Gene Expression Analysis using Microarrays Anne R Haake

















- Slides: 39

Gene Expression Analysis using Microarrays Anne R. Haake, Ph. D.

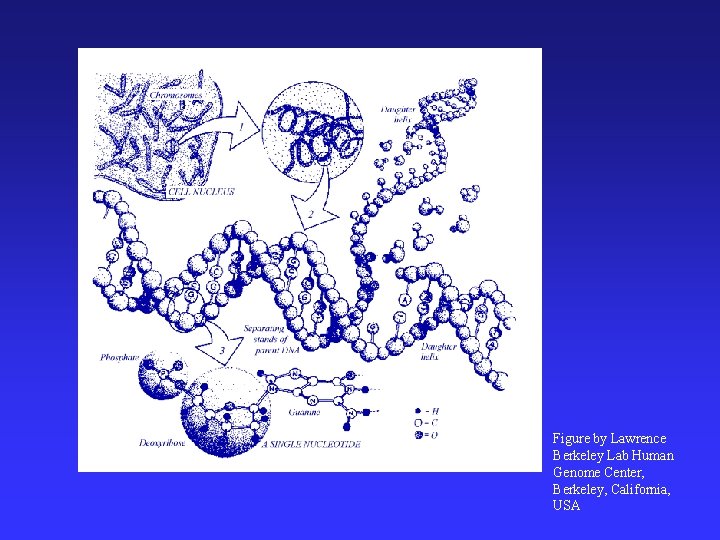
Figure by Lawrence Berkeley Lab Human Genome Center, Berkeley, California, USA

Post-Genomic Age ? A switch in focus from sequencing to understanding how genomes function

How do we relate gene identity to cell physiology, disease & drug discovery? Functional Genomics =“development and application of global (genome -wide or system-wide) experimental approaches to assess gene function by making use of the information and reagents provided by structural genomics”

Gene Expression Analysis • • What is gene expression? What can we learn from expression analysis? How is the analysis accomplished? What are the challenges for bioinformatics?

Gene Expression Flow of Information

• Individual cells in an organism have the same genes (DNA) but…. • It is the expression of thousands of genes and their products (RNA, proteins), functioning in a complicated and orchestrated way, that make that organism what it is.

Differential Gene Expression A Few Examples: • Cell type specific -e. g. skin cell vs. brain cell • Developmental stage -e. g. embryonic skin cell vs. adult skin cell • Disease state -e. g. normal skin cell vs. skin tumor cell • Environment-specific -e. g. skin cell untreated vs. treated drugs, toxins

What can we learn by analyzing complex patterns of gene expression? • Classifications: for diagnosis, prediction… Cell-type, stage-specific, disease-related, treatment-related patterns of gene expression? • Gene Networks/Pathways: Functional roles of genes in cellular processes? Gene regulation and gene interactions

