Genetic Engineering 1 Lecture 18 Pages 323 340







- Slides: 47

Genetic Engineering 1 Lecture 18 Pages 323 - 340

The Tools of Molecular Biology

Old fashioned way was to breed for what you wanted 10_01_experiment. DNA. jpg Mendel did it!

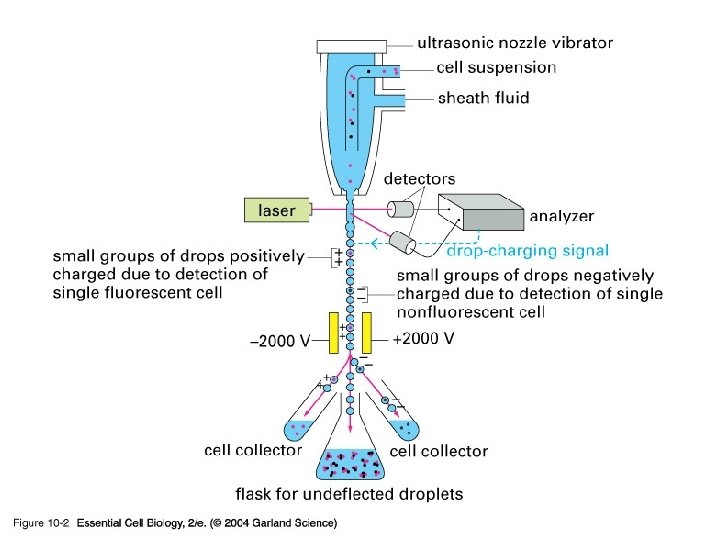
10_02_cell_sorter. jpg

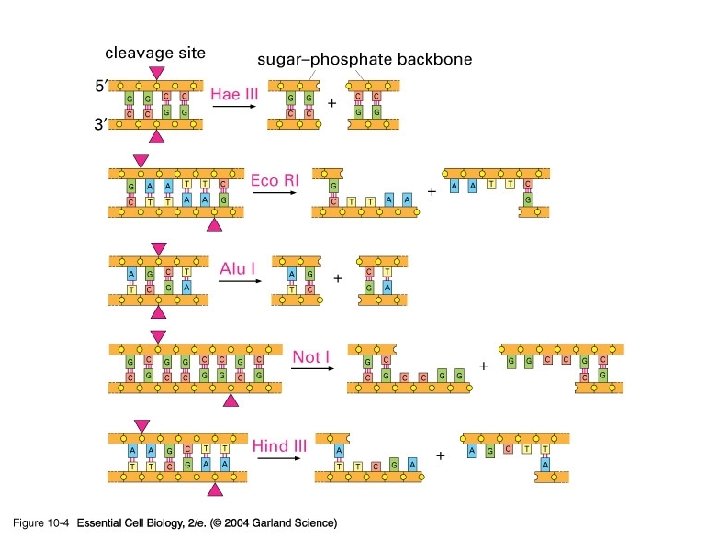
Once you have the cells what next? • Issue was to examine the DNA in a consistent manner • Best method is to use restriction enzymes – Come mainly from bacteria – Use individually or in a mix

10_04_Restrict. nuclease. jpg

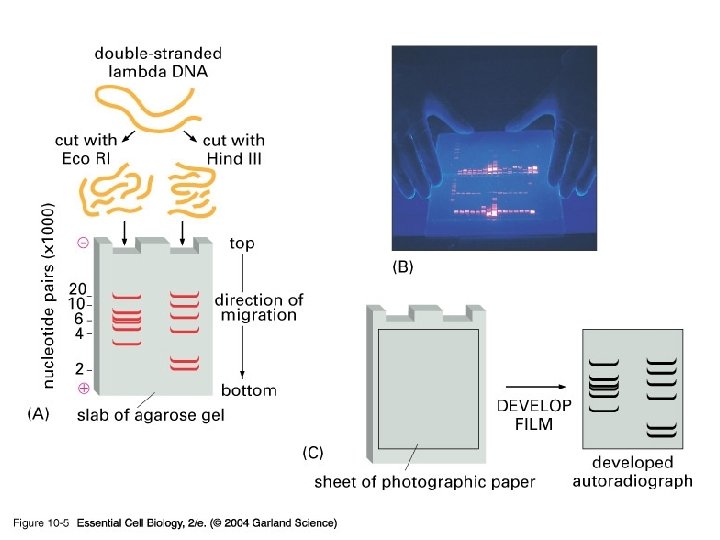
What do you do with these digested fragments of DNA? • Isolate those that you want to work on • How? – Best method is to use gel electrophoresis • Agarose • Polyacrylamide

10_05_gel. electrophor. jpg

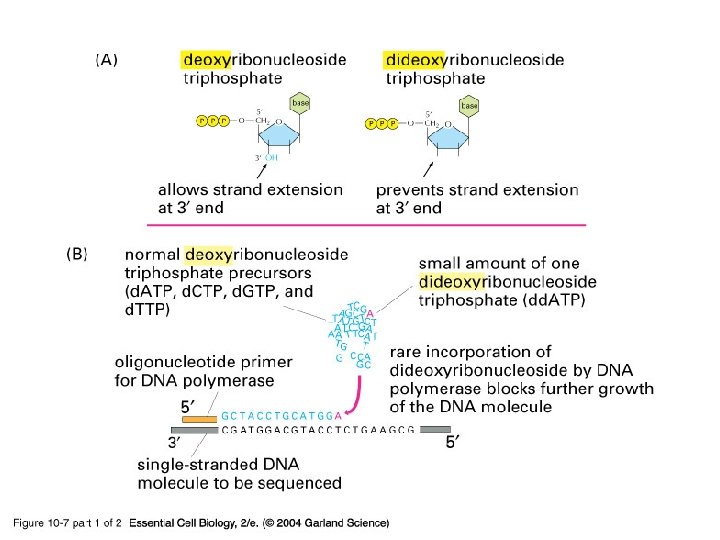
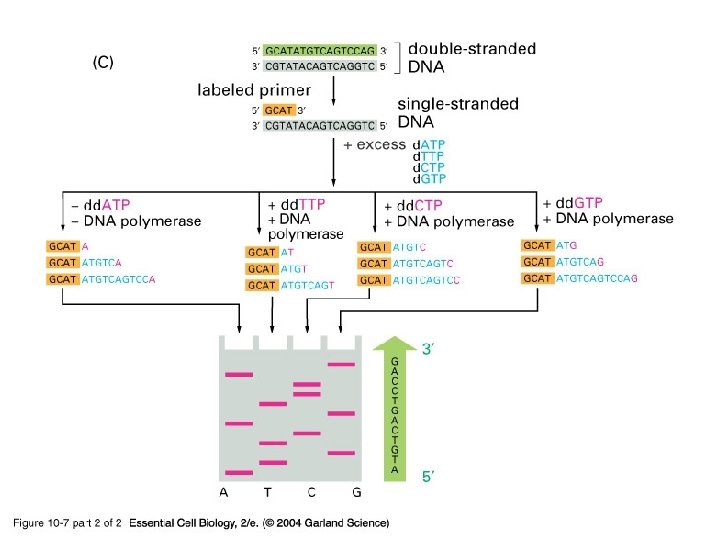
Then what do you do with this piece of DNA? • Clone • Sequence – Rely on the use of dideoxy nucleotides

