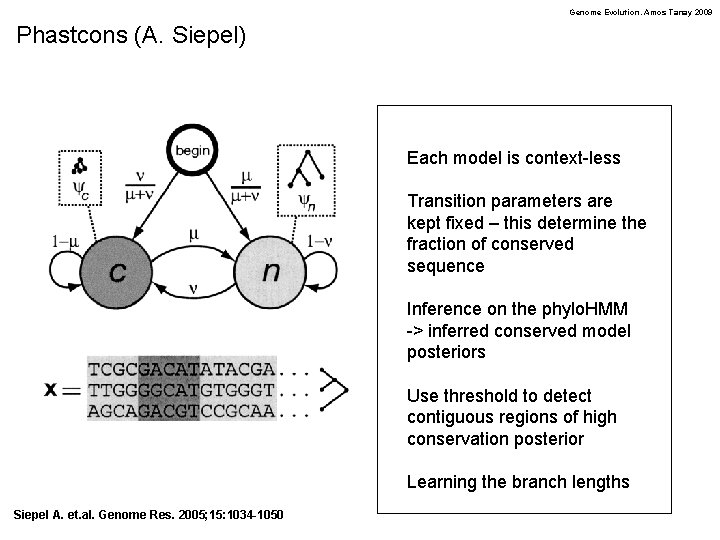
Genome Evolution. Amos Tanay 2009 Phastcons (A. Siepel) Each model is context-less Transition parameters are kept fixed – this determine the fraction of conserved sequence Inference on the phylo. HMM -> inferred conserved model posteriors Use threshold to detect contiguous regions of high conservation posterior Learning the branch lengths Siepel A. et. al. Genome Res. 2005; 15: 1034 -1050

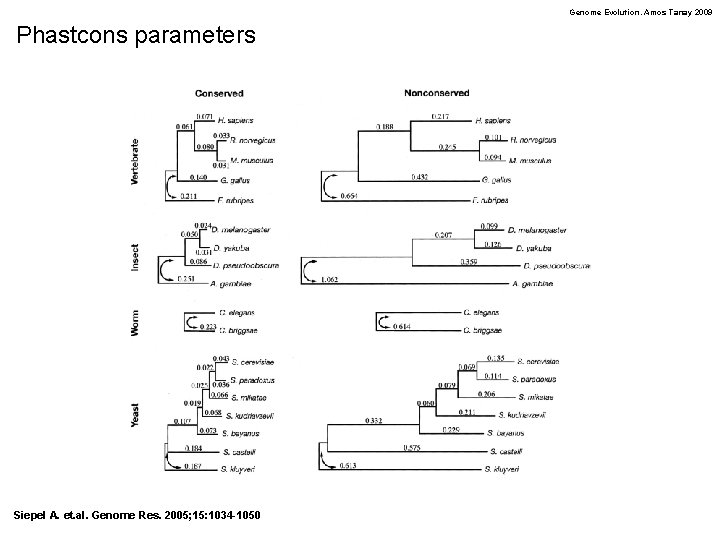
Genome Evolution. Amos Tanay 2009 Phastcons parameters Siepel A. et. al. Genome Res. 2005; 15: 1034 -1050

Genome Evolution. Amos Tanay 2009 Fixation probabilities and population size: what selection coefficient can drive a 70% decrease in substitution rate (if N_e = 10, 000)?

