DNA Computing Implications for Theoretical Computer Science Lila






![Reality Intrudes • News from the lab bench: [Nirenberg, Matthaei ’ 61] synthesize RNA, Reality Intrudes • News from the lab bench: [Nirenberg, Matthaei ’ 61] synthesize RNA,](https://slidetodoc.com/presentation_image_h2/4f19571aa8ae5913ef10182948859af9/image-10.jpg)



![Biomolecular (DNA) Computing • • • Hamiltonian Path Problem [Adleman, Science, 1994] DNA-based addition Biomolecular (DNA) Computing • • • Hamiltonian Path Problem [Adleman, Science, 1994] DNA-based addition](https://slidetodoc.com/presentation_image_h2/4f19571aa8ae5913ef10182948859af9/image-18.jpg)







- Slides: 56

DNA Computing: Implications for Theoretical Computer Science Lila Kari Dept. of Computer Science University of Western Ontario London, ON, Canada http: //www. csd. uwo. ca/~lila/ lila@csd. uwo. ca

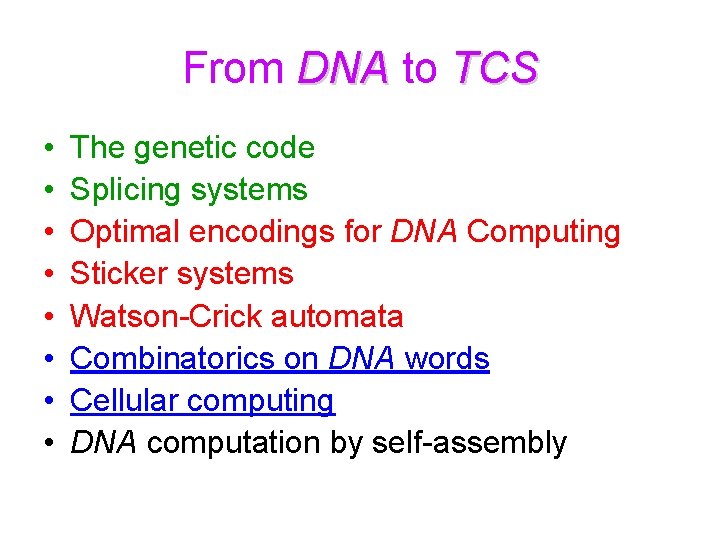
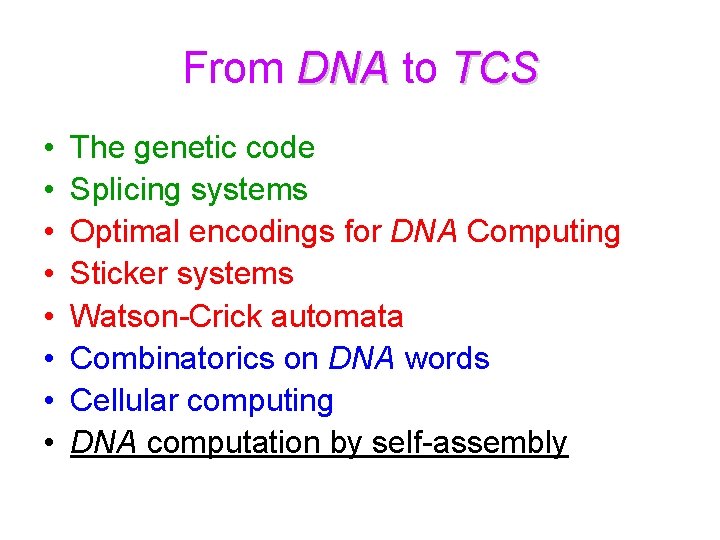
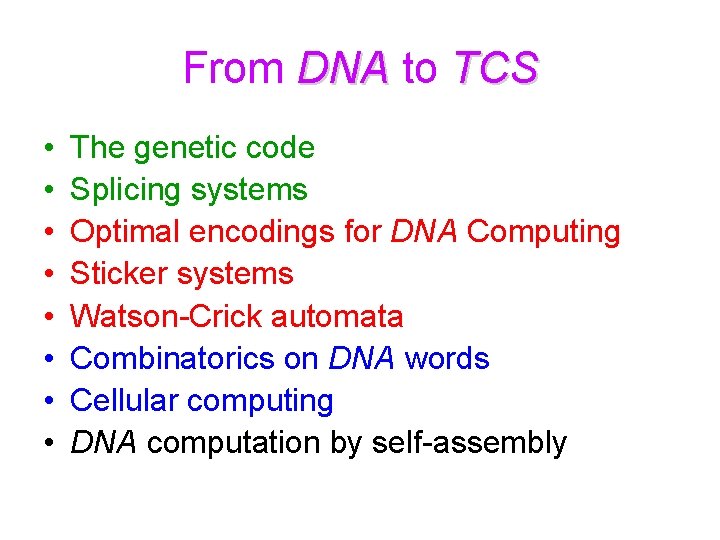
From DNA to TCS • • The genetic code Splicing systems Optimal encodings for DNA Computing Sticker systems Watson-Crick automata Combinatorics on DNA words Cellular computing DNA computation by self-assembly

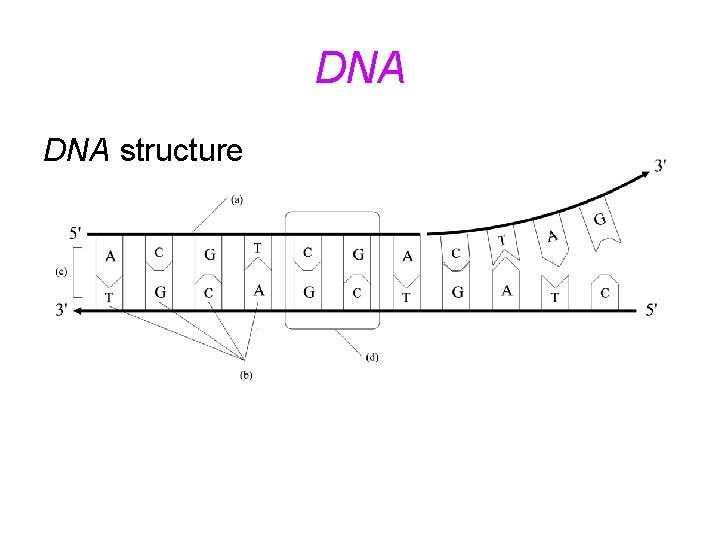
1953: Watson and Crick discover DNA structure

DNA structure

The RNA Tie Club • 1954 “Solve the riddle of the RNA structure and to understand how it builds proteins” (clockwise from upper left: Francis Crick, L. Orgel, James Watson, Al. Rich) • There are 20 aminoacids that build up proteins