10_07_1_enzym. dideoxy. jpg

10_07_2_enzym. dideoxy. jpg

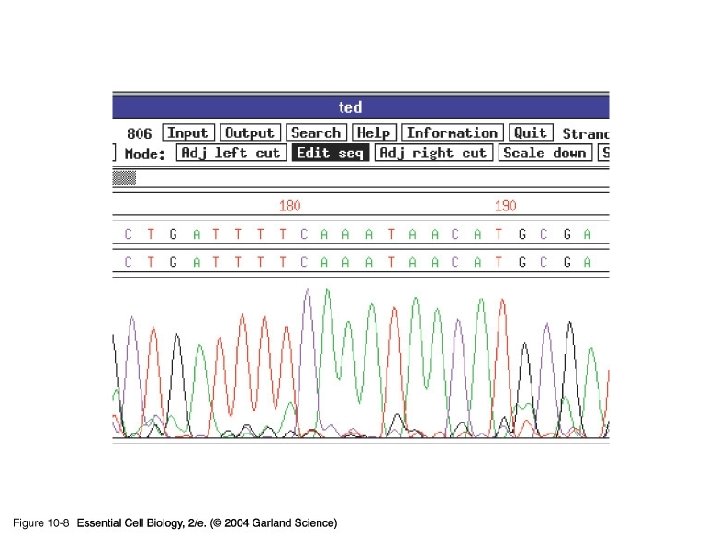
10_08_DNA. sequencing. jpg

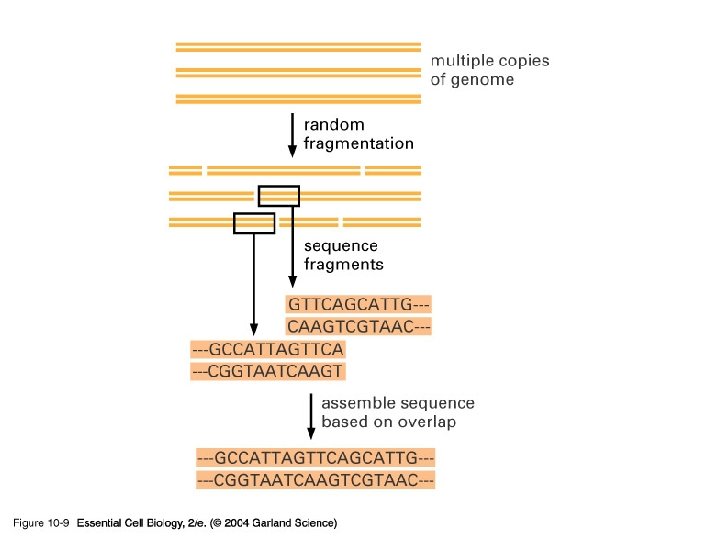
10_09_Shotgun. sequenc. jpg

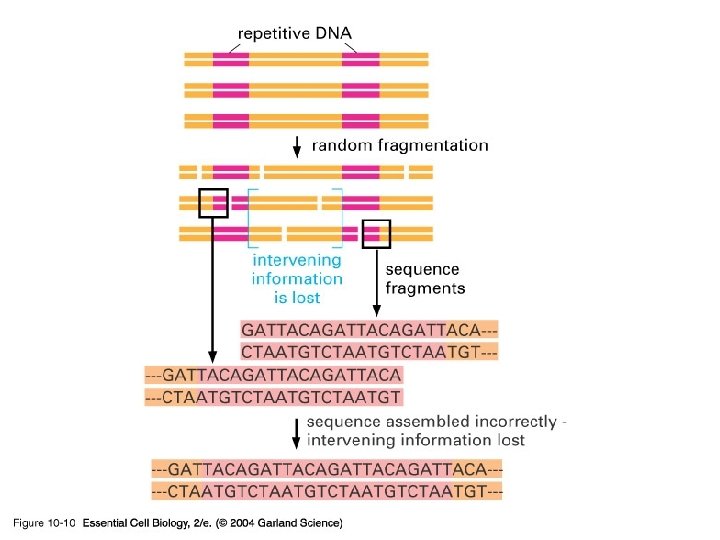
10_10_Repetit. sequence. jpg

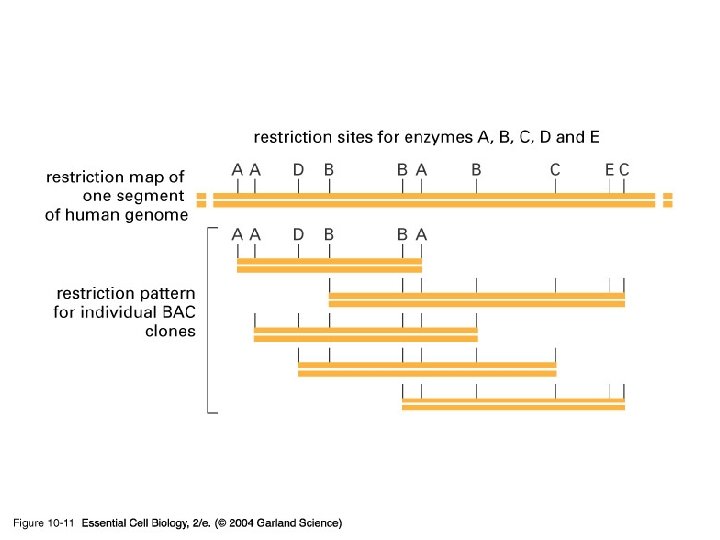
10_11_BAC. clones. jpg

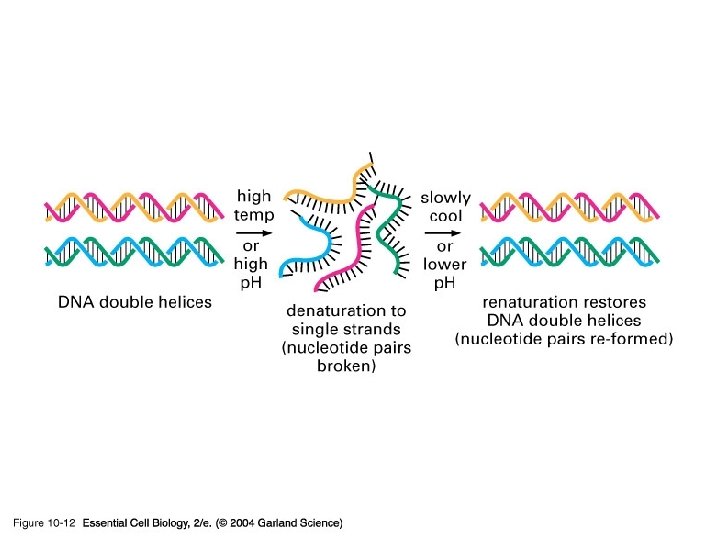
10_12_de_renaturation. jpg

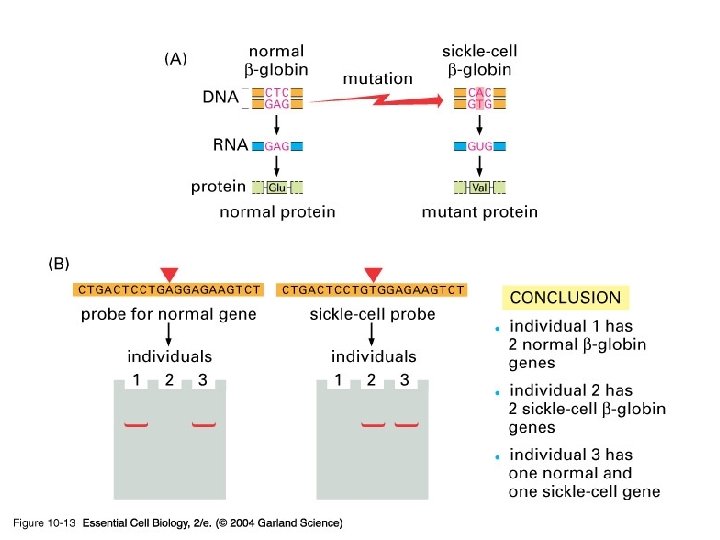
10_13_hybridization. jpg

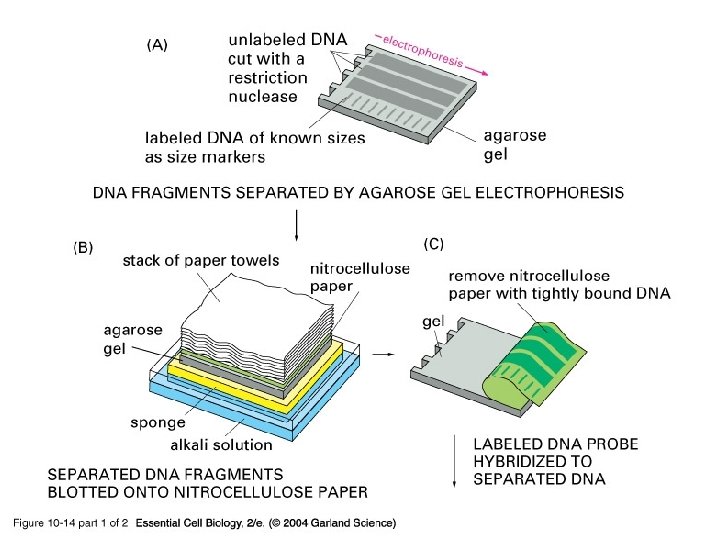
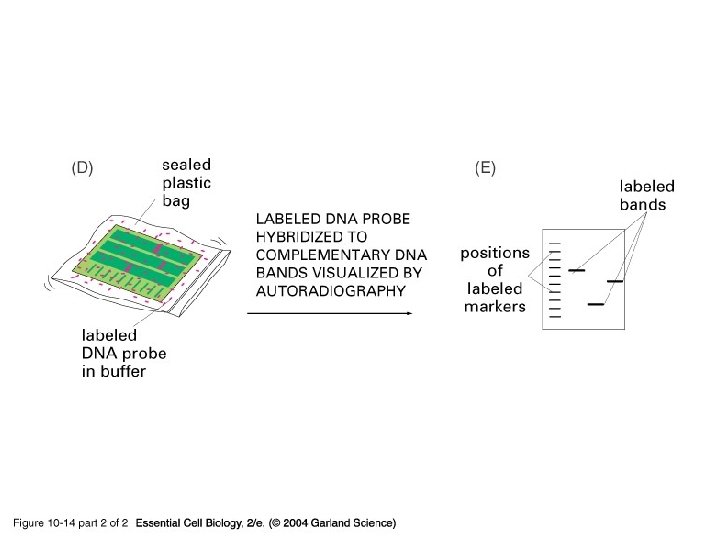
Blotting Purpose to make a permanent record of the results of a gel electrophoresis. The compass • Southern blots - DNA • Northern blots - RNA • Western blots - Proteins