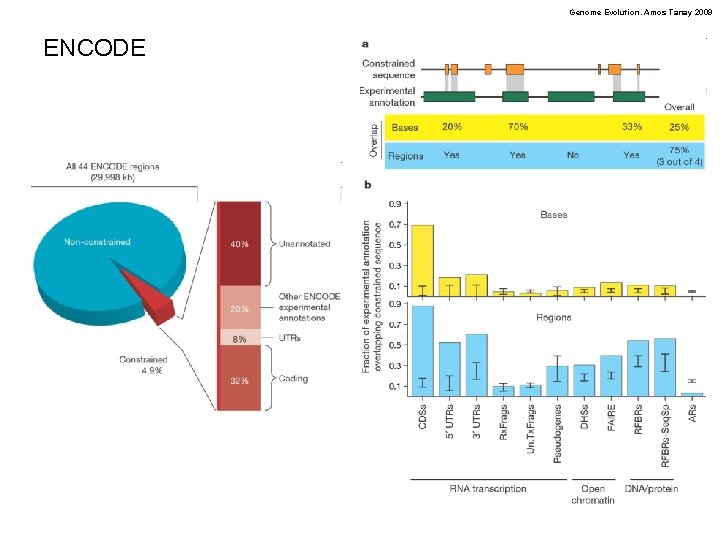
Genome Evolution. Amos Tanay 2009 ENCODE

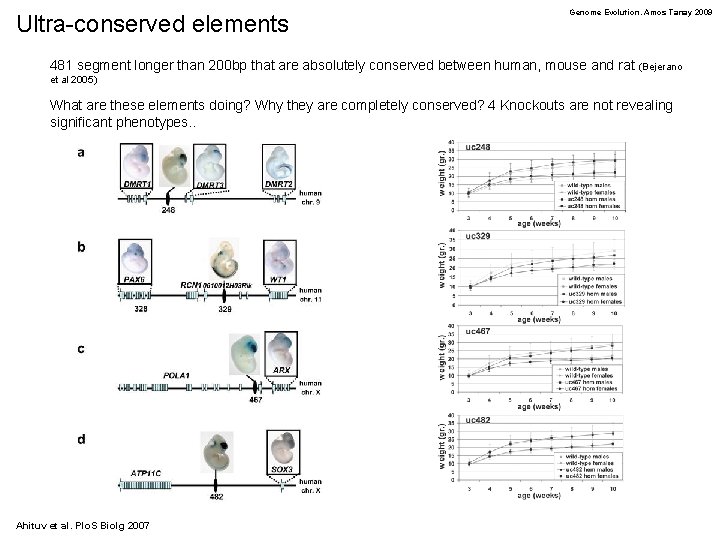
Ultra-conserved elements Genome Evolution. Amos Tanay 2009 481 segment longer than 200 bp that are absolutely conserved between human, mouse and rat (Bejerano et al 2005) What are these elements doing? Why they are completely conserved? 4 Knockouts are not revealing significant phenotypes. . Ahituv et al. Plo. S Biolg 2007

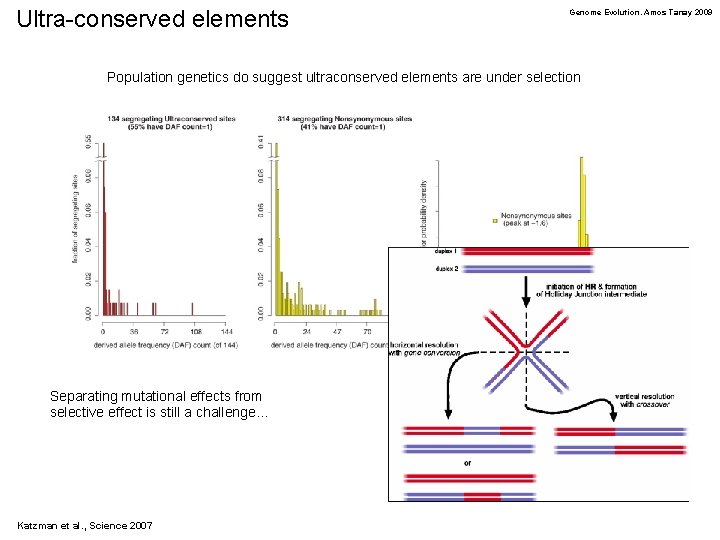
Ultra-conserved elements Genome Evolution. Amos Tanay 2009 Population genetics do suggest ultraconserved elements are under selection Separating mutational effects from selective effect is still a challenge… Katzman et al. , Science 2007