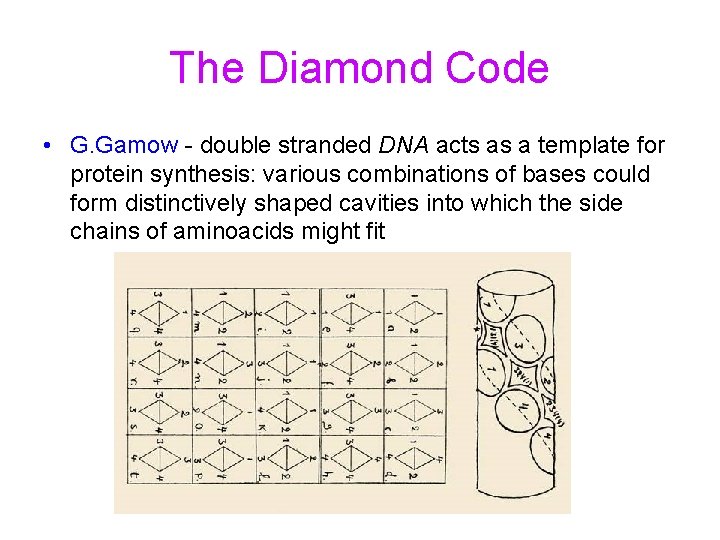
The Diamond Code • G. Gamow - double stranded DNA acts as a template for protein synthesis: various combinations of bases could form distinctively shaped cavities into which the side chains of aminoacids might fit

Comma-Free Codes (the prettiest wrong idea in 20 -th century science) • The RNA piglet model

The prettiest wrong idea in all of 20 th century science • Suckling-pig model of protein synthesis • Construct a code in which when two sense codons (triplets) are catenated, the subword codons are nonsense codons • If CGU and AAG are sense codons, then GUA and UAA must be nonsense because they appear in CGUAAG

Comma-free codes (Crick 1957) • How many words can a comma-free code include? • For n=4 and k=3 the size of a maximal comma-free code is the magic number 20 • For an alphabet of n letters grouped into kletter words, if k is prime, the number of maximal comma-free codes is (n^k –n)/k • For n=4 and k=3 this equals 408
![Reality Intrudes News from the lab bench Nirenberg Matthaei 61 synthesize RNA Reality Intrudes • News from the lab bench: [Nirenberg, Matthaei ’ 61] synthesize RNA,](https://slidetodoc.com/presentation_image_h2/4f19571aa8ae5913ef10182948859af9/image-10.jpg)
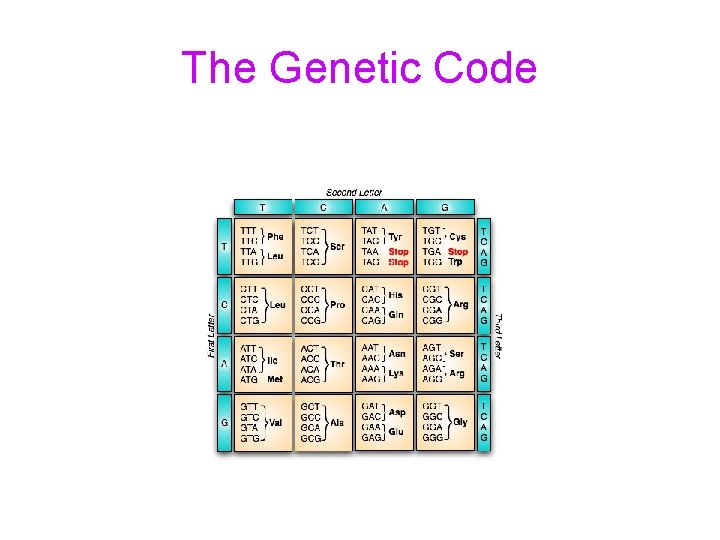
Reality Intrudes • News from the lab bench: [Nirenberg, Matthaei ’ 61] synthesize RNA, namely poly-U, coding for phenylalanine • By 1965 the genetic code was solved • The code resembled none of theoretical notions • The “extra” codons are merely redundant

The Genetic Code

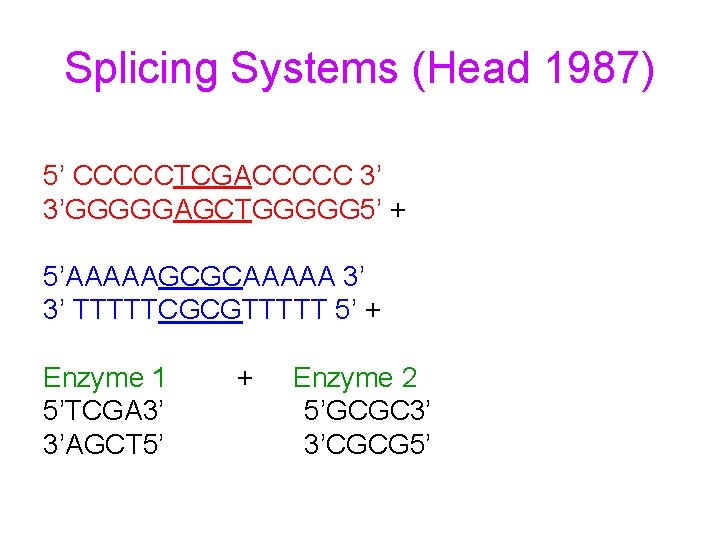
Splicing Systems (Head 1987) 5’ CCCCCTCGACCCCC 3’ 3’GGGGGAGCTGGGGG 5’ + 5’AAAAAGCGCAAAAA 3’ 3’ TTTTTCGCGTTTTT 5’ + Enzyme 1 5’TCGA 3’ 3’AGCT 5’ + Enzyme 2 5’GCGC 3’ 3’CGCG 5’

Splicing Systems 5’ CCCCCT CGACCCCC 3’ 3’GGGGGAGC TGGGGG 5’ + 5’AAAAAG CGCAAAAA 3’ 3’ TTTTTCGC GTTTTT 5’ DNA strands with compatible sticky ends recombine to produce two new strands

Splicing operation

Splicing system sample results Theorem (Paun’ 95, Freund, Kari, Paun , ’ 99) Every type-0 language can be generated by a splicing system with finitely many axioms and finitely many rules. Theorem (Freund, Kari, Paun ’ 99) For every given alphabet T there exists a splicing system, with finitely many axioms and finitely many rules, that is universal for the class of systems with terminal alphabet T.

