Genome Evolution Amos Tanay 2009 Genome evolution Lecture



- Slides: 32

Genome Evolution. Amos Tanay 2009 Genome evolution Lecture 7: Selection in protein coding genes

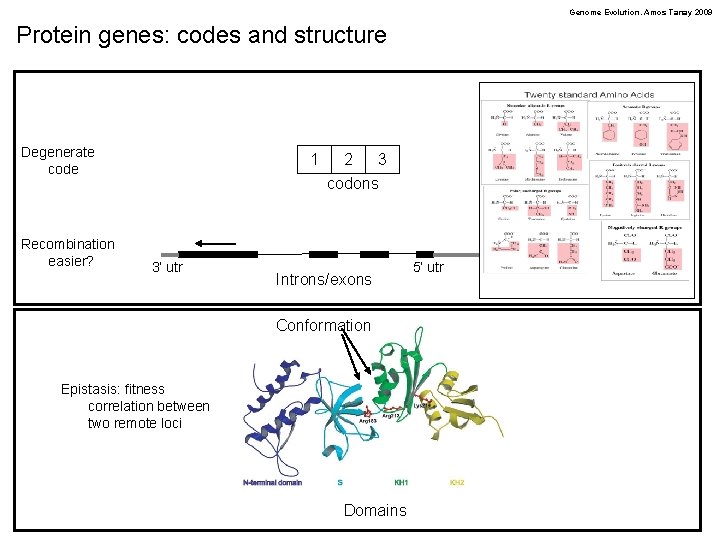
Genome Evolution. Amos Tanay 2009 Protein genes: codes and structure Degenerate code Recombination easier? 1 3’ utr 2 3 codons Introns/exons Conformation Epistasis: fitness correlation between two remote loci Domains 5’ utr

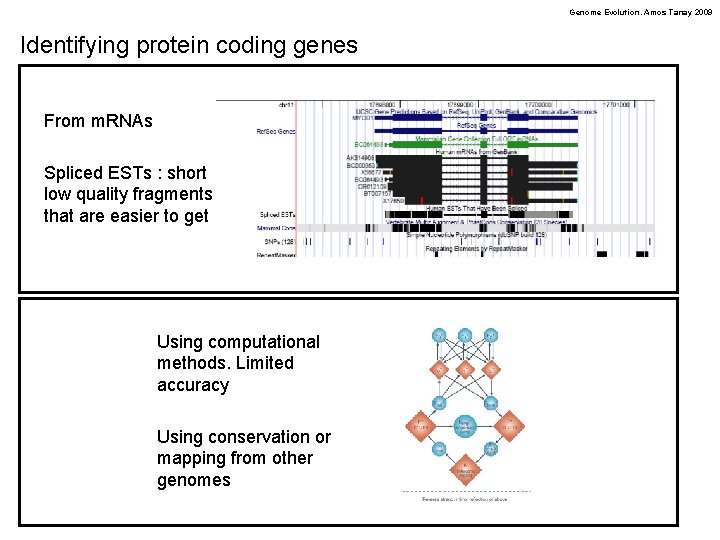
Genome Evolution. Amos Tanay 2009 Identifying protein coding genes From m. RNAs Spliced ESTs : short low quality fragments that are easier to get Using computational methods. Limited accuracy Using conservation or mapping from other genomes

Genome Evolution. Amos Tanay 2009 Questions on protein function and evolution Identification: • Identify protein coding genes – Not yet resolved for new species, but with new technology this question may be less important than before Structure/Function: • Define functional domains – Highly important for understanding protein function • Which parts of the proteins are “important” (e. g. , catalytic? ) – Difficult since structural modeling is hard and context dependent Evolution • Identify places and times where a new protein feature emerged – Positive selection • • Understand mutation/selection through codon degeneracy Understanding processes of duplication and diversitification

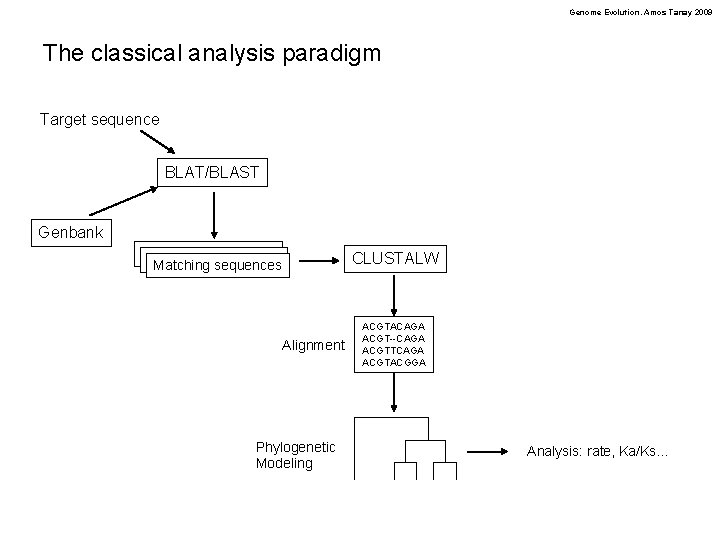
Genome Evolution. Amos Tanay 2009 The classical analysis paradigm Target sequence BLAT/BLAST Genbank CLUSTALW Matching sequences Alignment Phylogenetic Modeling ACGTACAGA ACGT--CAGA ACGTTCAGA ACGTACGGA Analysis: rate, Ka/Ks…