10_14_1_Southrn. blotting. jpg

10_14_2_Southrn. blotting. jpg

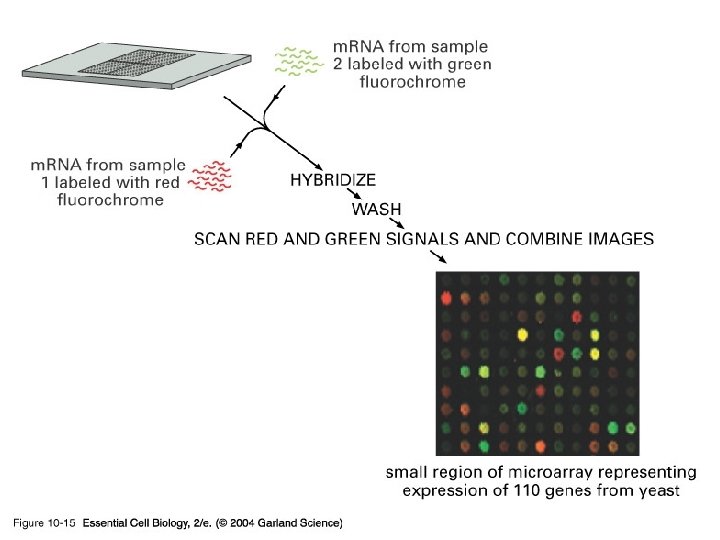
10_15_DNA. microarrays. jpg

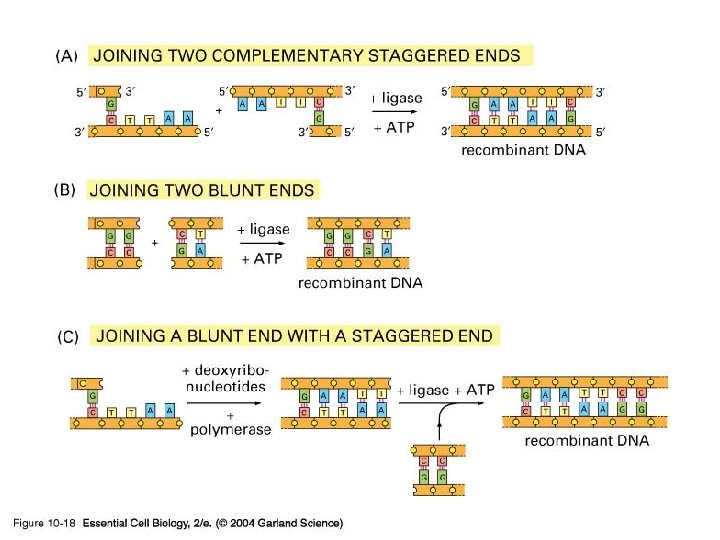
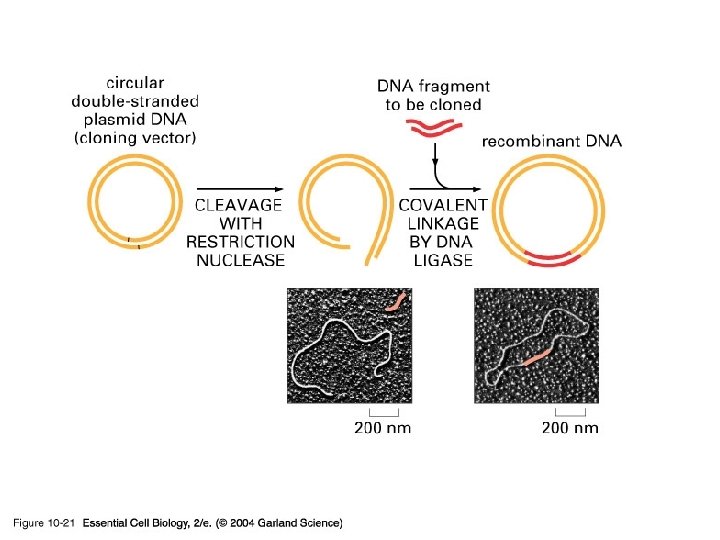
Cloning and growing • One can use the techniques of cell biology to manufacture artificial and real products, be they genes, proteins, or organisms • If you want to insert some DNA into another molecule then the best place to start is to use the same restriction enzyme to cut both - so they have the same ends.

10_18_ DNA. in. vitro. jpg

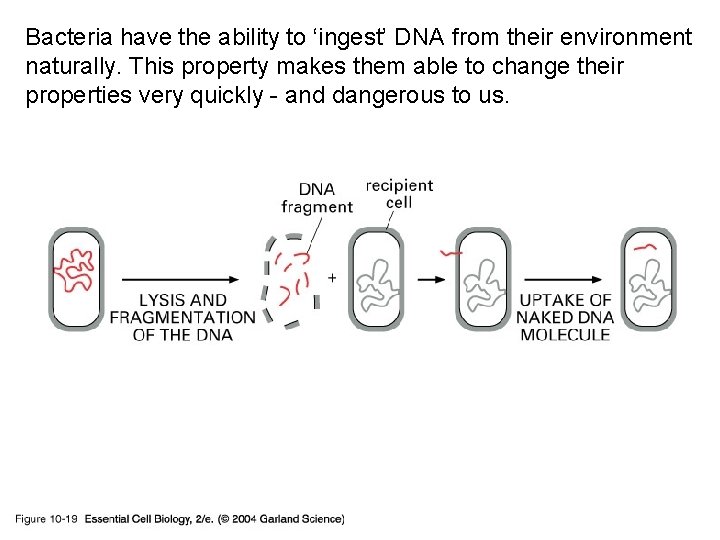
Bacteria have the ability to ‘ingest’ DNA from their environment naturally. This property makes them able to change their properties very quickly - and dangerous to us. 10_19_DNA. uptake. jpg

Bacteria are able to also pass between themselves, other small pieces of DNA known as plasmids. We can make use of plasmids to carry our test DNA into bacteria as shown on the next side… 10_20_Bacteria. plasmid. jpg

10_21_DNA ligase. jpg

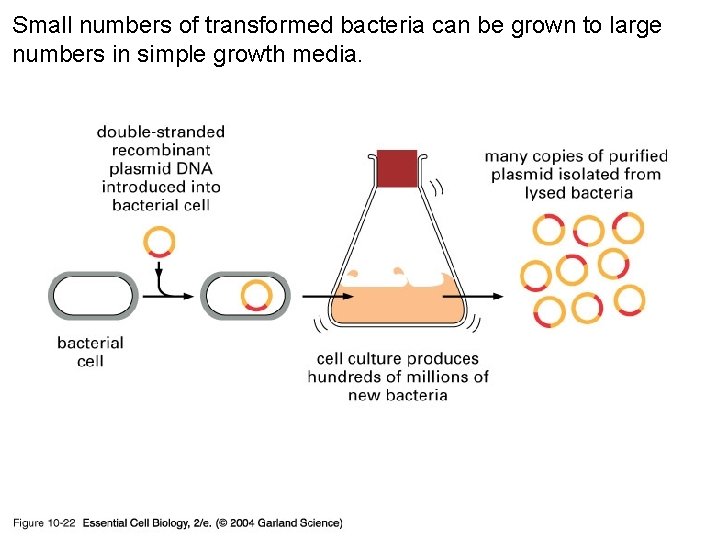
Small numbers of transformed bacteria can be grown to large numbers in simple growth media. 10_22_cloned. DNA. frag. jpg