From DNA to TCS • • The genetic code Splicing systems Optimal encodings for DNA Computing Sticker systems Watson-Crick automata Combinatorics on DNA words Cellular computing DNA computation by self-assembly

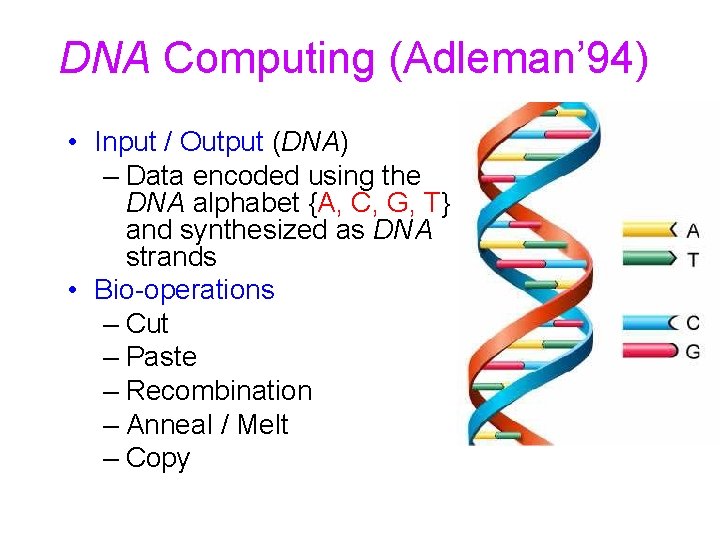
DNA Computing (Adleman’ 94) • Input / Output (DNA) – Data encoded using the DNA alphabet {A, C, G, T} and synthesized as DNA strands • Bio-operations – Cut – Paste – Recombination – Anneal / Melt – Copy
![Biomolecular DNA Computing Hamiltonian Path Problem Adleman Science 1994 DNAbased addition Biomolecular (DNA) Computing • • • Hamiltonian Path Problem [Adleman, Science, 1994] DNA-based addition](https://slidetodoc.com/presentation_image_h2/4f19571aa8ae5913ef10182948859af9/image-18.jpg)
Biomolecular (DNA) Computing • • • Hamiltonian Path Problem [Adleman, Science, 1994] DNA-based addition [Guarnieri et al, Science, 1996] Maximal Clique Problem [Ouyang et al, Science, 1997] DNA computing by self-assembly [Winfree et al, Nature 1998] Computations by circular insertions, deletions [Daley, Kari, Gloor, Siromoney, SPIRE’ 99] DNA computing on surfaces [Liu et al, Nature, 2000] Molecular computation by DNA hairpin formation[Sakamoto et al, Science, 2000] 20 -variable Satisfiability [Braich et al. , Science 2002] An autonomous molecular computer for logical control of gene expression [Benenson et al, Nature, 2004] Folding DNA to create nanoscale shapes and patterns [Rothemund, Nature, 2006] Efficient Turing-universal computation with DNA polymers [Qian, Soloveichik, Winfree, DNA Computing and Molecular Programming, 2010] Molecular robots guided by prescriptive landscapes [Lund et al. , Nature, 2010]

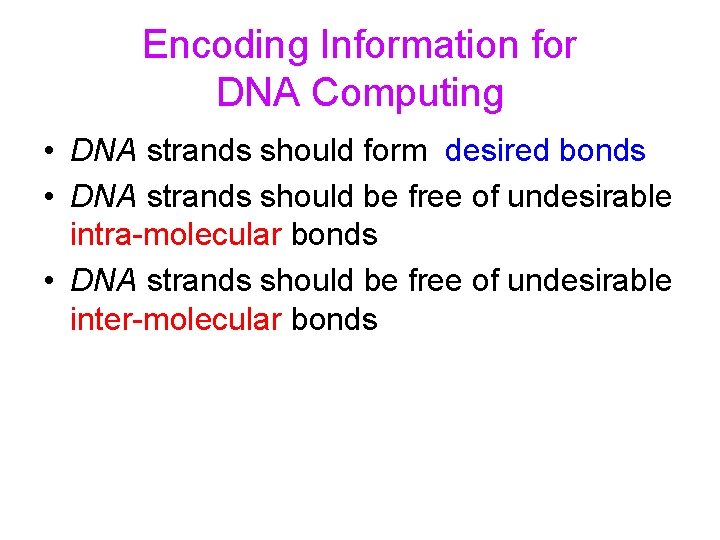
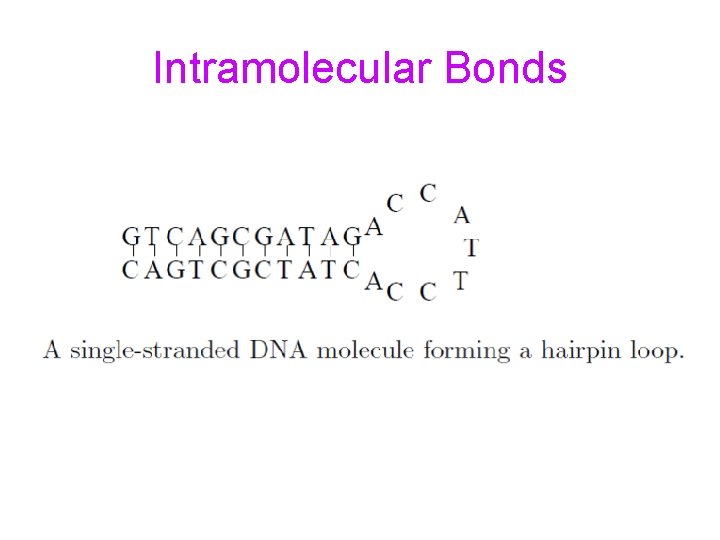
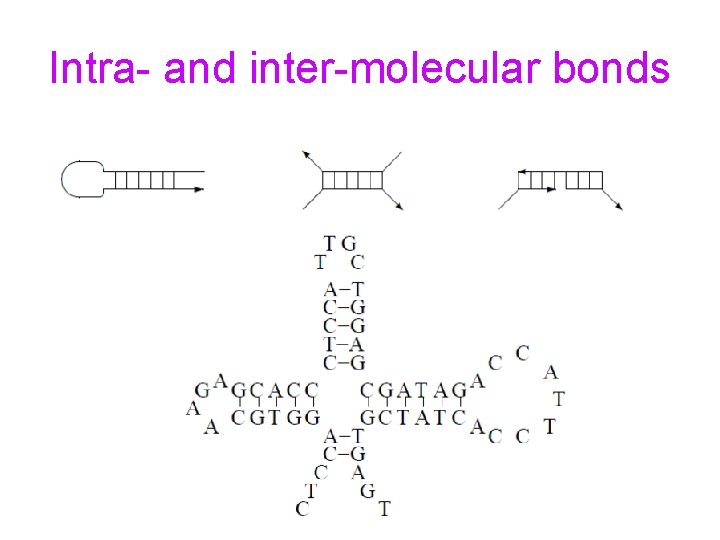
Encoding Information for DNA Computing • DNA strands should form desired bonds • DNA strands should be free of undesirable intra-molecular bonds • DNA strands should be free of undesirable inter-molecular bonds