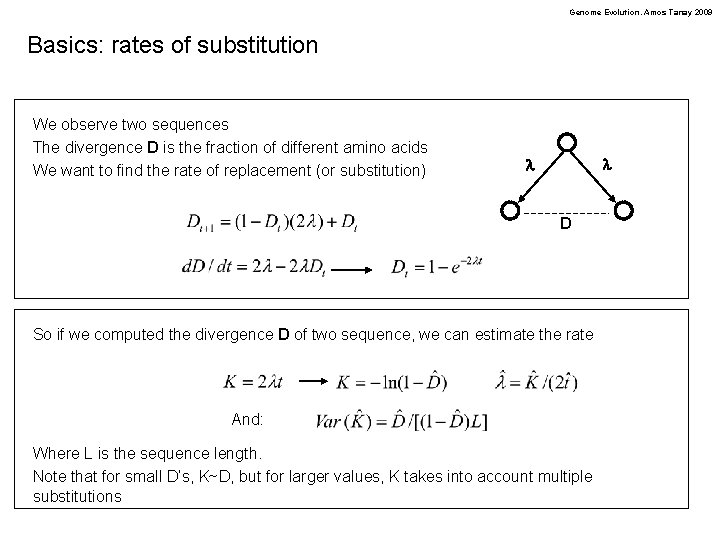
Genome Evolution. Amos Tanay 2009 Basics: rates of substitution We observe two sequences The divergence D is the fraction of different amino acids We want to find the rate of replacement (or substitution) l l D So if we computed the divergence D of two sequence, we can estimate the rate And: Where L is the sequence length. Note that for small D’s, K~D, but for larger values, K takes into account multiple substitutions

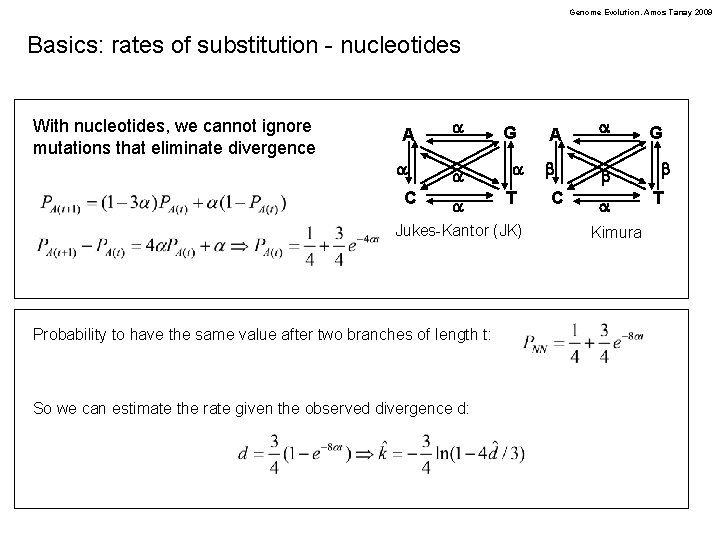
Genome Evolution. Amos Tanay 2009 Basics: rates of substitution - nucleotides With nucleotides, we cannot ignore mutations that eliminate divergence A a C a a a G a T Jukes-Kantor (JK) Probability to have the same value after two branches of length t: So we can estimate the rate given the observed divergence d: A a b b C a Kimura G b T

Genome Evolution. Amos Tanay 2009 Using universal matrices: PAM/BLOSSOM 62 Given a multiple alignment (of protein coding DNA) we can convert the DNA to proteins. We can then try to model the phylogenetic relations between the proteins using a fixed rate matrix Q, some phylogeney T and branch lengths ti When modeling hundreds/thousands amino acid sequences, we cannot learn from the data the rate matrix (20 x 20 parameters!) AND the branch lengths AND the phylogeny. Based on surveys of high quality aligned proteins, Margaret Dayhoff and colleuges generated the famous PAM (Point Accepted mutations): PAM 1 is for 1% substitution probability. Using conserved aligned blocks, Henikoff and Henikoff generated the BLOSUM family of matrices. Henikoff approach improved analysis of distantly related proteins, and is based on more sequence (lots of conserved blocks), but filtering away highly conserved positions (BLOSUM 62 filter anything that is more than 62% conserved) S. Henikoff

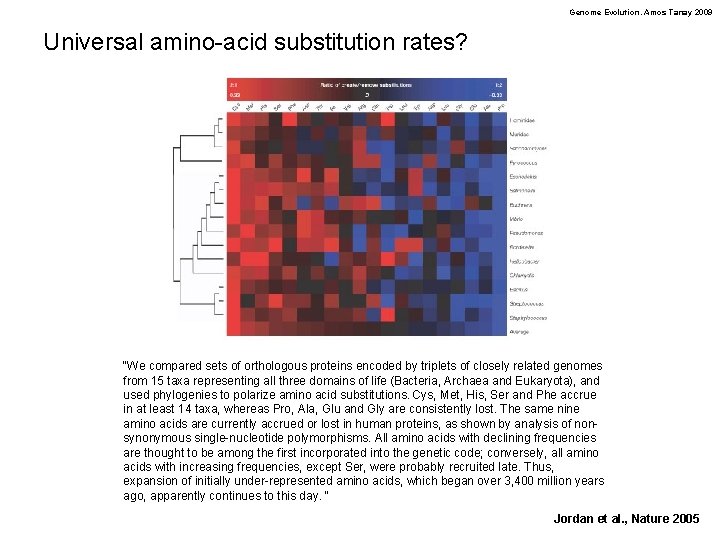
Genome Evolution. Amos Tanay 2009 Universal amino-acid substitution rates? “We compared sets of orthologous proteins encoded by triplets of closely related genomes from 15 taxa representing all three domains of life (Bacteria, Archaea and Eukaryota), and used phylogenies to polarize amino acid substitutions. Cys, Met, His, Ser and Phe accrue in at least 14 taxa, whereas Pro, Ala, Glu and Gly are consistently lost. The same nine amino acids are currently accrued or lost in human proteins, as shown by analysis of nonsynonymous single-nucleotide polymorphisms. All amino acids with declining frequencies are thought to be among the first incorporated into the genetic code; conversely, all amino acids with increasing frequencies, except Ser, were probably recruited late. Thus, expansion of initially under-represented amino acids, which began over 3, 400 million years ago, apparently continues to this day. “ Jordan et al. , Nature 2005