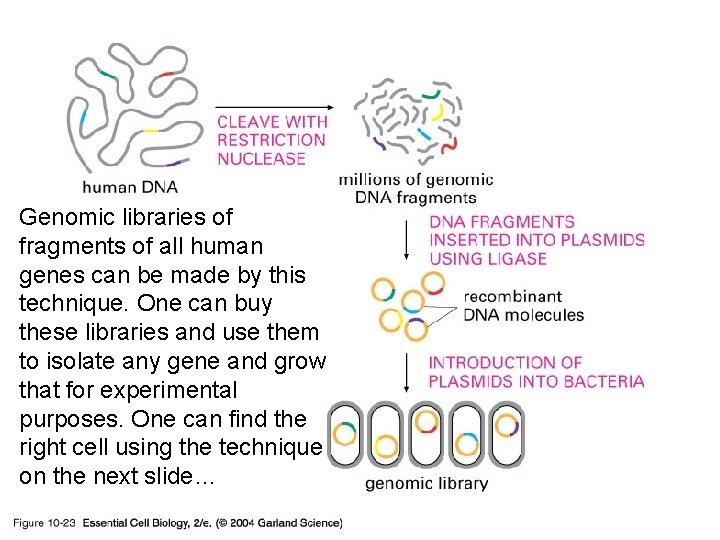
10_23_genomic. library. jpg Genomic libraries of fragments of all human genes can be made by this technique. One can buy these libraries and use them to isolate any gene and grow that for experimental purposes. One can find the right cell using the technique on the next slide…

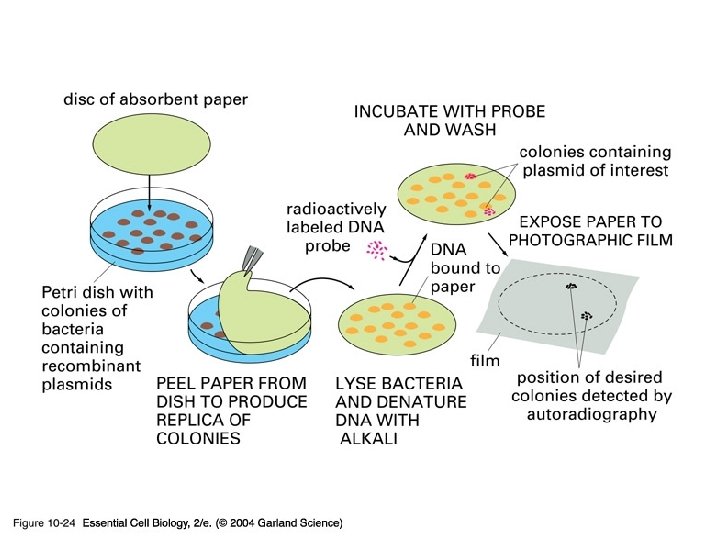
10_24_hybridization. jpg

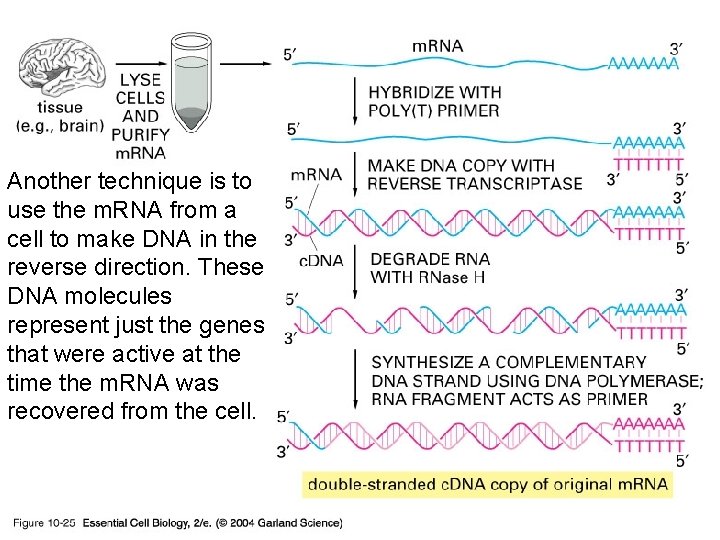
10_25_c. DNA. jpg Another technique is to use the m. RNA from a cell to make DNA in the reverse direction. These DNA molecules represent just the genes that were active at the time the m. RNA was recovered from the cell.

