Transgenic Development Plant Genetic Engineering Genetic Engineering The



























- Slides: 68

Transgenic Development (Plant Genetic Engineering)

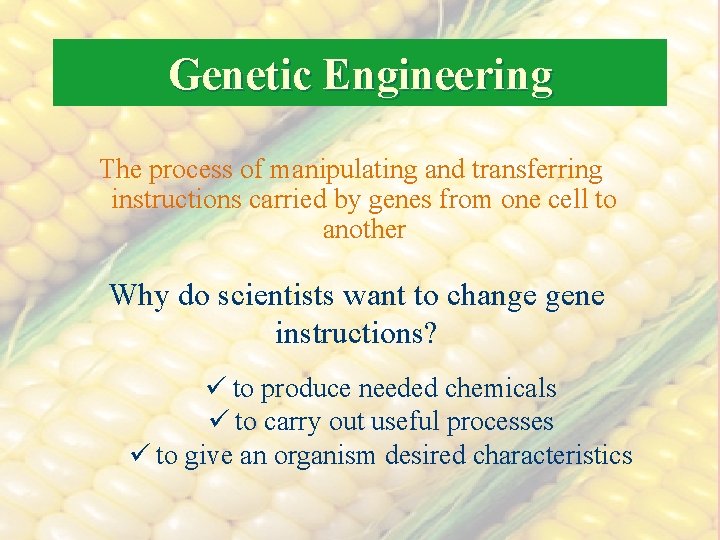
Genetic Engineering The process of manipulating and transferring instructions carried by genes from one cell to another Why do scientists want to change gene instructions? ü to produce needed chemicals ü to carry out useful processes ü to give an organism desired characteristics

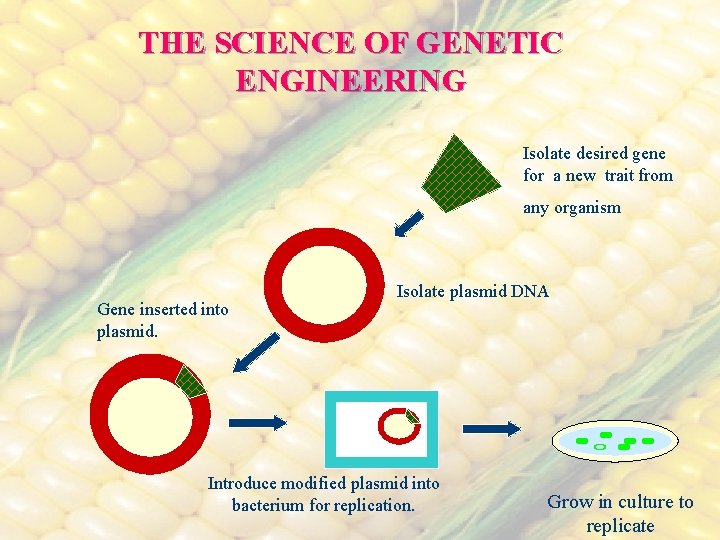
THE SCIENCE OF GENETIC ENGINEERING Isolate desired gene for a new trait from any organism Gene inserted into plasmid. Isolate plasmid DNA Introduce modified plasmid into bacterium for replication. Grow in culture to replicate


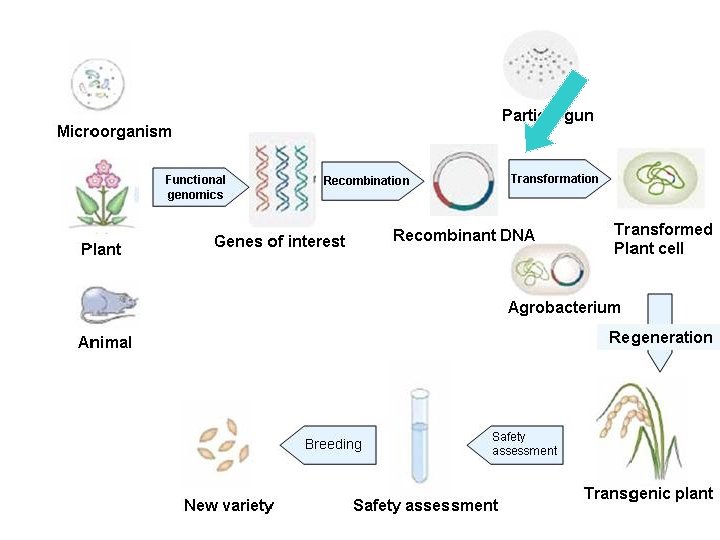
Plant transformation ügetting DNA into a cell ügetting it stably integrated ügetting a plant back from the cell Requirement 1. a suitable transformation method 2. a means of screening for transformants 3. an efficient regeneration system 4. genes/constructs vectors reporter genes ‘genes of interest’ Promoter/terminator selectable marker genes

Transformation technique n Biological. • Agrobacterium mediated transformation. n Mechanical. • • • n Particle bombardment. Electroporation. Microinjection. Chemical. • Polyethylene glycol.

Transformation methods DNA must be introduced into plant cells Indirect Agrobacterium tumefaciens Direct 1. Microprojectile bombardment 2. Electroporation 3. Microinjection Method depends on plant type, cost, application