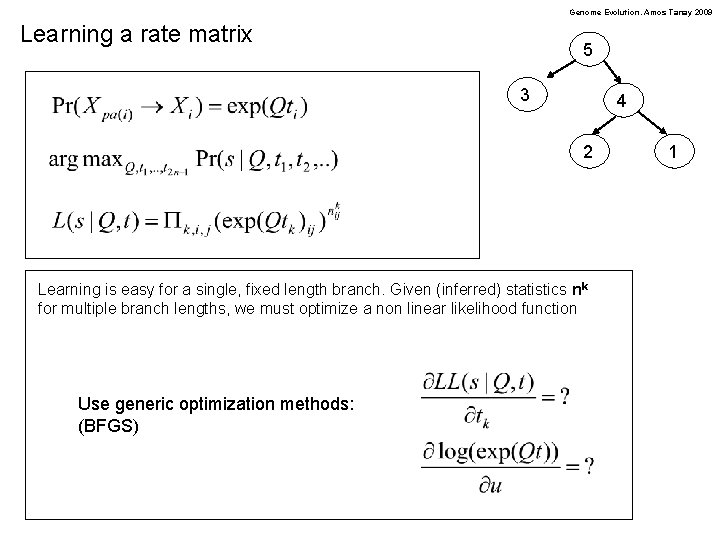
Genome Evolution. Amos Tanay 2009 Learning a rate matrix 5 3 4 2 Learning is easy for a single, fixed length branch. Given (inferred) statistics nk for multiple branch lengths, we must optimize a non linear likelihood function Use generic optimization methods: (BFGS) 1

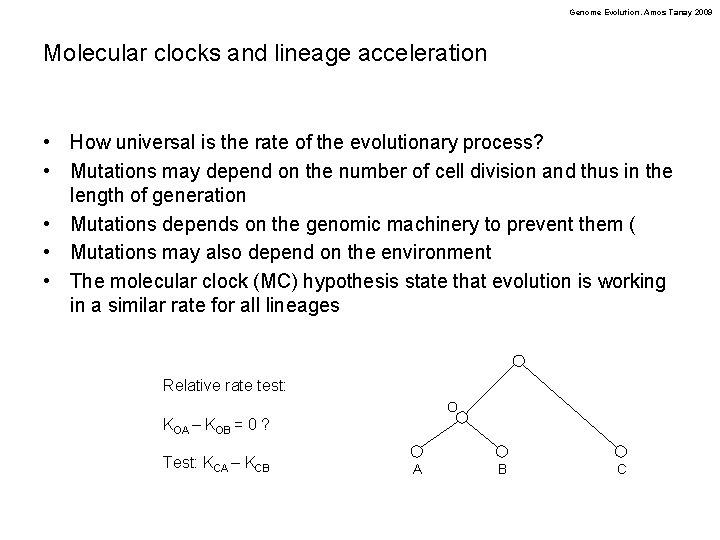
Genome Evolution. Amos Tanay 2009 Molecular clocks and lineage acceleration • How universal is the rate of the evolutionary process? • Mutations may depend on the number of cell division and thus in the length of generation • Mutations depends on the genomic machinery to prevent them ( • Mutations may also depend on the environment • The molecular clock (MC) hypothesis state that evolution is working in a similar rate for all lineages Relative rate test: O KOA – KOB = 0 ? Test: KCA – KCB A B C

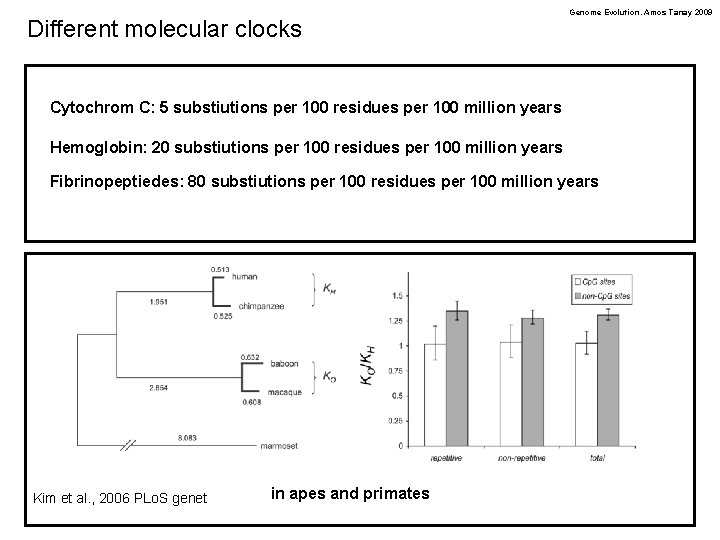
Different molecular clocks Genome Evolution. Amos Tanay 2009 Cytochrom C: 5 substiutions per 100 residues per 100 million years Hemoglobin: 20 substiutions per 100 residues per 100 million years Fibrinopeptiedes: 80 substiutions per 100 residues per 100 million years Kim et al. , 2006 PLo. S genet in apes and primates