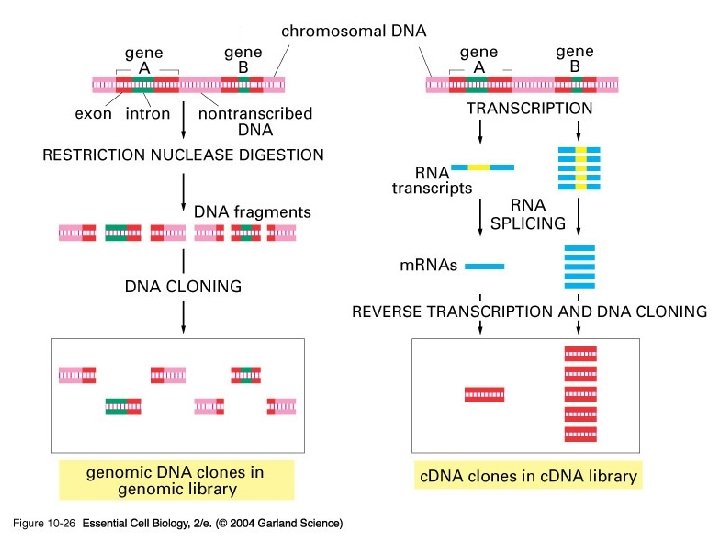
10_26_Genomic_c. DNA. jpg

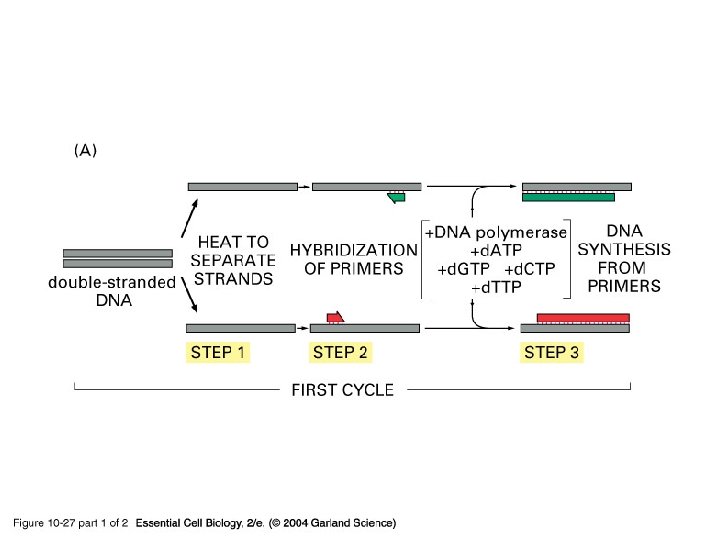
10_27_1_PCR_amplify. jpg

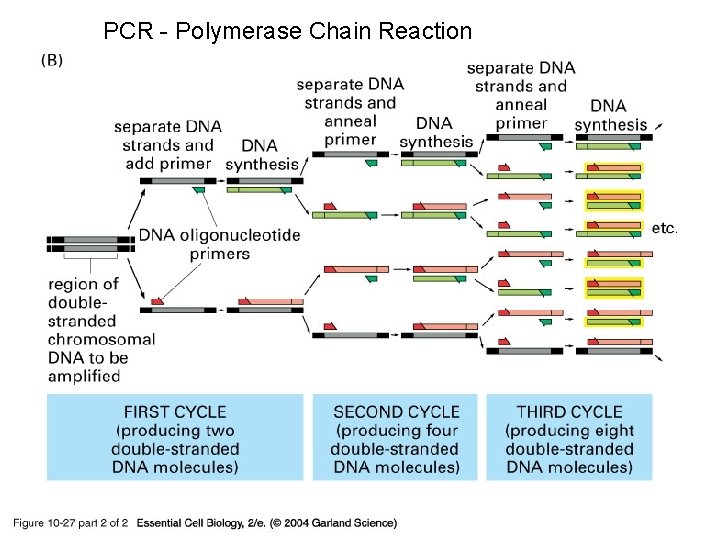
PCR - Polymerase Chain Reaction 10_27_2_PCR_amplify. jpg