Intramolecular Bonds

Intra- and inter-molecular bonds

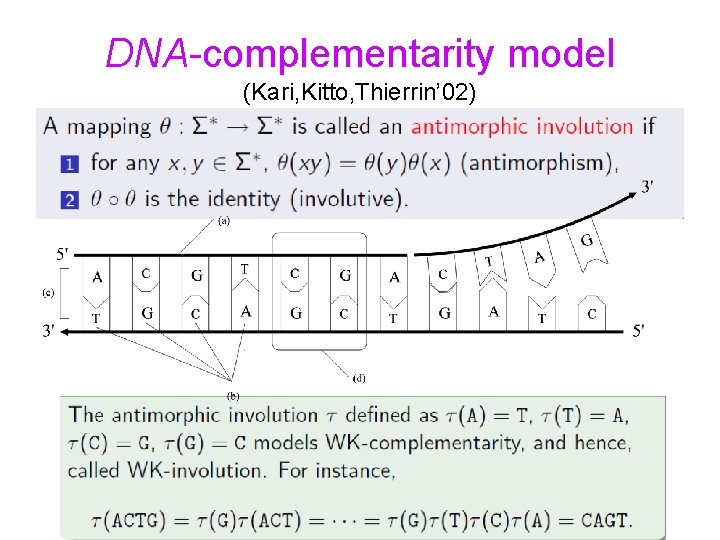
DNA-complementarity model (Kari, Kitto, Thierrin’ 02)

Bond-free languages Bonds between DNA strands

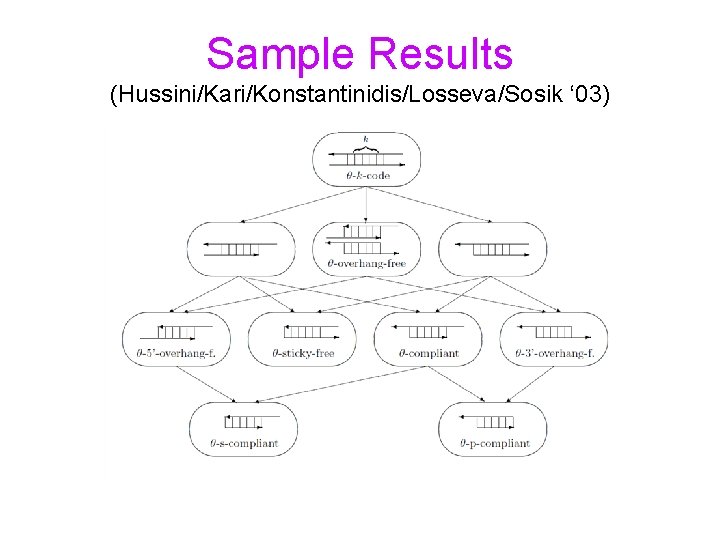
Sample Results (Hussini/Kari/Konstantinidis/Losseva/Sosik ‘ 03)

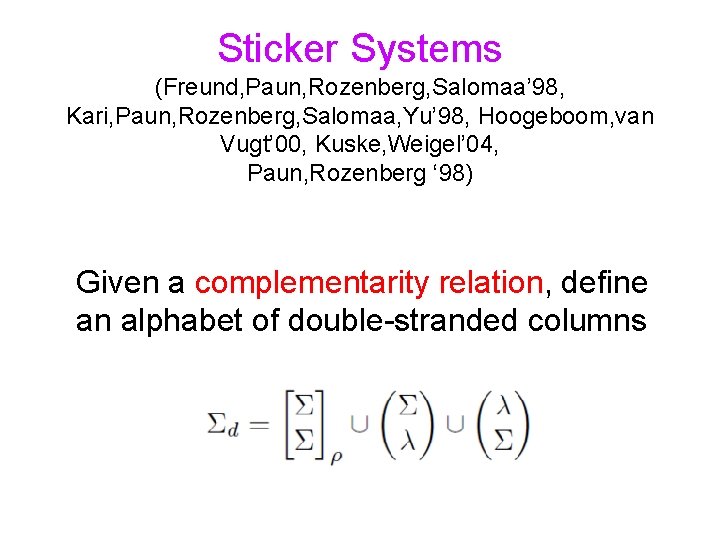
Sticker Systems (Freund, Paun, Rozenberg, Salomaa’ 98, Kari, Paun, Rozenberg, Salomaa, Yu’ 98, Hoogeboom, van Vugt’ 00, Kuske, Weigel’ 04, Paun, Rozenberg ‘ 98) Given a complementarity relation, define an alphabet of double-stranded columns