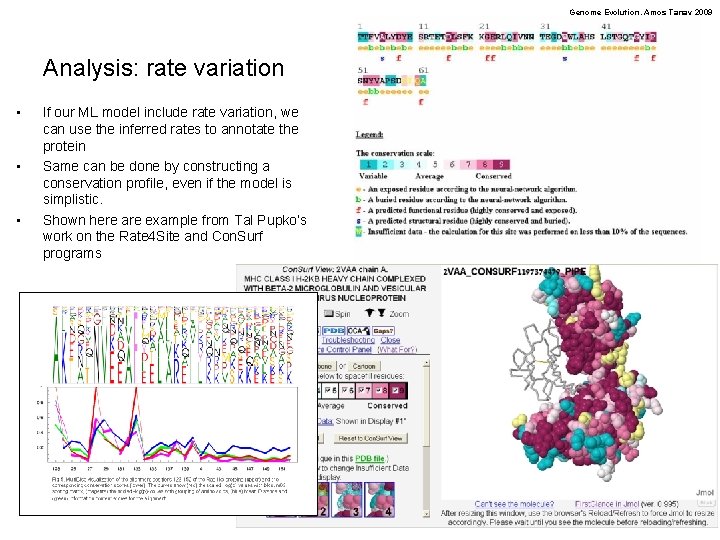
Genome Evolution. Amos Tanay 2009 Analysis: rate variation • • • If our ML model include rate variation, we can use the inferred rates to annotate the protein Same can be done by constructing a conservation profile, even if the model is simplistic. Shown here are example from Tal Pupko’s work on the Rate 4 Site and Con. Surf programs

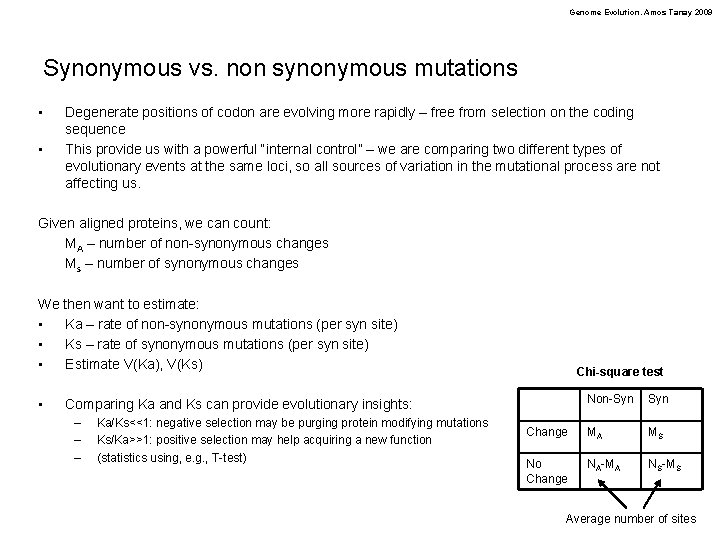
Genome Evolution. Amos Tanay 2009 Synonymous vs. non synonymous mutations • • Degenerate positions of codon are evolving more rapidly – free from selection on the coding sequence This provide us with a powerful “internal control” – we are comparing two different types of evolutionary events at the same loci, so all sources of variation in the mutational process are not affecting us. Given aligned proteins, we can count: MA – number of non-synonymous changes Ms – number of synonymous changes We then want to estimate: • Ka – rate of non-synonymous mutations (per syn site) • Ks – rate of synonymous mutations (per syn site) • Estimate V(Ka), V(Ks) • Chi-square test Non-Syn Change MA MS No Change NA-MA NS-MS Comparing Ka and Ks can provide evolutionary insights: – – – Ka/Ks<<1: negative selection may be purging protein modifying mutations Ks/Ka>>1: positive selection may help acquiring a new function (statistics using, e. g. , T-test) Average number of sites

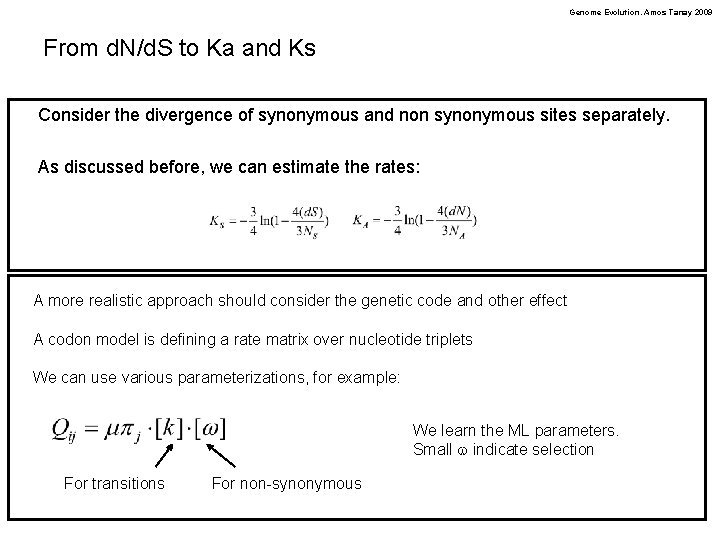
Genome Evolution. Amos Tanay 2009 From d. N/d. S to Ka and Ks Consider the divergence of synonymous and non synonymous sites separately. As discussed before, we can estimate the rates: A more realistic approach should consider the genetic code and other effect A codon model is defining a rate matrix over nucleotide triplets We can use various parameterizations, for example: We learn the ML parameters. Small w indicate selection For transitions For non-synonymous

Genome Evolution. Amos Tanay 2009 Codon bias • • Different codons appears in significantly different frequencies, which is not expected assuming neutrality Bias is measured in several ways, most popular is the codon adaptation index: Codon frequency divided by the frequency of the synonymous codon with maximal frequency • Possible sources of bias: – Selection for translational efficiency given different t. RNA abudnances • Highly expressed genes tend to have stronger codon adaptation indices – Sequence context mutational effects • E. g. Cp. Gs are highly mutable – Selection for low insertion/deletion potential • Weak selection for codon bias should be stronger for genomes with larger effective population size. In some cases this is true