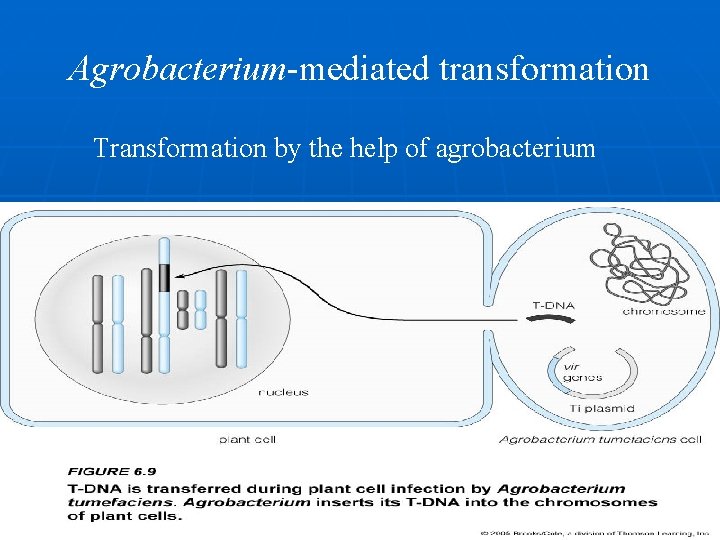
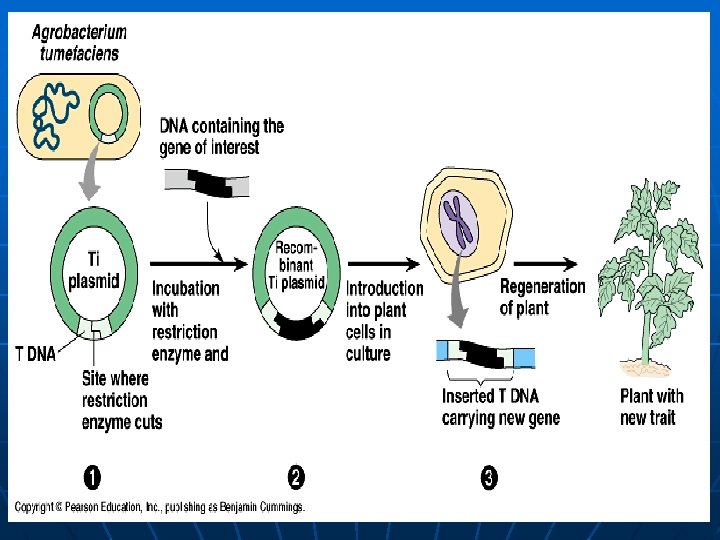
Agrobacterium-mediated transformation Transformation by the help of agrobacterium Agrobacterium is a ‘natural genetic engineer’ i. e. it transfers some of its DNA to plants

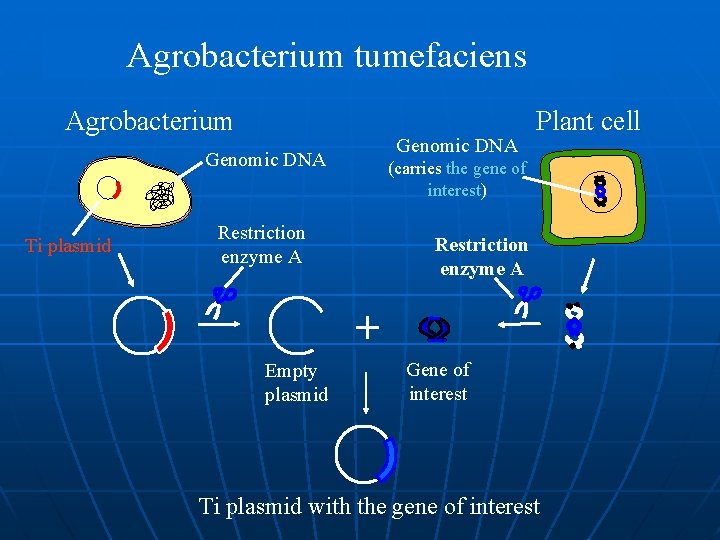
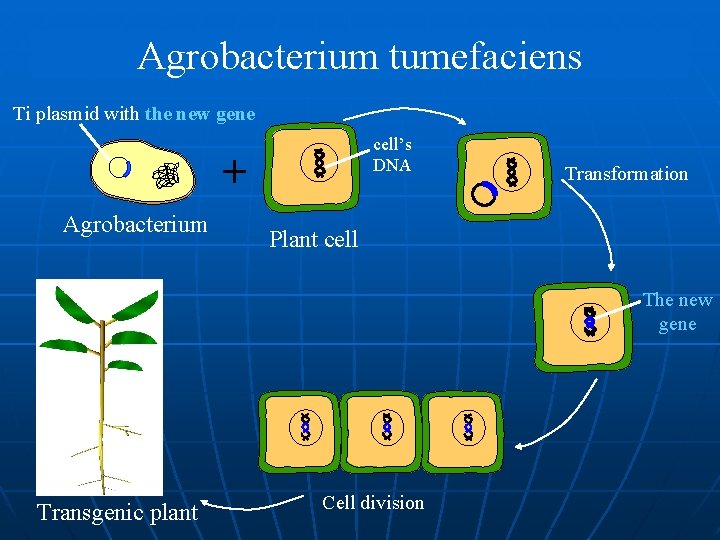
Agrobacterium tumefaciens Agrobacterium Genomic DNA Ti plasmid Plant cell (carries the gene of interest) interest Restriction enzyme A + Empty plasmid Gene of interest Ti plasmid with the gene of interest

Agrobacterium tumefaciens Ti plasmid with the new gene cell’s DNA + Agrobacterium Transformation Plant cell The new gene Transgenic plant Cell division