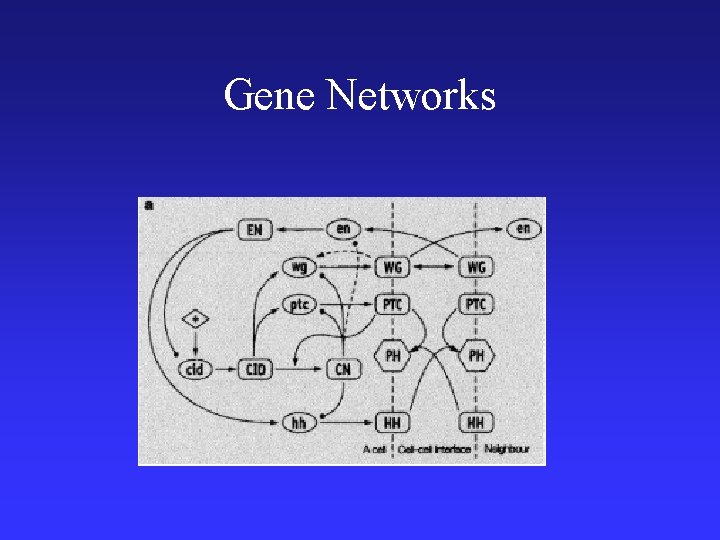
Gene Networks

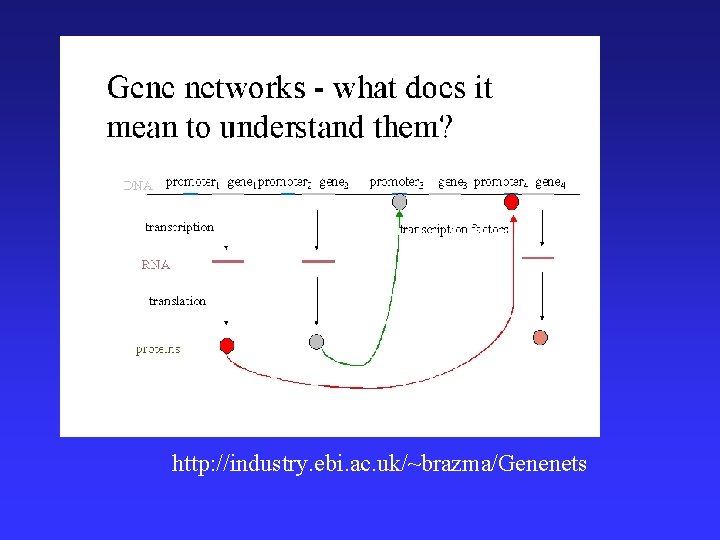
• http: //industry. ebi. ac. uk/~brazma/Genenets

Gene Expression Analysis Need efficient ways to study these complex patterns. 1) Techniques of Biochemistry/Molecular Biology Resolution of the patterns = expression data (RNA or protein) 2) Management of complex data sets 3) Mining of the data to gain useful information

Gene Expression Analysis High-Throughput Techniques • Microarray or Gene Chip = c. DNA arrays or oligo arrays (Affymetrix) • Filter Arrays • Differential Display • SAGE

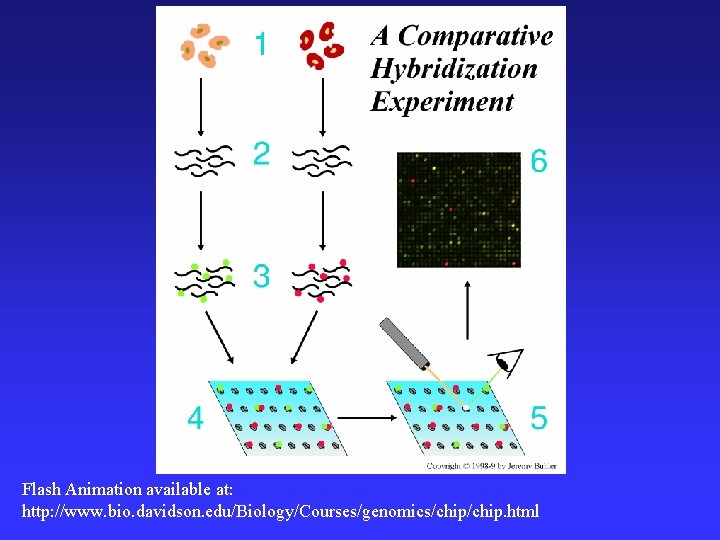
• Gene Chip technology DNA microarrays = hundreds to thousands of different DNA sequences spotted onto glass microscope slide • Compare binding (base-pairing) of two different sets of expressed gene sequences to the template DNA microarray • Allows simultaneous analysis of thousands of genes: Is the gene expressed? At what level? *expression levels are relative

Flash Animation available at: http: //www. bio. davidson. edu/Biology/Courses/genomics/chip. html

The Full Yeast Genome on a Chip 6116 Yeast Genes 96 Intergenic regions + lots of control samples – Primers purchased from Research Genetics • Total spots printed: 707, 520 • Total Arrays: 110 • Actual Time to print: 52 hours • Credits: Dr. Patrick O. Brown laboratory: pbrown@cmgm. stanford. edu