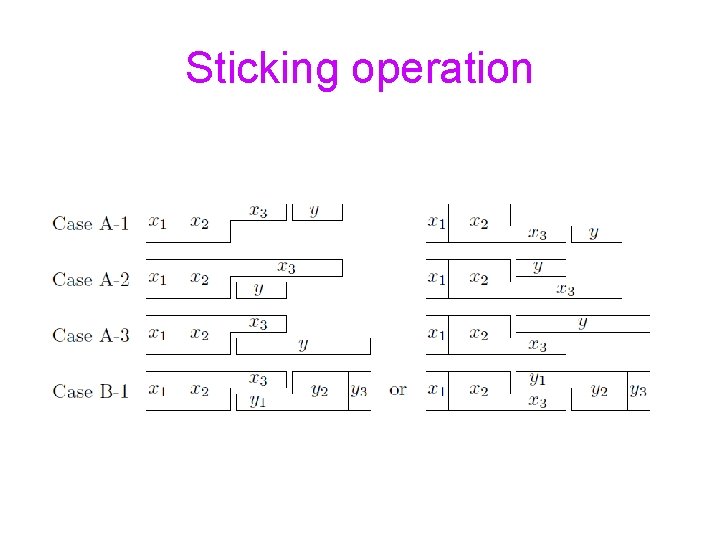
Sticking operation

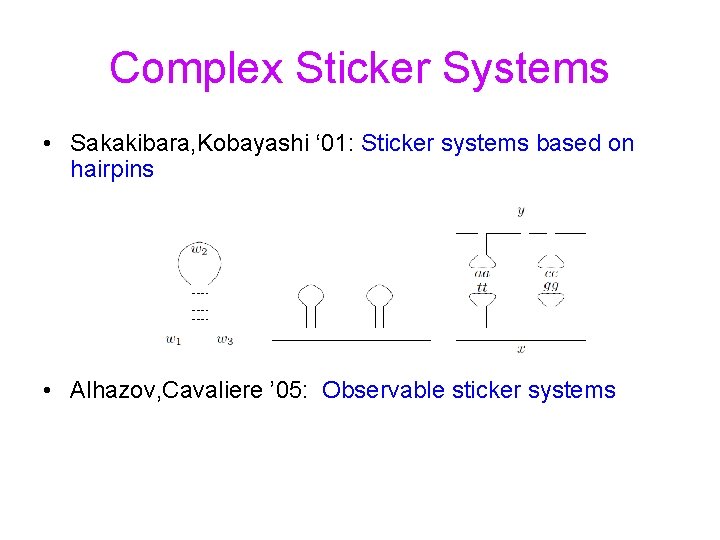
Complex Sticker Systems • Sakakibara, Kobayashi ‘ 01: Sticker systems based on hairpins • Alhazov, Cavaliere ’ 05: Observable sticker systems

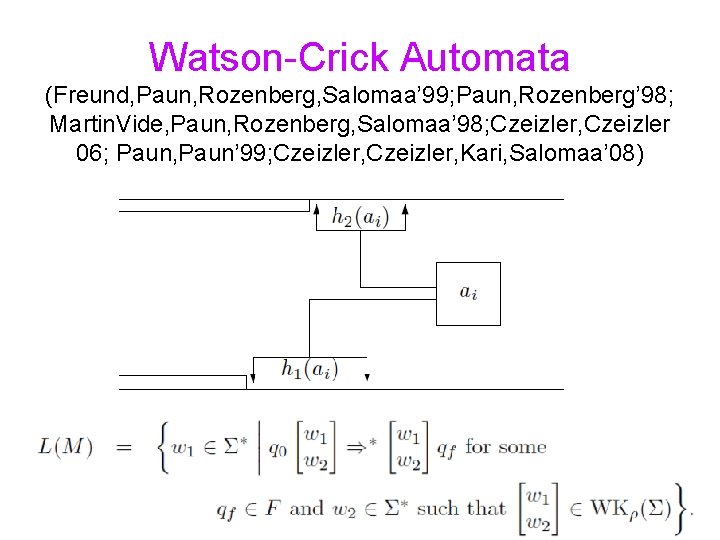
Watson-Crick Automata (Freund, Paun, Rozenberg, Salomaa’ 99; Paun, Rozenberg’ 98; Martin. Vide, Paun, Rozenberg, Salomaa’ 98; Czeizler, Czeizler 06; Paun, Paun’ 99; Czeizler, Kari, Salomaa’ 08)

From DNA to TCS • • The genetic code Splicing systems Optimal encodings for DNA Computing Sticker systems Watson-Crick automata Combinatorics on DNA words Cellular computing DNA computation by self-assembly

Combinatorics on DNA Words • IDEA: Consider the word w and its WKcomplement, WK(w), as equivalent • The word ACTG CAGT can be considered repetitive (periodic) because it can be written as ACGT WK(ACGT)2 • Generalize classical notions such as power of a word, border, primitive word, palindrome, conjugacy, commutativity

Identity => Antimorphic involution f Pseudo-palindrome (de Luca, De Luca’ 06, Kari, Mahalingam’ 09) u = f(u) Pseudo-commutativity(Kari, Mahalingam’ 08) u v = f(v) u Pseudo-bordered word (Kari, Mahalingam’ 07) w = v x = y f(v) Pseudoknot-bordered word (Kari, Seki’ 09) w = u v x = y f(u) f(v) Pseudo-conjugacy of u, v (Kari, Mahalingam’ 08) u x = f(x) v