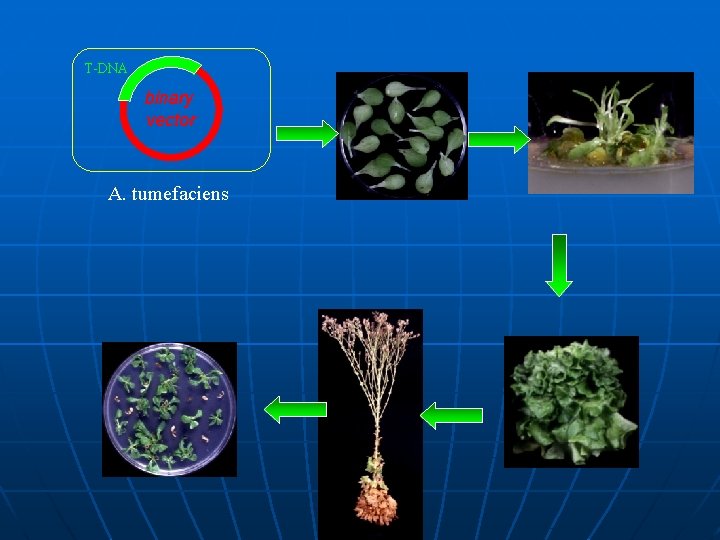
T-DNA binary vector A. tumefaciens

Success Factor Species n Genotypes n Explant Agrobacterium strains n Plasmid n n

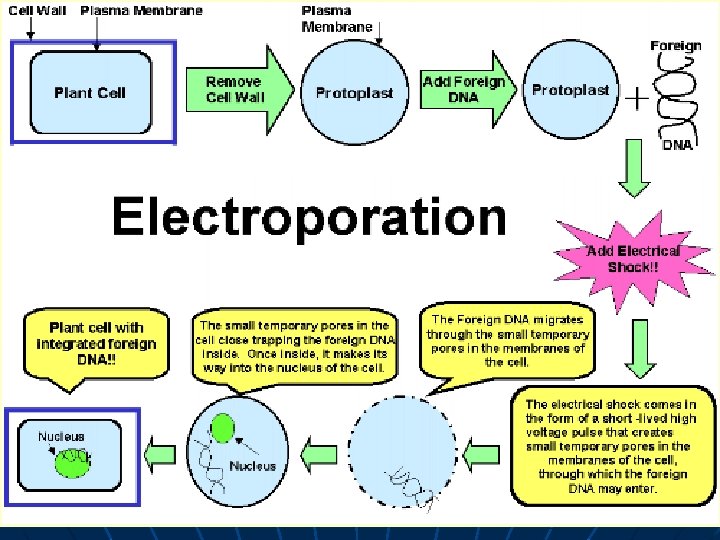
Direct gene transfer Introducing gene directly to the target cell 1. Electroporation 2. Microinjection 3. Particle Bombardment

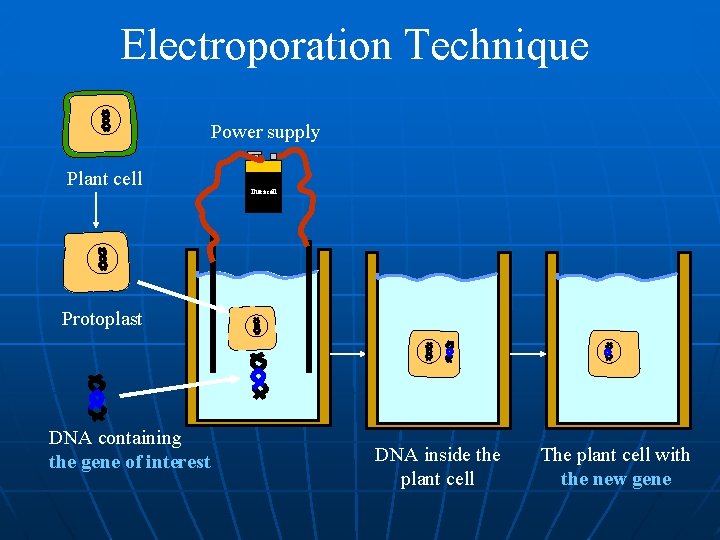
Electroporation ü Explants: cells and protoplasts ü Most direct way to introduce foreign DNA into the nucleus ü Achieved by electromechanically operated devices ü Transformation frequency is high


Electroporation Technique Power supply Plant cell Duracell Protoplast DNA containing the gene of interest DNA inside the plant cell The plant cell with the new gene

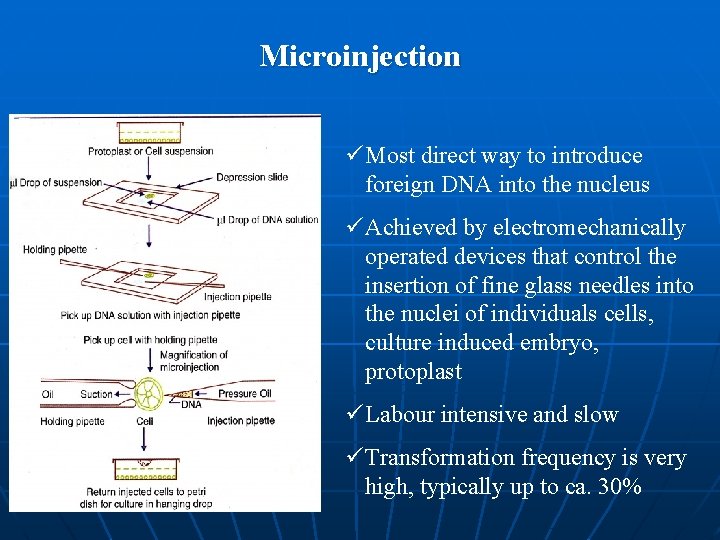
Microinjection ü Most direct way to introduce foreign DNA into the nucleus ü Achieved by electromechanically operated devices that control the insertion of fine glass needles into the nuclei of individuals cells, culture induced embryo, protoplast ü Labour intensive and slow ü Transformation frequency is very high, typically up to ca. 30%