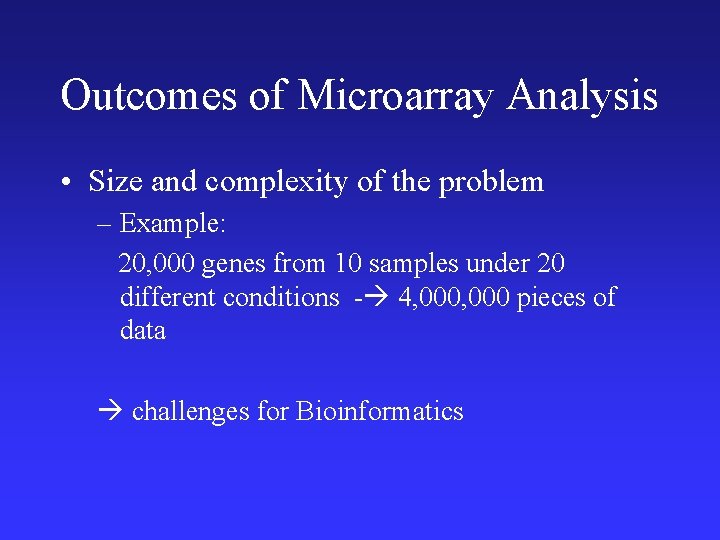
Outcomes of Microarray Analysis • Size and complexity of the problem – Example: 20, 000 genes from 10 samples under 20 different conditions - 4, 000 pieces of data challenges for Bioinformatics

Outcomes of Microarray Analysis • Large, complex data sets • Wide availability of technology large number of distributed databases Current state: data scattered among many independent sites (accessible via Internet) or not publicly available at all.

Current Problems Facing Bioinformatics • Standardization & Quality Control In the Experiments (data quality at several levels) • Management of the Data -Standardization of the databases -Public access to the databases • Information from the Data -Need for data mining algorithms customized for gene expression analysis

Microarray Databases • Need public repository with standardized annotation Issues : - difficulty in describing expression experiments; remember that measurements are relative (complicates comparisons) – Structure of the database itself – Internet-based tools for searching and using semantic context to allow comparisons

Public Microarry Repositories 4 Major Efforts: Gene. X at US National Center for Genome Resources http: //www. ncgr. org/research/genex/ Array. Express at European Bioinformatics Institute http: //www. ebi. ac. uk/arrayexpress/

Public Repositories • Stanford University Database http: //genomewww 4. stanford. edu/Micro. Array/SMD/inde x. html

Public Repositories Gene Expression Omnibus at US National Center for Biotechnology Information Example at: http: //www. ncbi. nlm. nih. gov/geo/query/ acc. cgi? acc=GSM 39

Mining of the Expression Databases • A gene expression pattern derived from a single microarray experiment is simply a snapshot (one experimental sample vs reference) • Usually want to understand a process or changes in expression over a collection of samples gene expression profile Example?

Mining of the Expression Databases General Approaches • Raw data from multiple experiments converted to a gene expression matrix - Rows: Different genes - Columns: Different samples - Numerical values encoded by color (red=positive green=negative blue=n. a. )

Typical approach Look for similarities (or differences) in patterns e. g. Compare rows to find evidence for co-regulation of genes 1) Need ways to measure similarity (distance) among the objects being compared 2) Then, group together objects (genes or samples) with similar properties.