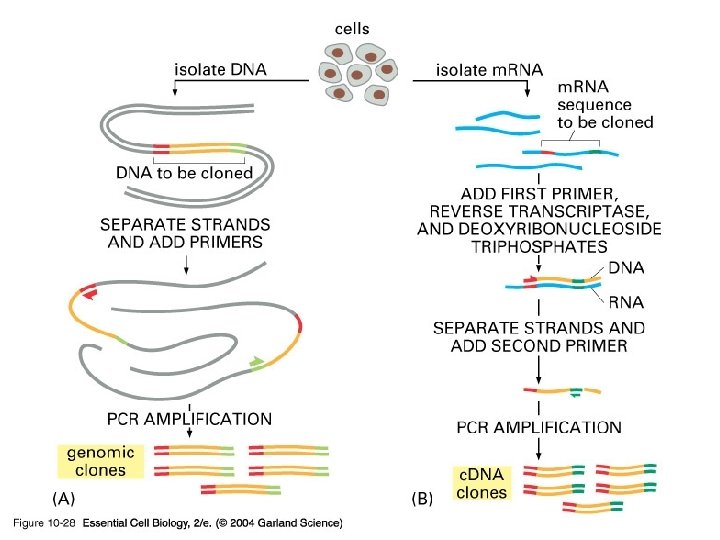
10_28_PCR_clones. jpg

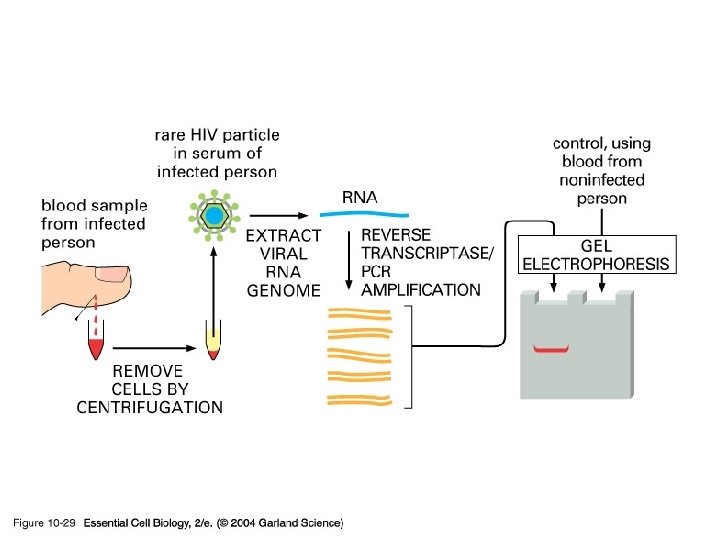
10_29_PCR_viral. jpg

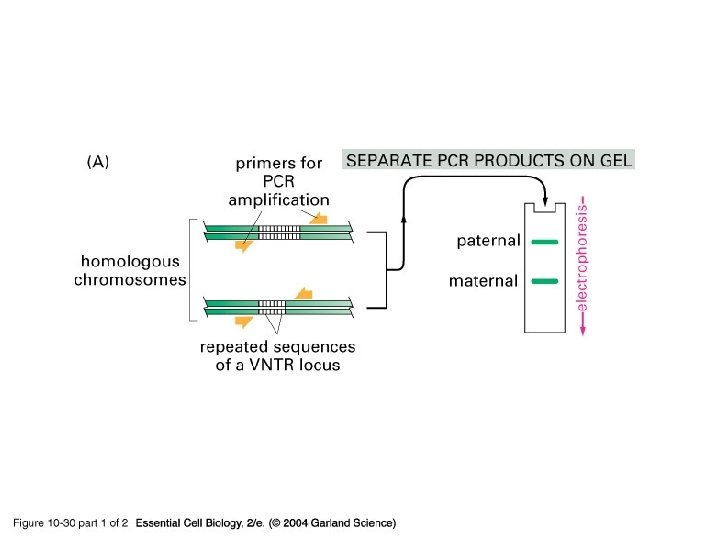
10_30_1_PCR_forensic. jpg

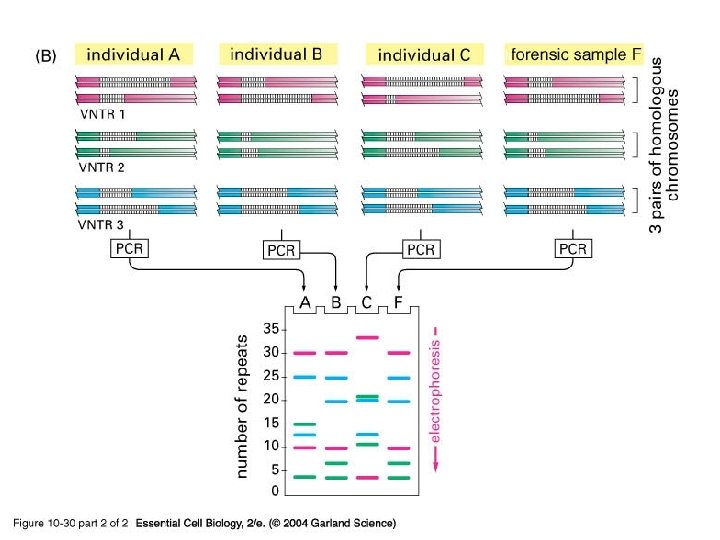
10_30_2_PCR_forensic. jpg

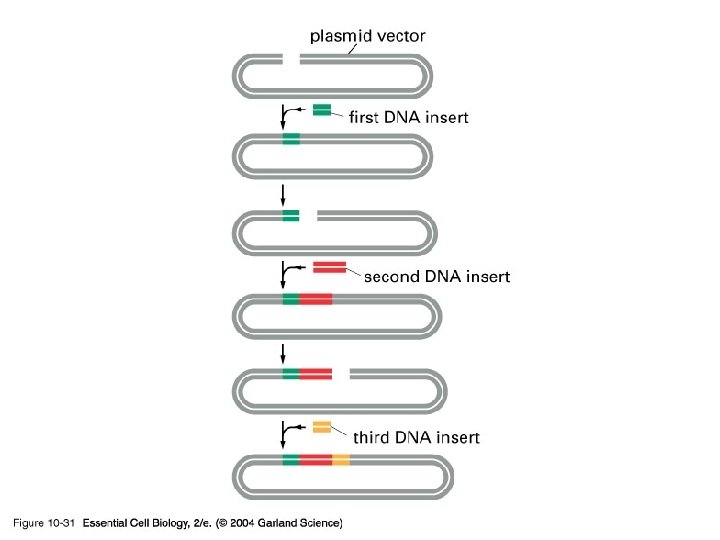