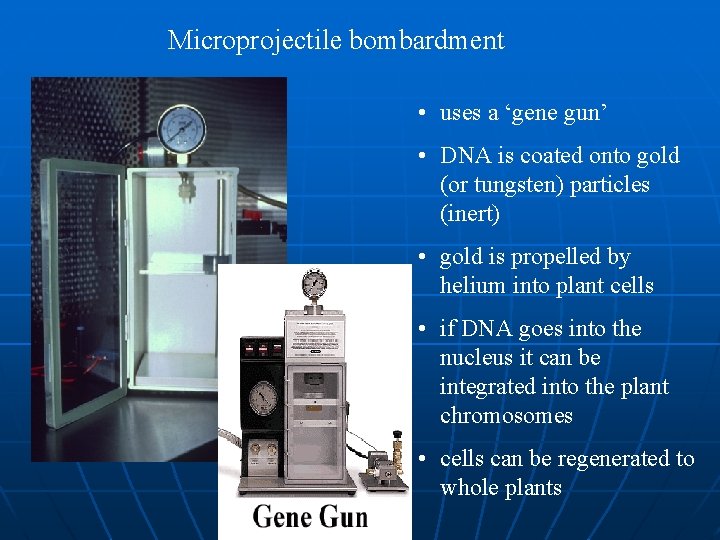
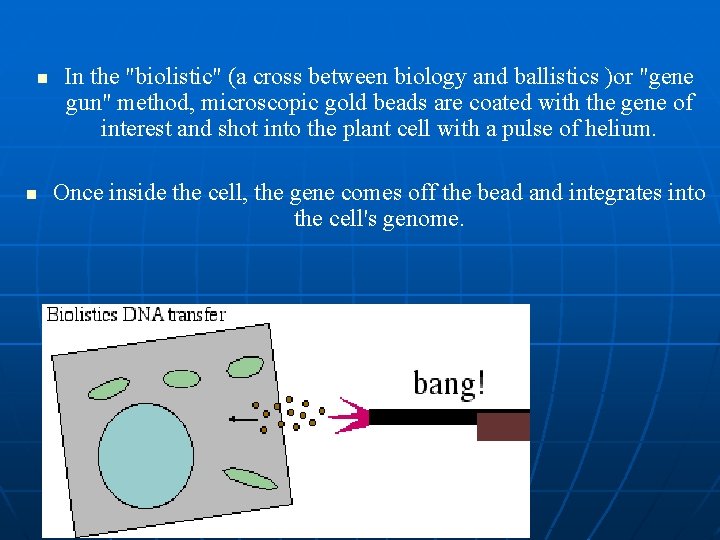
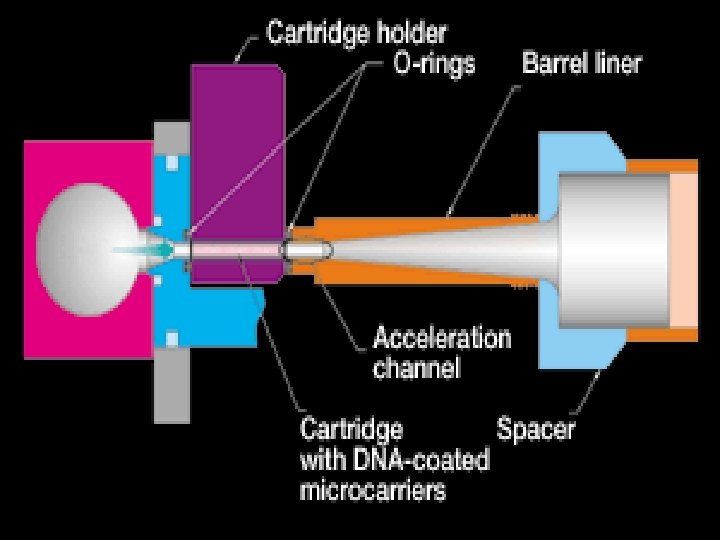
Microprojectile bombardment • uses a ‘gene gun’ • DNA is coated onto gold (or tungsten) particles (inert) • gold is propelled by helium into plant cells • if DNA goes into the nucleus it can be integrated into the plant chromosomes • cells can be regenerated to whole plants

n n In the "biolistic" (a cross between biology and ballistics )or "gene gun" method, microscopic gold beads are coated with the gene of interest and shot into the plant cell with a pulse of helium. Once inside the cell, the gene comes off the bead and integrates into the cell's genome.

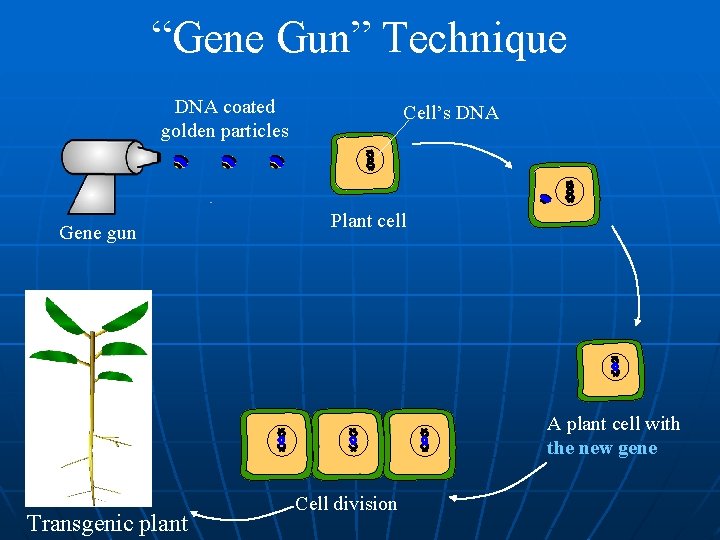
“Gene Gun” Technique DNA coated golden particles Gene gun Cell’s DNA Plant cell A plant cell with the new gene Transgenic plant Cell division


Model from Bio. Rad: Biorad's Helios Gene Gun

In Planta Transformation ♣ ♣ ♣ Meristem transformation Floral dip method Pollen transformation

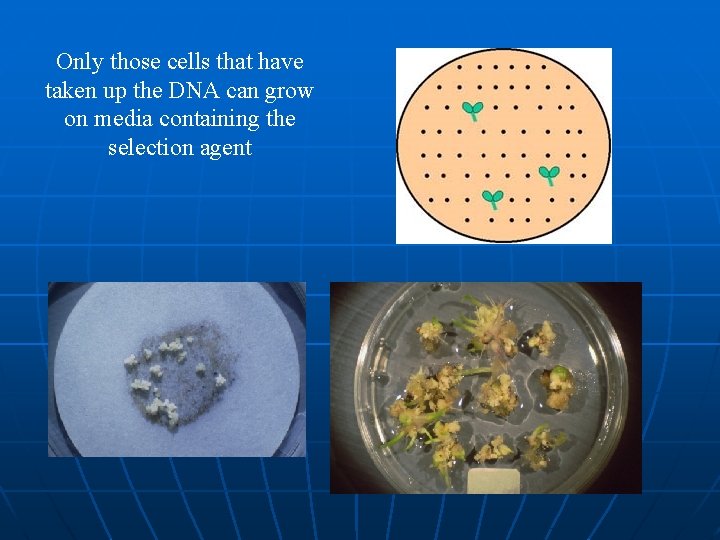
Screening technique Technique which is exploited to screen the transformation product (transformant Cell) Reason: There are many thousands of cells in a leaf disc or callus clump only a proportion of these will have taken up the DNA, therefore can get hundreds of plants back - maybe only 1% will be transformed

Screening (selection) ü ü Select at the level of the intact plant Select in culture • single cell is selection unit • possible to plate up to 1, 000 cells on a Petridish. • Progressive selection over a number of phases

Selection Strategies ü ü ü Positive Negative Visual Selectable marker gene Reporter gene

Positive selection ü Only individuals with characters satisfying the breeders are selected from population to be used as parents of the next generation ü Seed from selected individuals are mixed, then progenies are grown together ü ü ü Add into medium a toxic compound e. g. antibiotic, herbicide Only those cells able to grow in the presence of the selective agent give colonies Plate out and pick off growing colonies. Possible to select one colony from millions of plated cells in a days work. Need a strong selection pressure - get escapes

Negative selection ü The most primitive and least widely used method which can lead to improvement only in exceptional cases ü It implies culling out of all poorly developed and less productive individuals in a population whose productivity is to be genetically improved n n Add in an agent that kills dividing cells Plate out leave for a suitable time, wash out agent then put on growth medium. All cells growing on selective agent will die leaving only nongrowing cells to now grow. Useful for selecting auxotrophs.