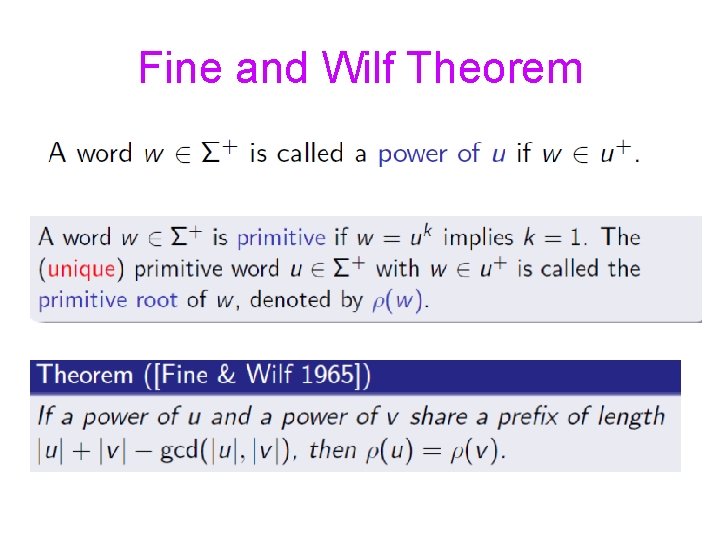
Fine and Wilf Theorem

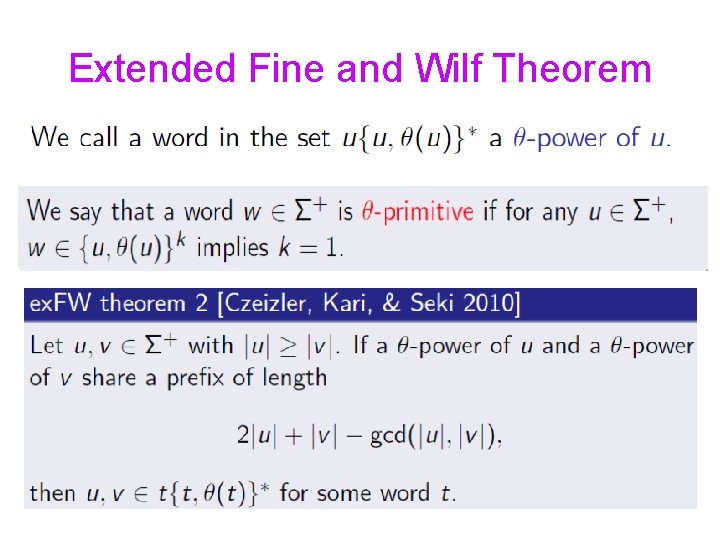
Extended Fine and Wilf Theorem

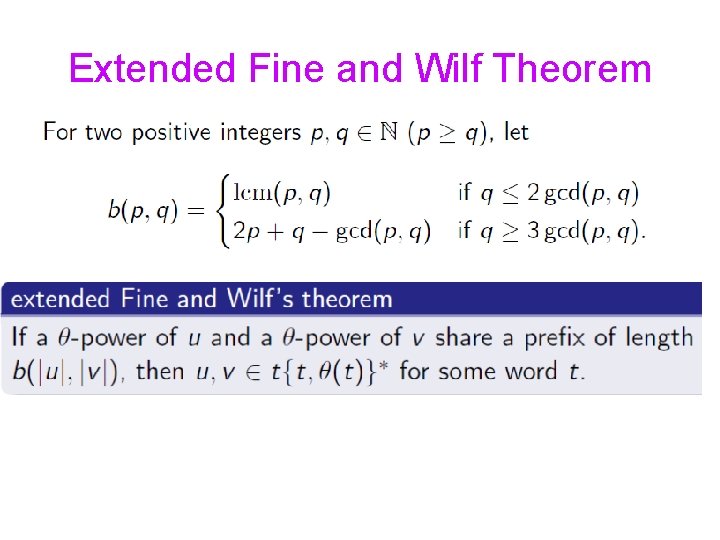
Extended Fine and Wilf Theorem

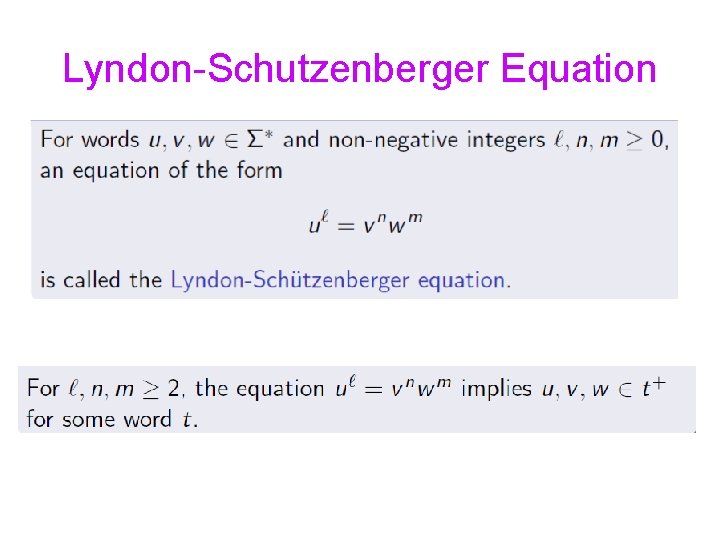
Lyndon-Schutzenberger Equation

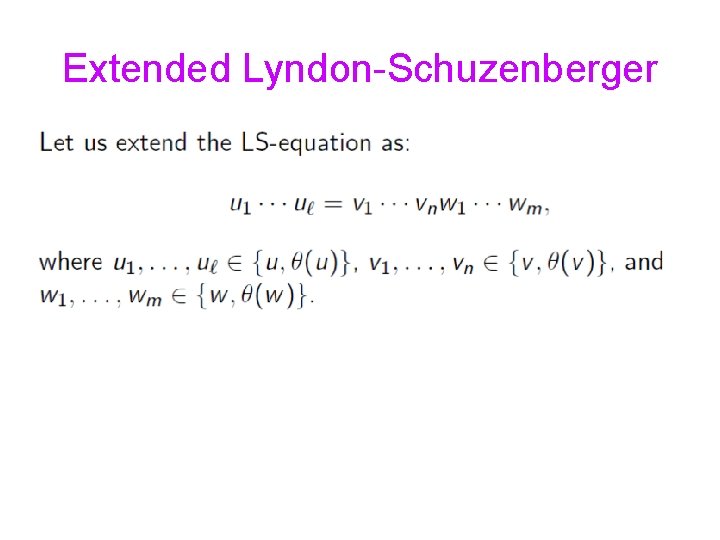
Extended Lyndon-Schuzenberger

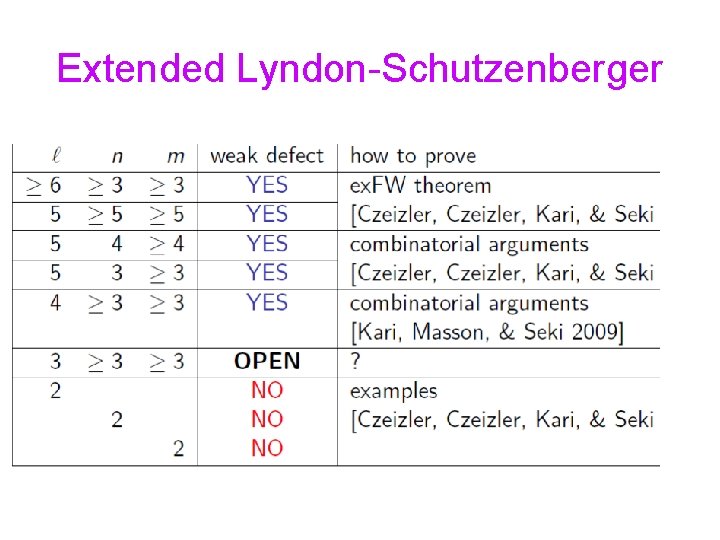
Extended Lyndon-Schutzenberger

Cellular Computing Photo courtesy of L. F. Landweber

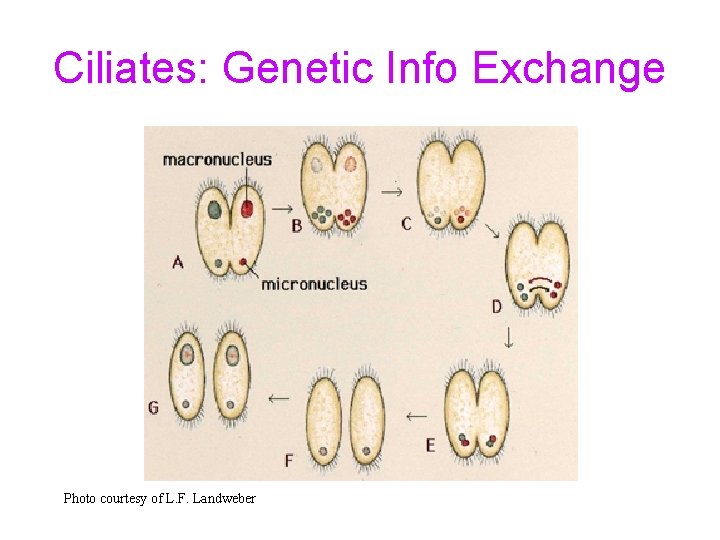
Ciliates: Genetic Info Exchange Photo courtesy of L. F. Landweber