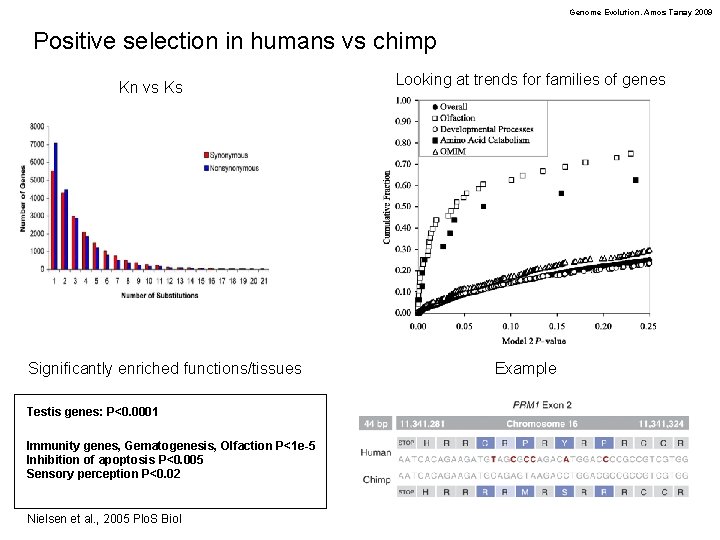
Genome Evolution. Amos Tanay 2009 Positive selection in humans vs chimp Kn vs Ks Significantly enriched functions/tissues Testis genes: P<0. 0001 Immunity genes, Gematogenesis, Olfaction P<1 e-5 Inhibition of apoptosis P<0. 005 Sensory perception P<0. 02 Nielsen et al. , 2005 Plo. S Biol Looking at trends for families of genes Example

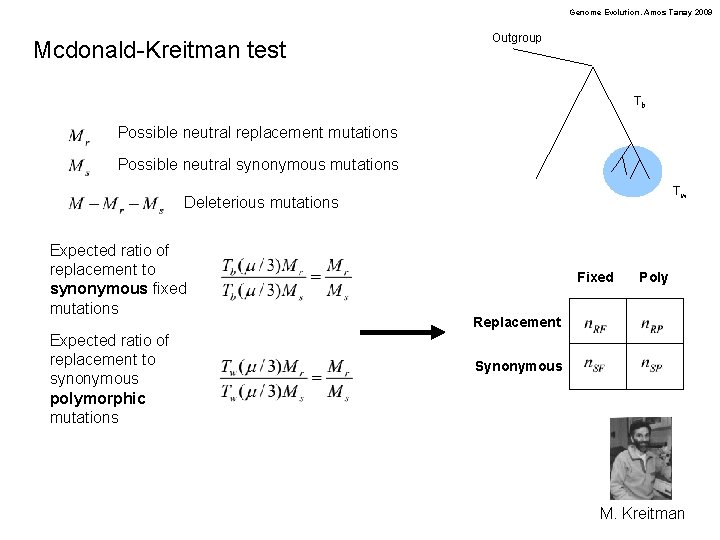
Genome Evolution. Amos Tanay 2009 Mcdonald-Kreitman test Outgroup Tb Possible neutral replacement mutations Possible neutral synonymous mutations Tw Deleterious mutations Expected ratio of replacement to synonymous fixed mutations Expected ratio of replacement to synonymous polymorphic mutations Fixed Poly Replacement Synonymous M. Kreitman

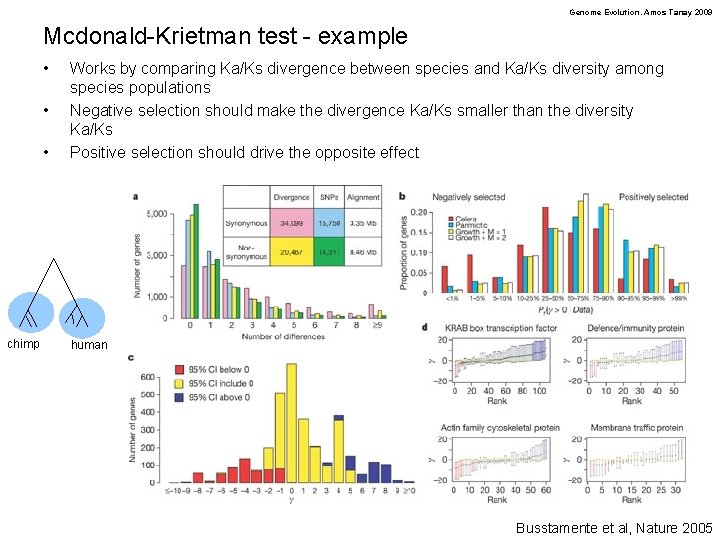
Genome Evolution. Amos Tanay 2009 Mcdonald-Krietman test - example • • • chimp Works by comparing Ka/Ks divergence between species and Ka/Ks diversity among species populations Negative selection should make the divergence Ka/Ks smaller than the diversity Ka/Ks Positive selection should drive the opposite effect human Busstamente et al, Nature 2005

Genome Evolution. Amos Tanay 2009 Reminder: the coalescent Past Theorem: The amount of time during which there are k lineages, tk has approximately an exponential distribution with mean 2 N * (2/(k(k-1))) 1 2 3 4 5 Present In the infinite sites model, mutations occur at distinct sites. Back in time Coalescent: mutation:

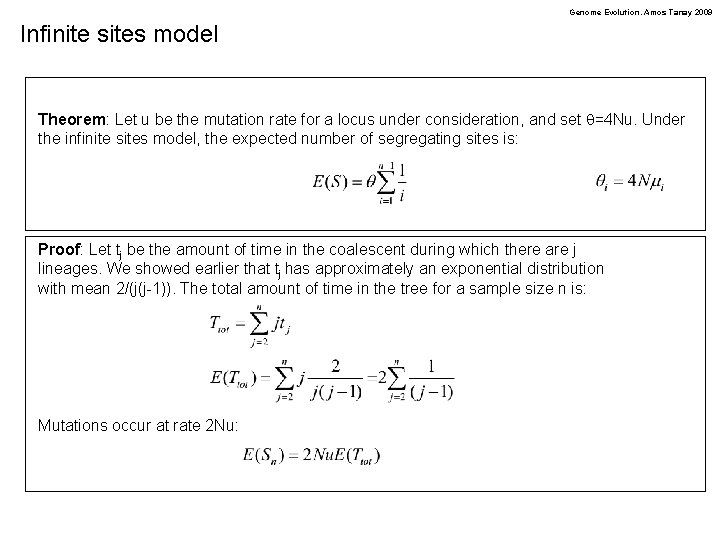
Genome Evolution. Amos Tanay 2009 Infinite sites model Theorem: Let u be the mutation rate for a locus under consideration, and set q=4 Nu. Under the infinite sites model, the expected number of segregating sites is: Proof: Let tj be the amount of time in the coalescent during which there are j lineages. We showed earlier that tj has approximately an exponential distribution with mean 2/(j(j-1)). The total amount of time in the tree for a sample size n is: Mutations occur at rate 2 Nu:

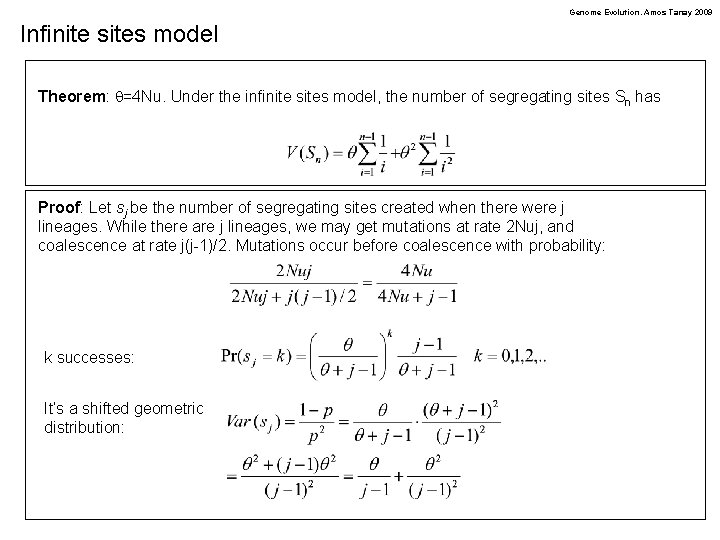
Genome Evolution. Amos Tanay 2009 Infinite sites model Theorem: q=4 Nu. Under the infinite sites model, the number of segregating sites Sn has Proof: Let sj be the number of segregating sites created when there were j lineages. While there are j lineages, we may get mutations at rate 2 Nuj, and coalescence at rate j(j-1)/2. Mutations occur before coalescence with probability: k successes: It’s a shifted geometric distribution:

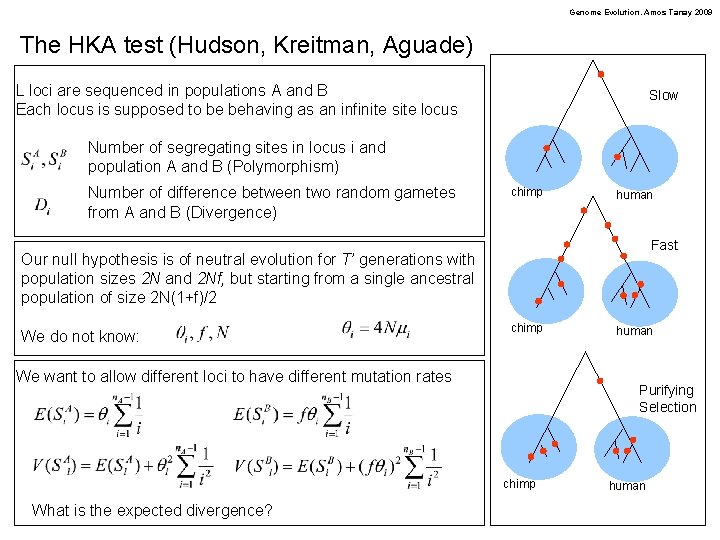
Genome Evolution. Amos Tanay 2009 The HKA test (Hudson, Kreitman, Aguade) L loci are sequenced in populations A and B Each locus is supposed to be behaving as an infinite site locus Slow Number of segregating sites in locus i and population A and B (Polymorphism) Number of difference between two random gametes from A and B (Divergence) chimp human Fast Our null hypothesis is of neutral evolution for T’ generations with population sizes 2 N and 2 Nf, but starting from a single ancestral population of size 2 N(1+f)/2 We do not know: chimp We want to allow different loci to have different mutation rates Purifying Selection chimp What is the expected divergence? human

Genome Evolution. Amos Tanay 2009 The HKA test (Hudson, Kreitman, Aguade) Number of segregating sites in locus i and population A and B (Polymorphism) Number of difference between two random gametes from A and B (Divergence) B Divergence Is a Poisson variable Coalescent in ancestral population Variance of Poisson variable A Variance of S with n=2