Positive and Visual Selection

Regeneration System How do we get plants back from cells? We use tissue culture techniques to regenerate whole plants from single cells Getting a plant back from a single cell is important so that every cell has the new DNA

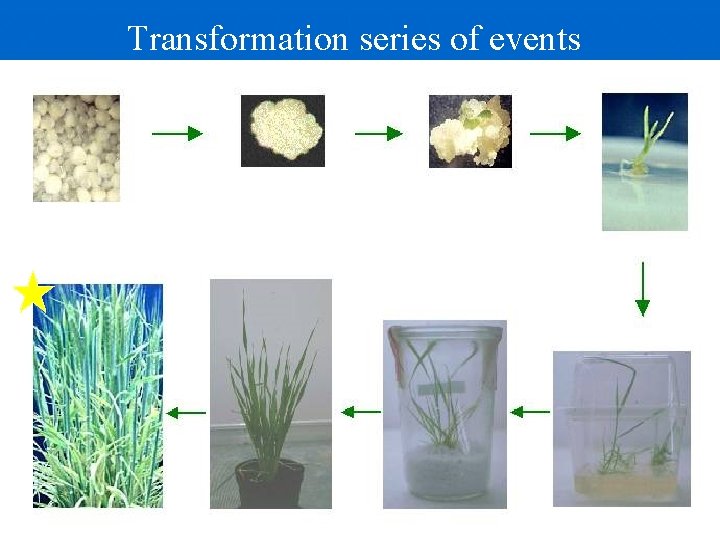
Transformation series of events Transform individual cells Callus formation Remove from sterile conditions Auxins Cytokinins

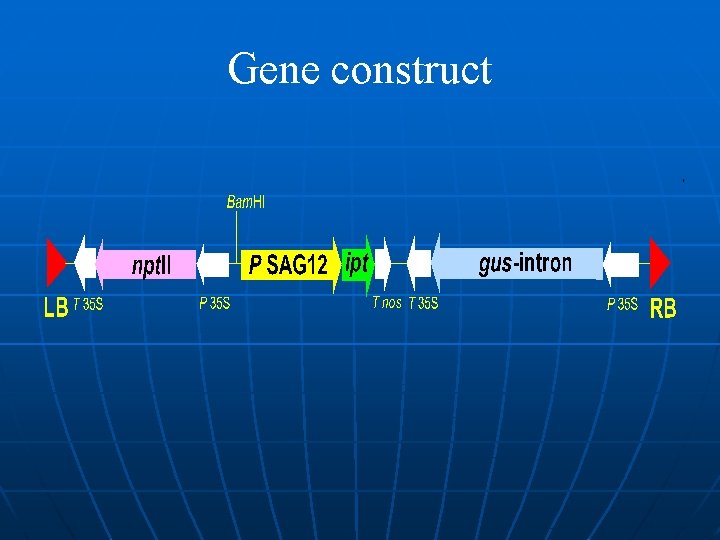
Gene construct

Gene construct Vectors Promoter/terminator Reporter genes Selectable marker genes ‘Genes of interest’.

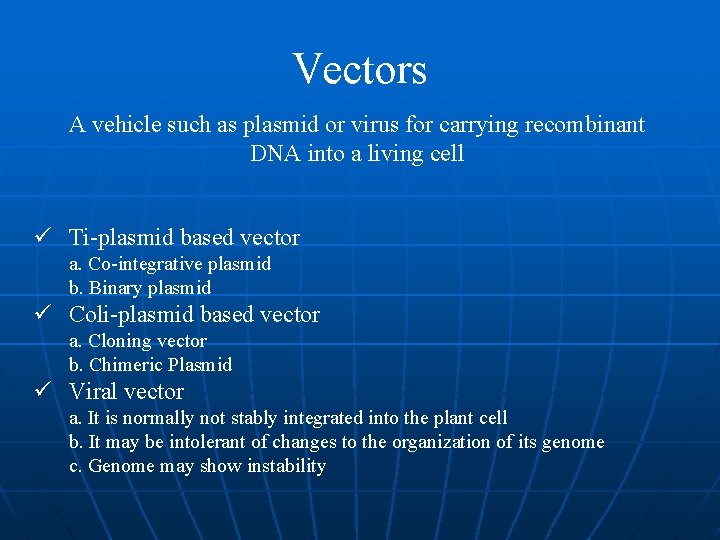
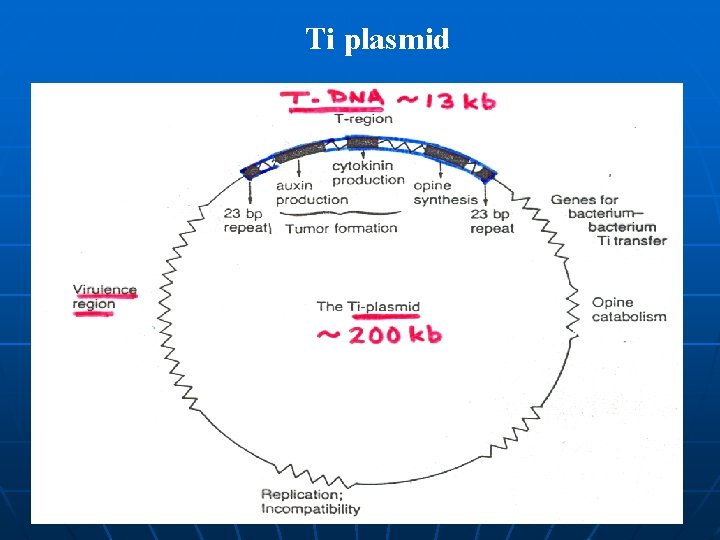
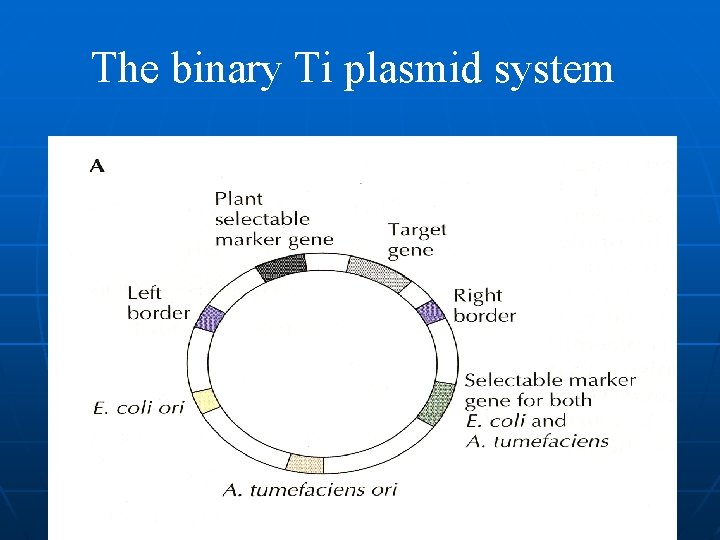
Vectors A vehicle such as plasmid or virus for carrying recombinant DNA into a living cell ü Ti-plasmid based vector a. Co-integrative plasmid b. Binary plasmid ü Coli-plasmid based vector a. Cloning vector b. Chimeric Plasmid ü Viral vector a. It is normally not stably integrated into the plant cell b. It may be intolerant of changes to the organization of its genome c. Genome may show instability