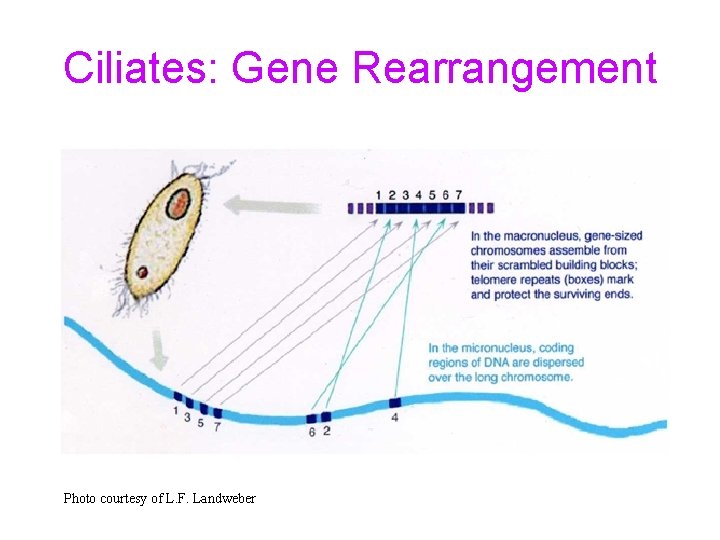
Ciliates: Gene Rearrangement Photo courtesy of L. F. Landweber

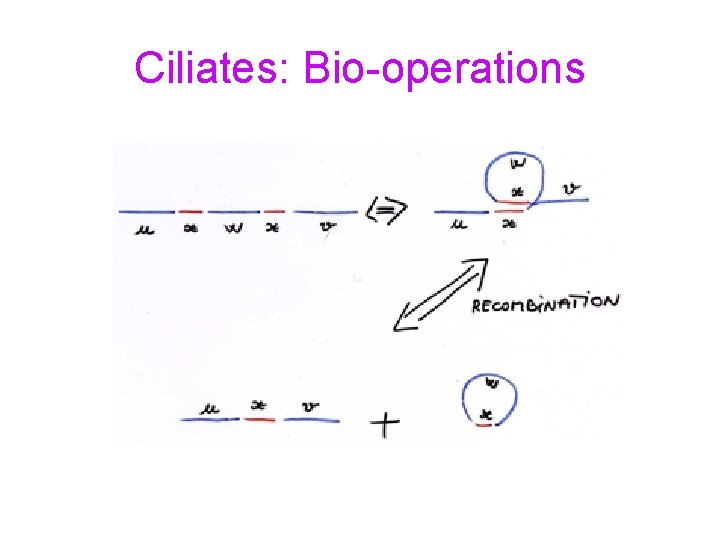
Ciliates: Bio-operations

Ciliate Computing • Guided Recombination System = A formal computational model based on contextual circular insertions and deletions • Such systems have the computational power of Turing Machines (Landweber, Kari ’ 99, Kari’ 99)

Other ciliate computing models * Ld, hi, dlad model (Harju, Rozenberg ’ 03, Harju, Petre, Rozenberg ’ 03, Prescott, Ehrenfeucht, Rozenberg’ 03) * Template guided recombination model (Angeleska, Jonoska, Saito, Landweber’ 07, Daley, Mc. Quillan ’ 06, Kari, Rahman ’ 10) * RNA guided recombination model (Nowacki et. al, ’ 07)

From DNA to TCS • • The genetic code Splicing systems Optimal encodings for DNA Computing Sticker systems Watson-Crick automata Combinatorics on DNA words Cellular computing DNA computation by self-assembly

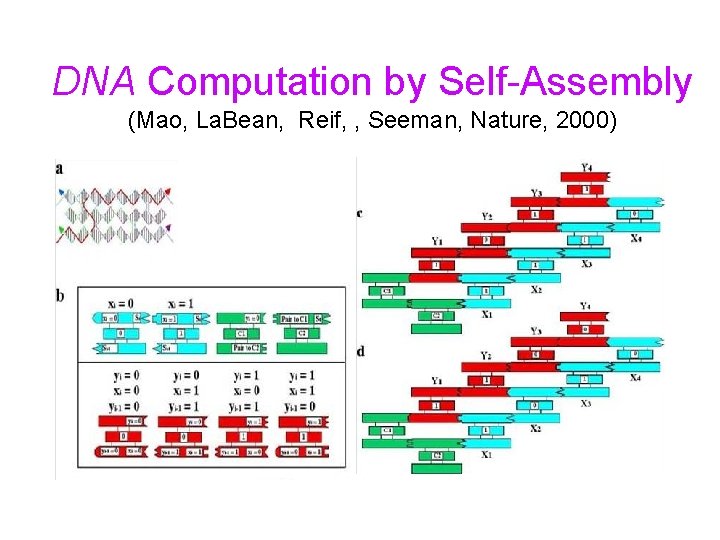
DNA Computation by Self-Assembly (Mao, La. Bean, Reif, , Seeman, Nature, 2000)

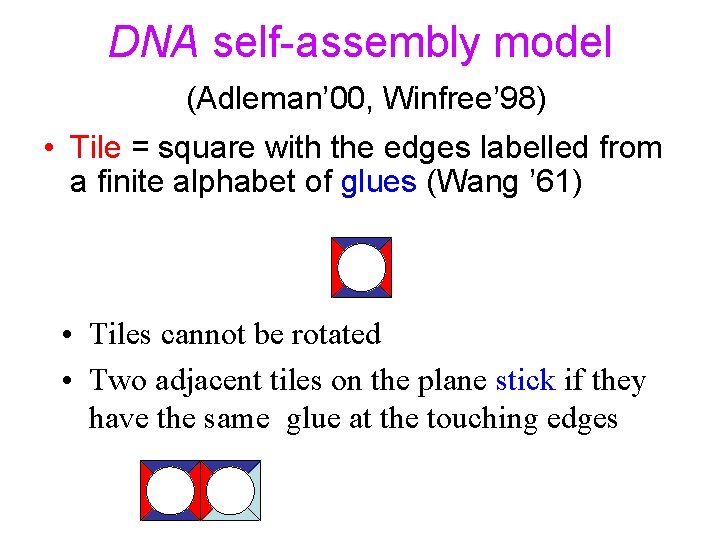
DNA self-assembly model (Adleman’ 00, Winfree’ 98) • Tile = square with the edges labelled from a finite alphabet of glues (Wang ’ 61) • Tiles cannot be rotated • Two adjacent tiles on the plane stick if they have the same glue at the touching edges