10_31_Serial. DNA. clone. jpg

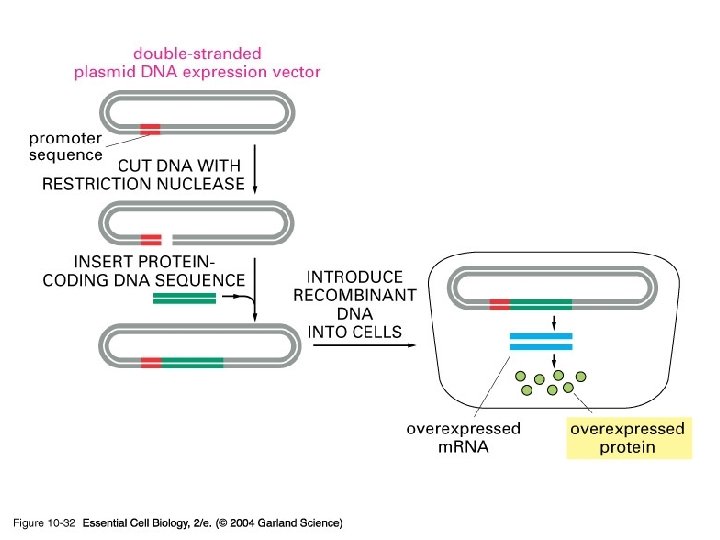
10_32_expressionvector. jpg

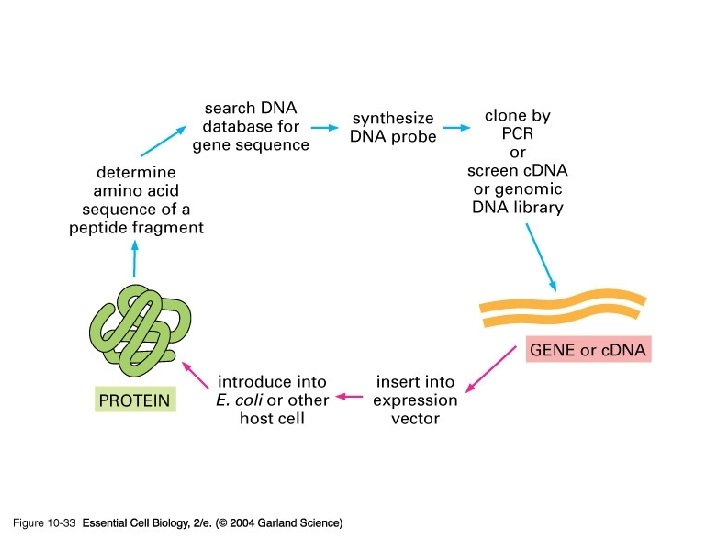
10_33_gene_protein. jpg

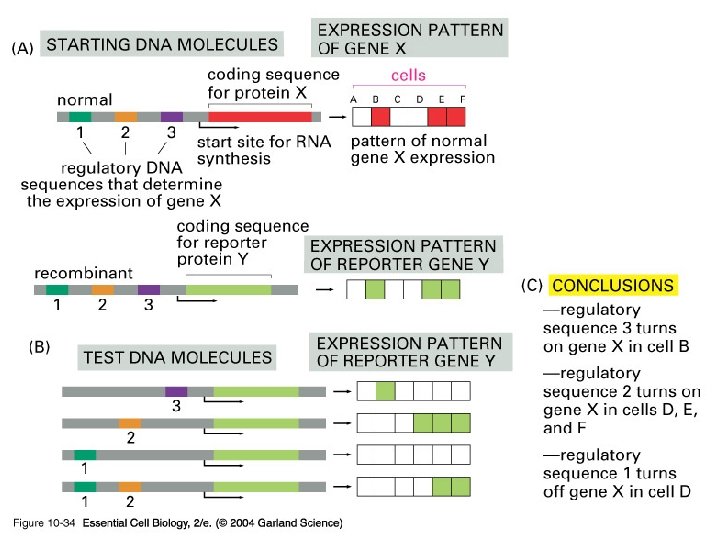
10_34_Reporter. genes. jpg

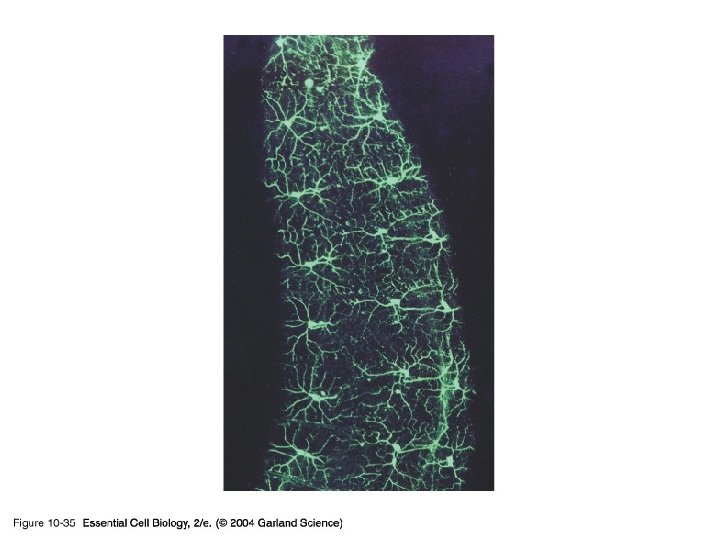
10_35_GFP. jpg

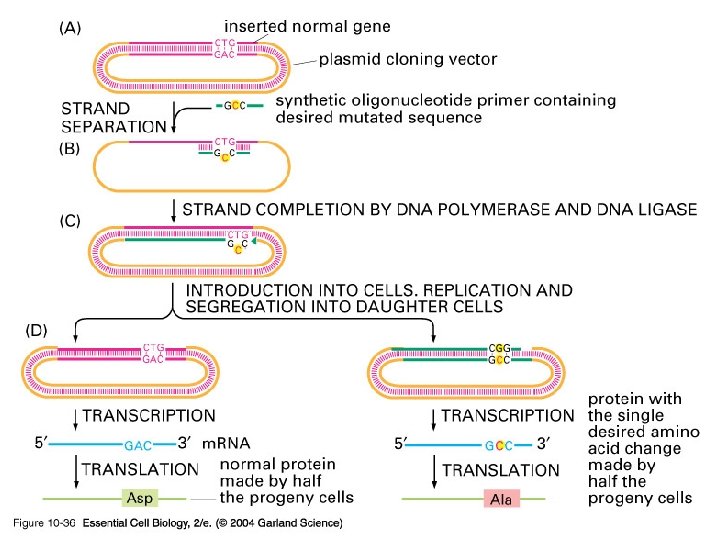