Ti plasmid

The binary Ti plasmid system

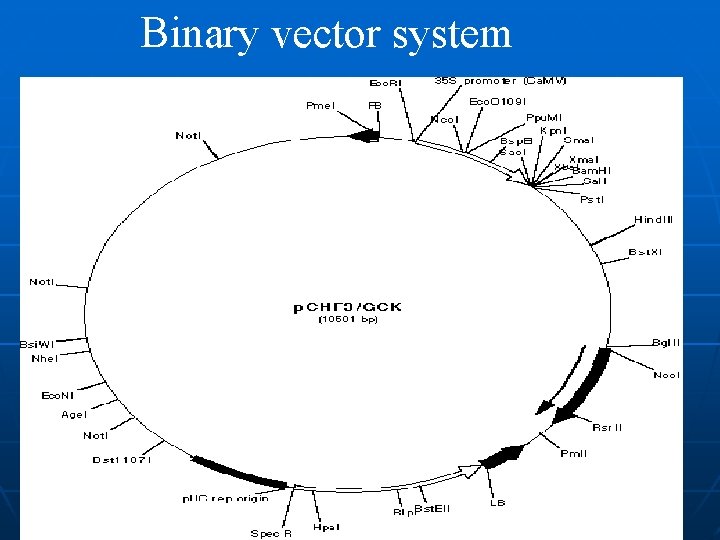
Binary vector system

Binary vector system

Promoter 1. 2. 3. 4. 5. 6. A nucleotide sequence within an operon Lying in front of the structural gene or genes Serves as a recognition site and point of attachment for the RNA polymerase It is starting point for transcription of the structural genes It contains many elements which are involved in producing specific pattern and level of expression It can be derived from pathogen, virus, plants themselves, artificial promoter

Types of Promoter ü Promoter always expressed in most tissue (constitutive) -. 35 s promoter from Ca. MV Virus -. Nos, Ocs and Mas Promoter from bacteria -. Actin promoter from monocot -. Ubiquitin promoter from monocot -. Adh 1 promoter from monocot -. p. EMU promoter from monocot ü Tissue specific promoter -. Haesa promoter -. Agl 12 promoter ü ü Inducible promoter -. Aux promoter Artificial promoter -. Mac promoter (Mas and 35 s promoter)

Reporter gene Easy to visualise or assay - ß-glucuronidase (GUS) (E. coli) -green fluorescent protein (GFP) (jellyfish) - luciferase (firefly)

GUS The Uid. A gene encoding activity is commonly used. Gives a blue colour from a colourless substrate (X-glu) for a qualitative assay. Also causes fluorescence from Methyl Umbelliferyl Glucuronide (MUG) for a quantitative assay. Cells that are transformed with GUS will form a blue precipitate when tissue is soaked in the GUS substrate and incubated at 37 o. C this is a destructive assay (cells die)

5 - - glucuronidase Genes n n very stable enzyme cleaves -D glucuronide linkage simple biochemical reaction • It must take care to stay in linear range detection sensitivity depends on substrate used in enzymatic assay (fast) • colorimetric and fluorescent substrates available

5 - -glucuronidase Genes n n n Advantages • low background • can require little equipment (spectrophotometer) • stable enzyme at 37ºC Disadvantages • sensitive assays require expensive substrates or considerable equipment • stability of the enzyme makes it a poor choice for reporter in transient transfections (high background = low dynamic range) Primary applications • typically used in transgenic plants with X-gus colorimetric reporter

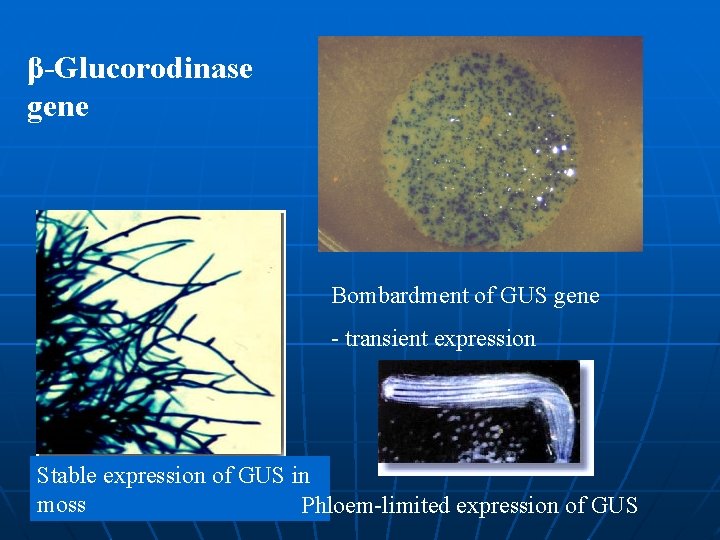
β-Glucorodinase gene Bombardment of GUS gene - transient expression Stable expression of GUS in moss Phloem-limited expression of GUS

GFP (Green Fluorescent Protein) GFP glows bright green when irradiated by blue or UV light This is a non destructive assay so the same cells can be monitored all the way through ü It fluoresces green under UV illumination ü It has been used for selection on its own