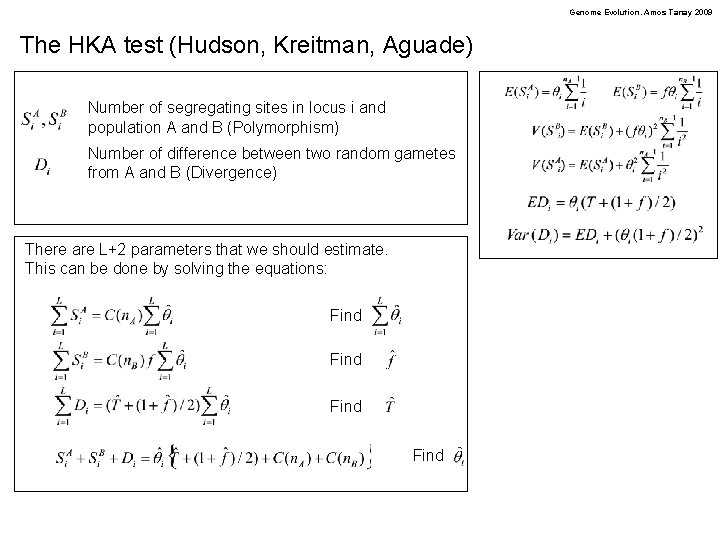
Genome Evolution. Amos Tanay 2009 The HKA test (Hudson, Kreitman, Aguade) Number of segregating sites in locus i and population A and B (Polymorphism) Number of difference between two random gametes from A and B (Divergence) There are L+2 parameters that we should estimate. This can be done by solving the equations: Find

Genome Evolution. Amos Tanay 2009 The HKA test (Hudson, Kreitman, Aguade) Number of segregating sites in locus i and population A and B (Polymorphism) Number of difference between two random gametes from A and B (Divergence) The goodness of fit can be expressed as: Significance is best tested using simulations (although we can assume normality and independence and use chi square with 3 L-(L+2) degrees of freedom)

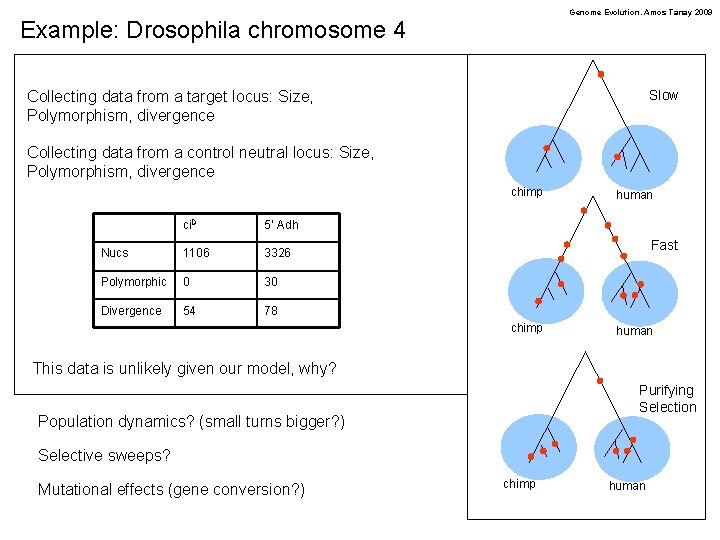
Genome Evolution. Amos Tanay 2009 Example: Drosophila chromosome 4 Slow Collecting data from a target locus: Size, Polymorphism, divergence Collecting data from a control neutral locus: Size, Polymorphism, divergence chimp ci. D 5’ Adh Nucs 1106 3326 Polymorphic 0 30 Divergence 54 78 human Fast chimp human This data is unlikely given our model, why? Purifying Selection Population dynamics? (small turns bigger? ) Selective sweeps? Mutational effects (gene conversion? ) chimp human

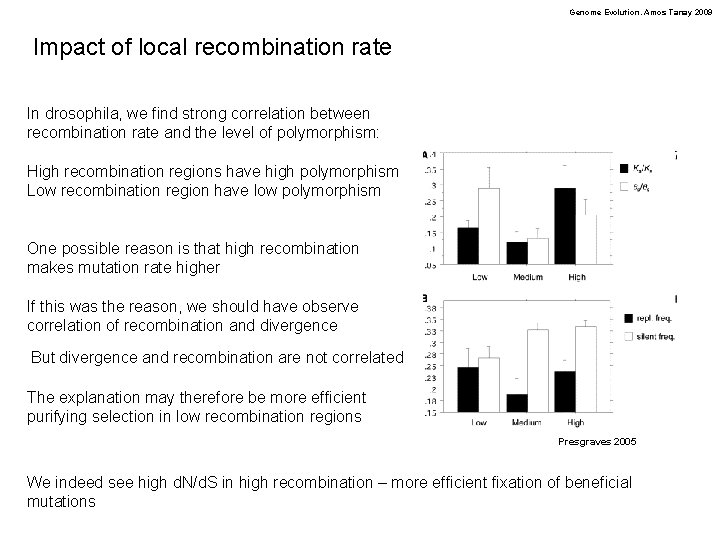
Genome Evolution. Amos Tanay 2009 Impact of local recombination rate In drosophila, we find strong correlation between recombination rate and the level of polymorphism: High recombination regions have high polymorphism Low recombination region have low polymorphism One possible reason is that high recombination makes mutation rate higher If this was the reason, we should have observe correlation of recombination and divergence But divergence and recombination are not correlated The explanation may therefore be more efficient purifying selection in low recombination regions Presgraves 2005 We indeed see high d. N/d. S in high recombination – more efficient fixation of beneficial mutations

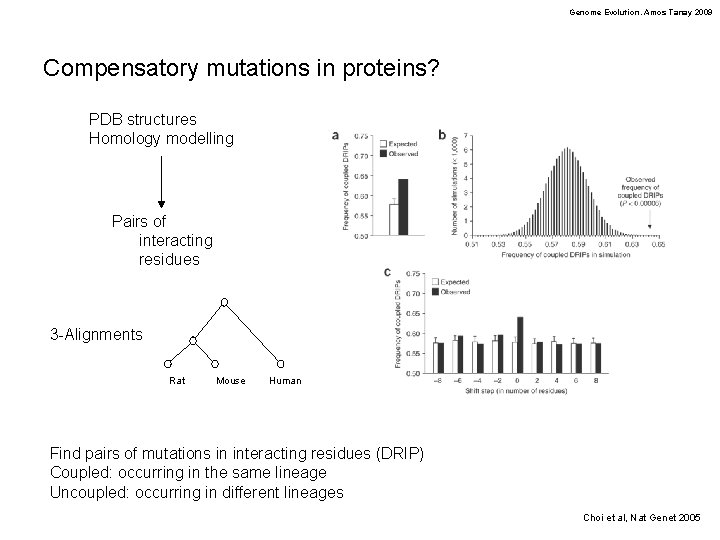
Genome Evolution. Amos Tanay 2009 Compensatory mutations in proteins? PDB structures Homology modelling Pairs of interacting residues 3 -Alignments Rat Mouse Human Find pairs of mutations in interacting residues (DRIP) Coupled: occurring in the same lineage Uncoupled: occurring in different lineages Choi et al, Nat Genet 2005