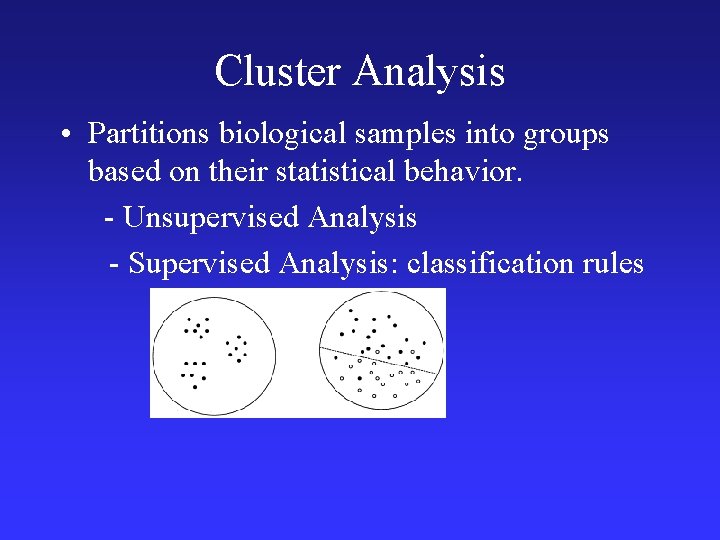
Cluster Analysis • Partitions biological samples into groups based on their statistical behavior. - Unsupervised Analysis - Supervised Analysis: classification rules

Analytic Approaches • Clustering Algorithms – Hierarchical – K-mean – Self-organizing maps – Others

Eisen et al. http: //www. pnas. org/cgi/content/ full/95/25/14863

Success Story Gene Clustering Approach • Yeast genome – Complete set of genes used to study diauxic shift time course – Cluster analysis of data identified group of genes with similar expression profiles – Upstream regulatory sites of these genes compared to identify transcription factor binding sites (see Brazma & Vilo reference)

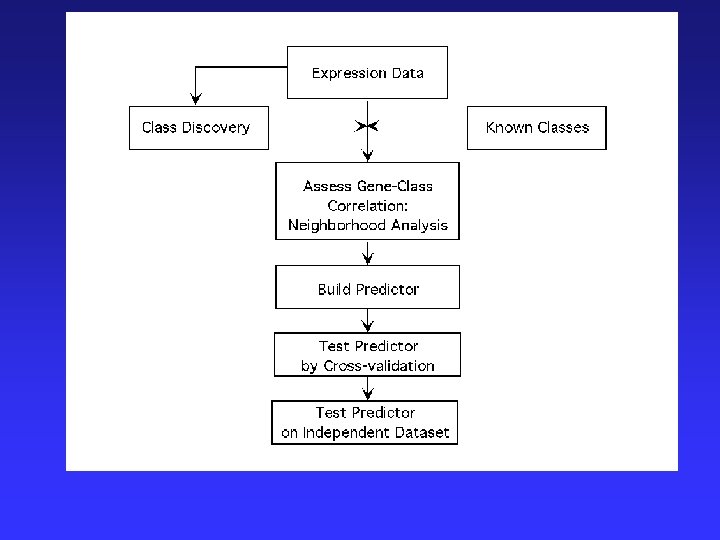
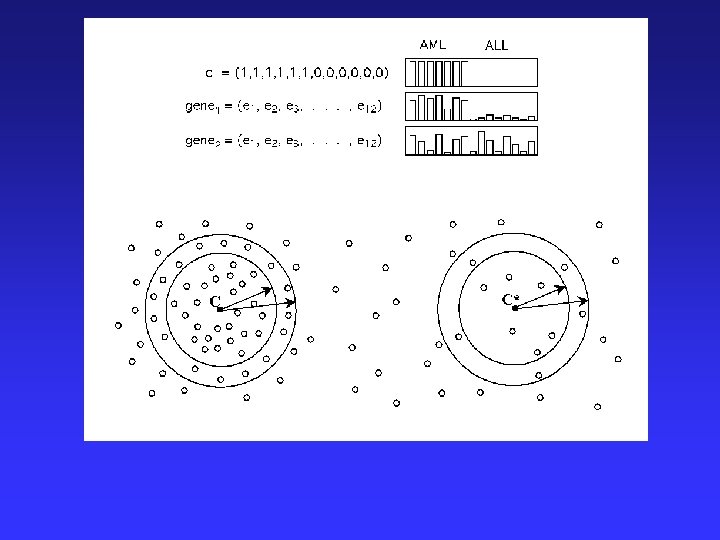
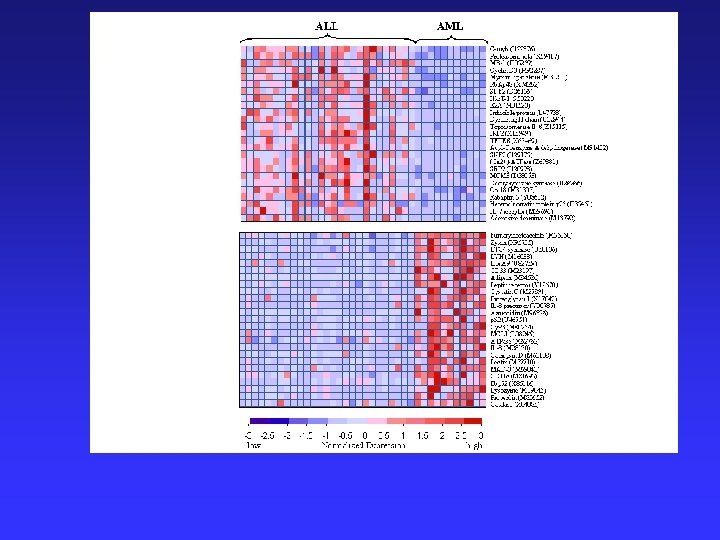
Example-Sample Clustering Classification of cancers – Comparing 2 acute leukemias (AML and ALL) Biological/Clinical Problems: • Previously, no single reliable test to distinguish • Differ greatly in clinical course & response to treatments. http: //waldo. wi. mit. edu/MPR/figures_ALL_AML. html