Green fluorescent protein (GFP) ü ü Source is bioluminescent jellyfish Aequora victoria ü GFP is an intermediate in the bioluminescent reaction Absorbs UV (~360 nm) and emits visible light. ü has been engineered to produce many different colors (green, blue, yellow, red) ü These are useful in fluorescent resonance energy transfer experiments Simply express in target cells and detect with fluorometer or fluorescence microscope Sensitivity is low ü GFP is non catalytic, 1 M concentration in cells is required to exceed auto-fluorescence

Green fluorescent protein (GFP) n n n Advantages • can detect in living cells • inexpensive (no substrate) Disadvantages • low sensitivity and dynamic range • equipment requirements Primary applications • lineage tracer and reporter in transgenic embryos

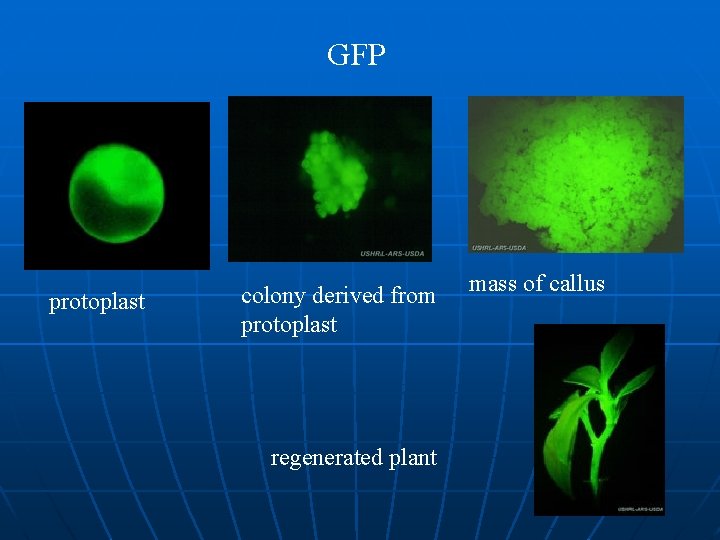
GFP protoplast colony derived from protoplast regenerated plant mass of callus

Luciferase n n n luc gene encodes an enzyme that is responsible for bioluminescence in the firefly. This is one of the few examples of a bioluminescent reaction that only requires enzyme, substrate and ATP. Rapid and simple biochemical assay. Read in minutes Two phases to the reaction, flash and glow. These can be used to design different types of assays. • Addition of substrates and ATP causes a flash of light that decays after a few seconds when [ATP] drops • after the flash, a stable, less intense “glow” reaction continues for many hours - AMP is responsible for this

Luciferase n n flash reaction is ~20 x more sensitive than glow reaction is more stable • allows use of scintillation counter • no injection of substrates required • potential for simple automation in microplate format

Luciferase n Advantages • large dynamic range up to 7 decades, depending on instrument and chemistry • rapid, suitable for automation • instability of luciferase at 37 °C (1/2 life of <1 hr) • inexpensive • widely used n disadvantages • Equipment requirement • luminometer (very big differences between models) • liquid scintillation counter (photon counter)

Selectable Marker Gene which confer tolerance to a phytotoxic substance Most common: 1. antibiotic resistance kanamycin (geneticin), hygromycin Kanamycin arrest bacterial cell growth by blocking various steps in protein synthesis 2. herbicide resistance phosphinothricin (bialapos); glyphosate

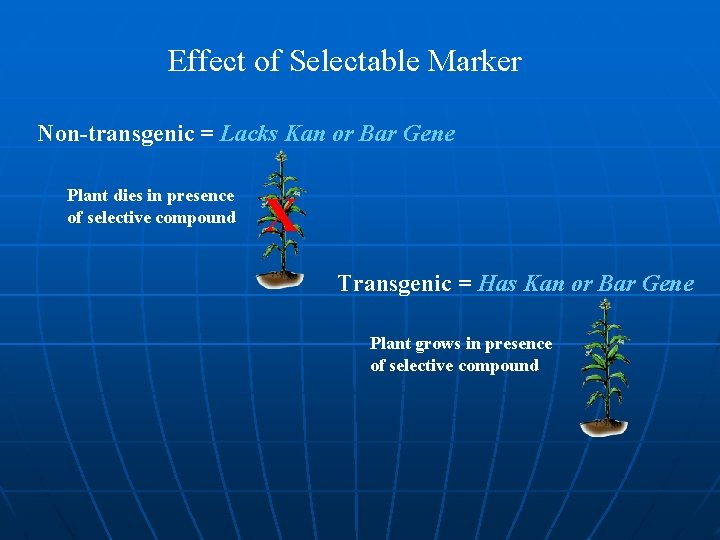
Effect of Selectable Marker Non-transgenic = Lacks Kan or Bar Gene Plant dies in presence of selective compound X Transgenic = Has Kan or Bar Gene Plant grows in presence of selective compound

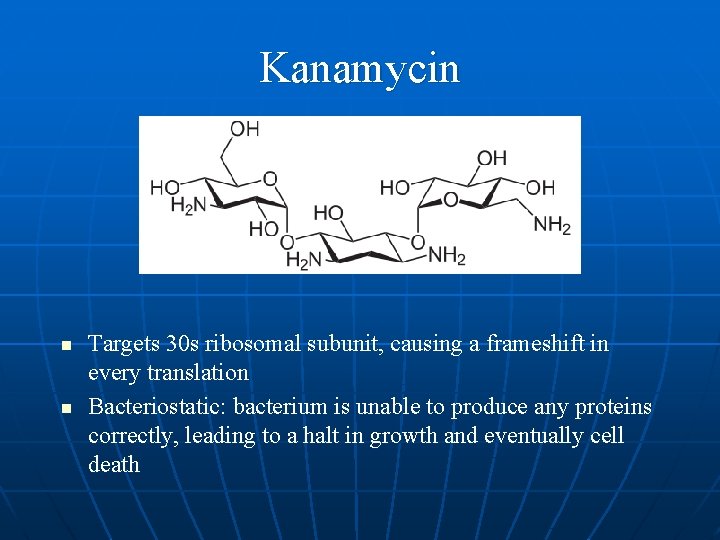
Kanamycin n n Targets 30 s ribosomal subunit, causing a frameshift in every translation Bacteriostatic: bacterium is unable to produce any proteins correctly, leading to a halt in growth and eventually cell death

Kanamycin use/resistance n n n Over-use of kanamycin has led to many wild bacteria possessing resistance plasmids As a result of this (as well as a lot of side effects in humans), kanamycin is widely used for genetic purposes rather than medicinal purposes, especially in transgenic plants Resistance is often to a family of related antibiotics, and can include antibiotic-degrading enzymes or proteins protecting the 30 s subunit

G 418 -Gentamycin n source: aminoglycoside antibiotic related to gentamycin n activity: broad action against prokaryotic and eukaryotic cells • inhibits protein synthesis by blocking initiation n resistance - bacterial neo gene (neomycin phosphotransferase, encoded by Tn 5 encodes resistance to kanamycin, neomycin, G 418 • but also cross protects against bleomycin and relatives.