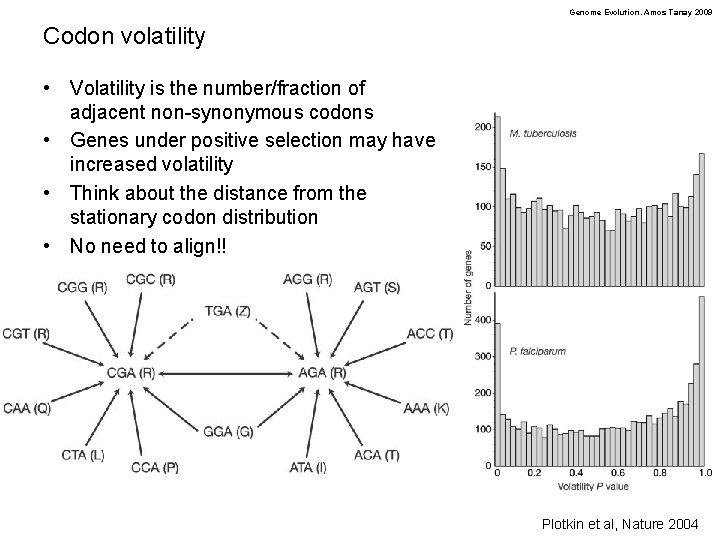
Genome Evolution. Amos Tanay 2009 Codon volatility • Volatility is the number/fraction of adjacent non-synonymous codons • Genes under positive selection may have increased volatility • Think about the distance from the stationary codon distribution • No need to align!! Plotkin et al, Nature 2004

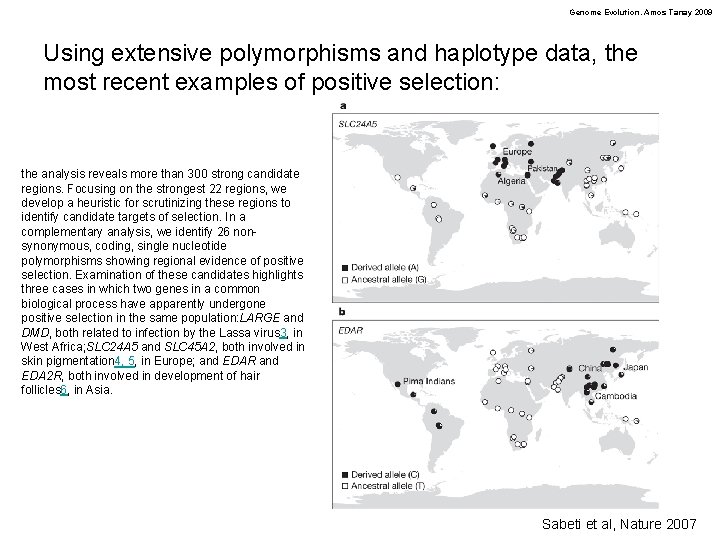
Genome Evolution. Amos Tanay 2009 Using extensive polymorphisms and haplotype data, the most recent examples of positive selection: the analysis reveals more than 300 strong candidate regions. Focusing on the strongest 22 regions, we develop a heuristic for scrutinizing these regions to identify candidate targets of selection. In a complementary analysis, we identify 26 nonsynonymous, coding, single nucleotide polymorphisms showing regional evidence of positive selection. Examination of these candidates highlights three cases in which two genes in a common biological process have apparently undergone positive selection in the same population: LARGE and DMD, both related to infection by the Lassa virus 3, in West Africa; SLC 24 A 5 and SLC 45 A 2, both involved in skin pigmentation 4, 5, in Europe; and EDAR and EDA 2 R, both involved in development of hair follicles 6, in Asia. Sabeti et al, Nature 2007

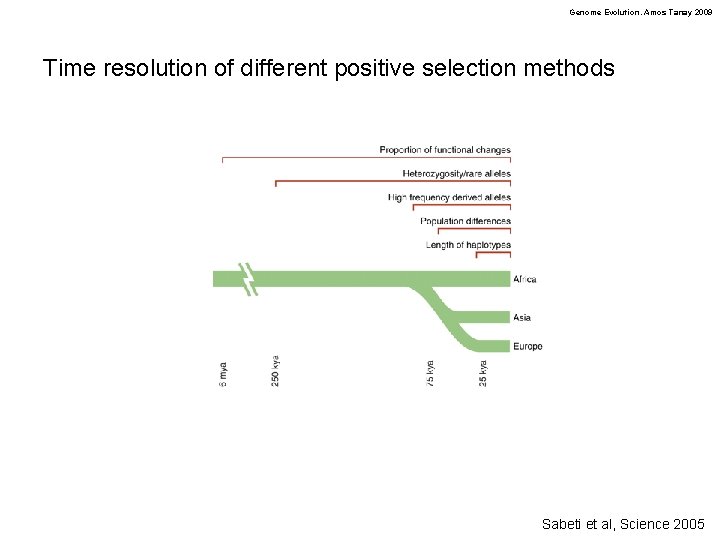
Genome Evolution. Amos Tanay 2009 Time resolution of different positive selection methods Sabeti et al, Science 2005