10_36_mutagenesis. jpg

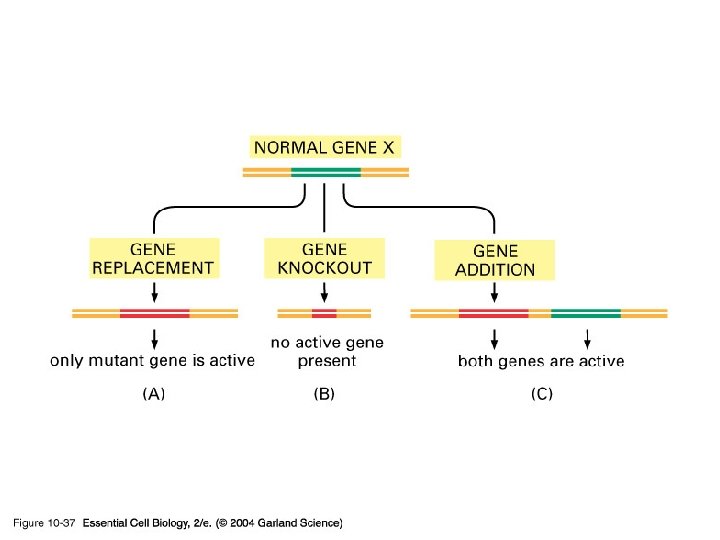
10_37_engineered. org. jpg

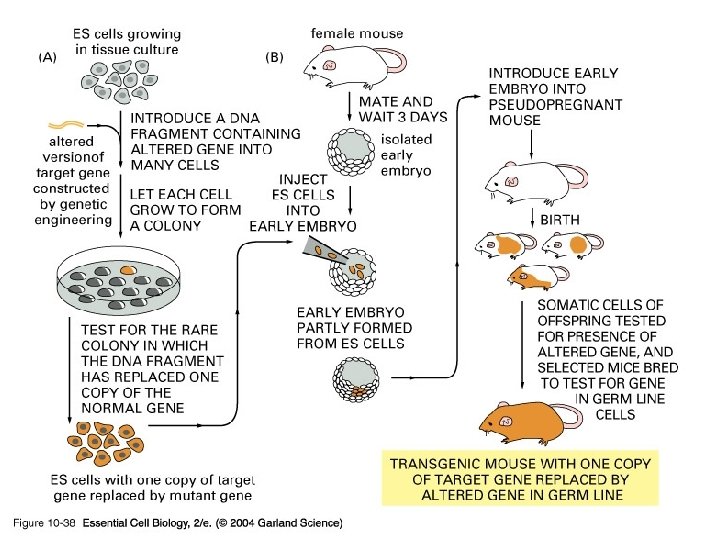
10_38_ES. cells. jpg

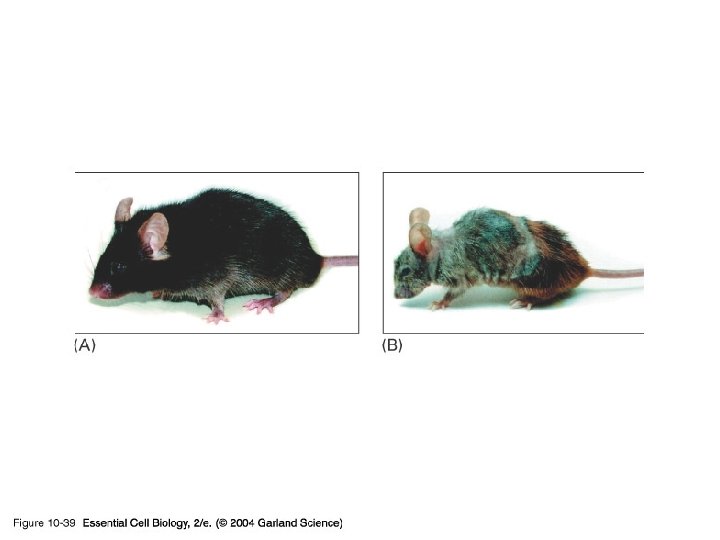
10_39_Transgenic. mice. jpg

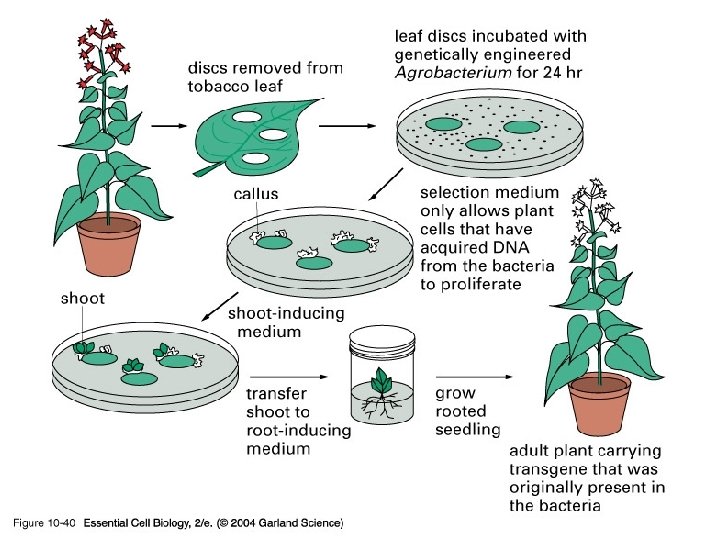
10_40_Transgenic. plant. jpg