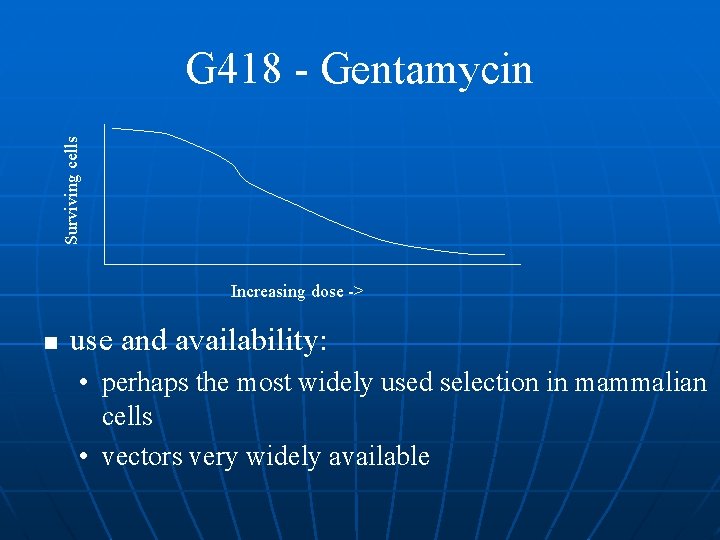
G 418 - Gentamycin n Stability: • 6 months frozen n selection conditions: • E. coli: 5 g/ml • Eukaryotic cells: 300 -1000 g/ml. G 418 requires careful optimization for cell types and lot to lot variations n Kill curves required n It requires at least seven days to obtain resistant colonies, two weeks is more typical n

Surviving cells G 418 - Gentamycin Increasing dose -> n use and availability: • perhaps the most widely used selection in mammalian cells • vectors very widely available

Hygromycin n n source: aminoglycoside antibiotic from Streptomyces hygroscopicus. Activity: kills bacteria, fungi and higher eukaryotic cells by inhibiting protein synthesis • interferes with translocation causing misreading of m. RNA n resistance: conferred by the bacterial gene hph • no cross resistance with other selective antibiotics

Hygromycin n stability: • one year at 4 ºC, 1 month at 37 ºC n selection conditions: • E. coli: 50 g/ml • Eukaryotic cell lines: n 50 - 1000 g/ml (must be optimized) n 10 days- 3 weeks required to generate effect n use and availability: • vectors containing hygromycin resistance gene are widely available • in use for many years

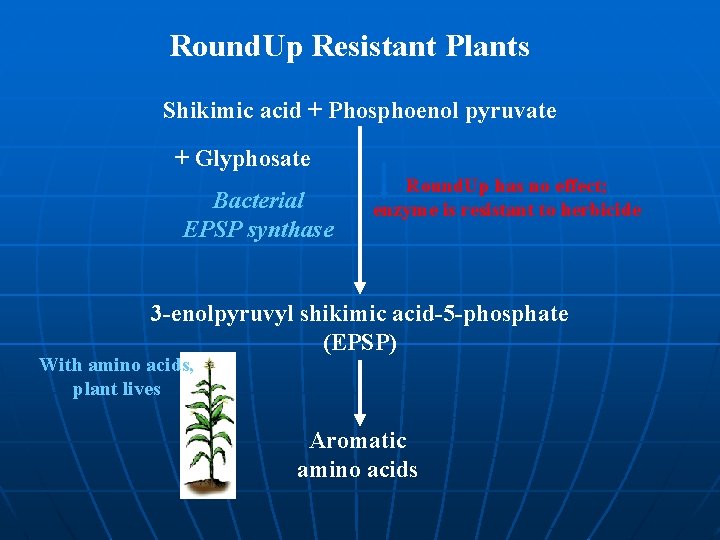
Glyphosate resistance üGlyphosate = “Roundup”, “Tumbleweed” = Systemic herbicide üGlyphosate inhibits EPSP synthase (Senolpyruvlshikimate-3 phosphate – involved in chloroplast amino acid synthesis) üEscherichia coli EPSP synthase = mutant form less sensitive to glyphosate n Cloned via Ti plasmid into soybeans, tobacco, petunias • Increased crop yields of crops treated with herbicides

Round. Up Sensitive Plants Shikimic acid + Phosphoenol pyruvate + Glyphosate X Plant EPSP synthase X 3 -Enolpyruvyl shikimic acid-5 -phosphate (EPSP) Without amino acids, plant dies X X Aromatic amino acids

Round. Up Resistant Plants Shikimic acid + Phosphoenol pyruvate + Glyphosate Bacterial EPSP synthase Round. Up has no effect; enzyme is resistant to herbicide 3 -enolpyruvyl shikimic acid-5 -phosphate (EPSP) With amino acids, plant lives Aromatic amino acids

Bialaphos n n Glufosinate – active substance of a broad-spectrumherbicide = synthetical copy of the aminoacid phosphinothricin produced by Streptomyces viridochomogenes Inhibit glutamine-synthetase (important enzyme in nitrogen-cycle of plants) caused plant dies Herbicide-tolerance is reached by gene-transfer from the bacterium to the plant The transfered gene encodes for the enzyme phophinothricin-acetyl-transferase degrade glufosinate

Bialaphos *Bialaphos (Phosphinothricin-alanyl-alanine) is an herbicide that inhibits a key enzyme in the nitrogen assimilation pathway, glutamine synthetase, leading to accumulation of toxic levels of ammonia in both bacteria and plant cells

Only those cells that have taken up the DNA can grow on media containing the selection agent