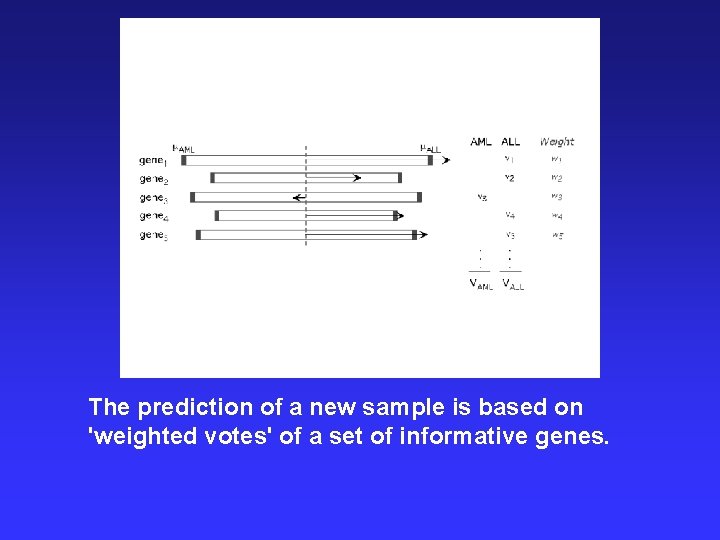
The prediction of a new sample is based on 'weighted votes' of a set of informative genes.

Analytic Approach: 1) Class discovery = classification by clustering of microarray data using tumors of known type Found 1100 of 6817 genes correlated with class distinction 2) Formation of a class predictor = 50 most informative genes class discovery of unknown tumors


Analytic Approaches Limitation of cluster analysis: similarity in expression pattern suggests co-regulation but doesn’t reveal cause-effect relationships • Bayesian Networks – Represent the dependence structure between multiple interacting quantities (e. g. expression levels of genes) – gene interactions & models of causal influence • Others? many

Check the Web: Free Software Available Some useful links: • Expression Profiler http: //ep. ebi. ac. uk/ • Gene. X (NCGR) http: //genex. sourceforge. net/ www. ncgr. org/research/genex/other_tools. html • http: //www. kdnuggets. com/software/suites. html

Additional References: • R. Ekins and F. W. Chu : Microarrays: their origins and applications. Trends in Biotechnology, 17: 217 -218, 1999. • Brazma et al. , One-stop shop for microarray data. Nature 403: 699 – 700, 2000. • Brazma A. and Vilo, J: Minireview. Gene expression data analysis. FEBS Letters, 480: 17 -24, 2000.