Dynamic Self-Assembly • Tile System T = Finite set of tiles, unlimited supply of each “tile type” • Supertiles self-assemble with tiles from T § Start with an arbitrary single tile: “seed” § Proceed by incremental additions of single tiles that stick A B C C A D A B

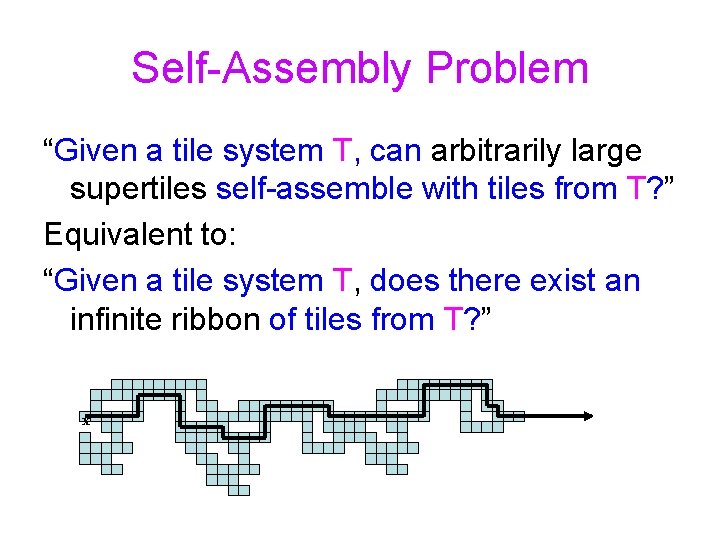
Self-Assembly Problem “Given a tile system T, can arbitrarily large supertiles self-assemble with tiles from T? ” Equivalent to: “Given a tile system T, does there exist an infinite ribbon of tiles from T? ” x

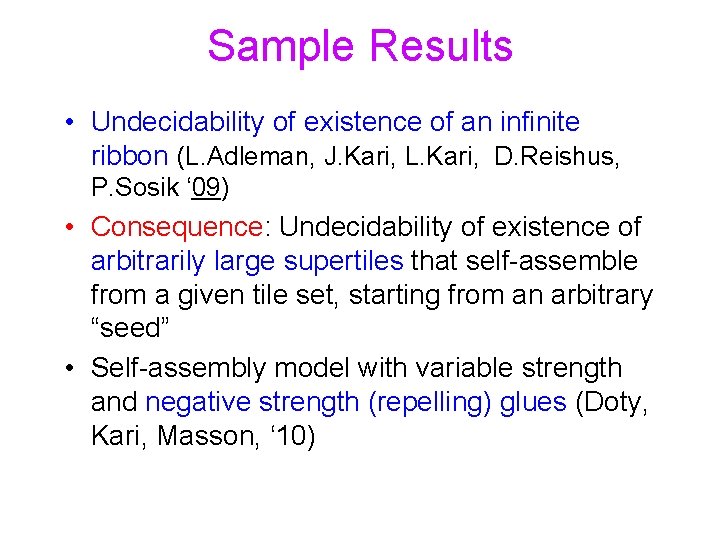
Sample Results • Undecidability of existence of an infinite ribbon (L. Adleman, J. Kari, L. Kari, D. Reishus, P. Sosik ‘ 09) • Consequence: Undecidability of existence of arbitrarily large supertiles that self-assemble from a given tile set, starting from an arbitrary “seed” • Self-assembly model with variable strength and negative strength (repelling) glues (Doty, Kari, Masson, ‘ 10)

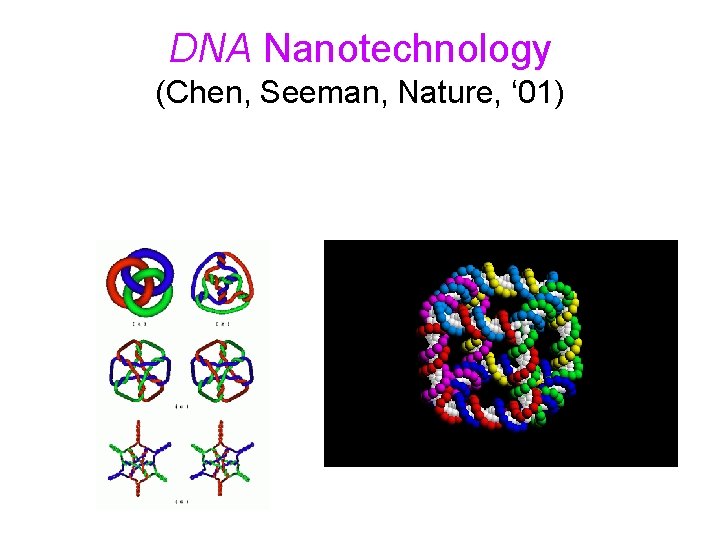
DNA Nanotechnology (Chen, Seeman, Nature, ‘ 01)

DNA Clonable Octahedron (Shih, Joyce, Nature ‘ 04)

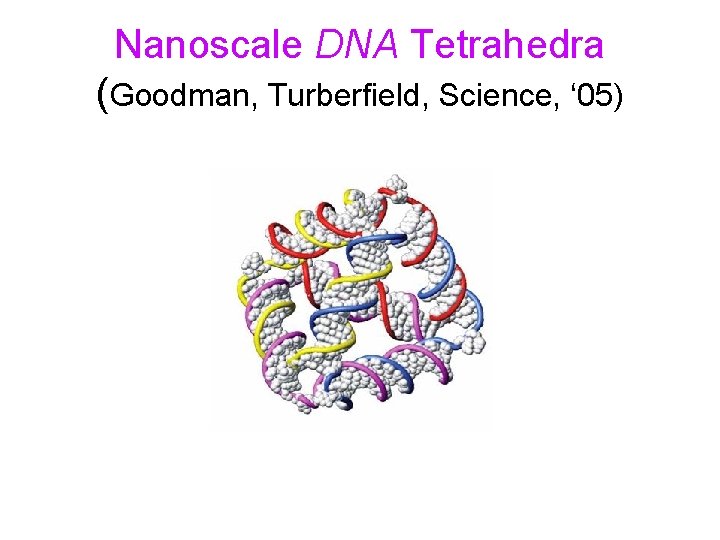
Nanoscale DNA Tetrahedra (Goodman, Turberfield, Science, ‘ 05)

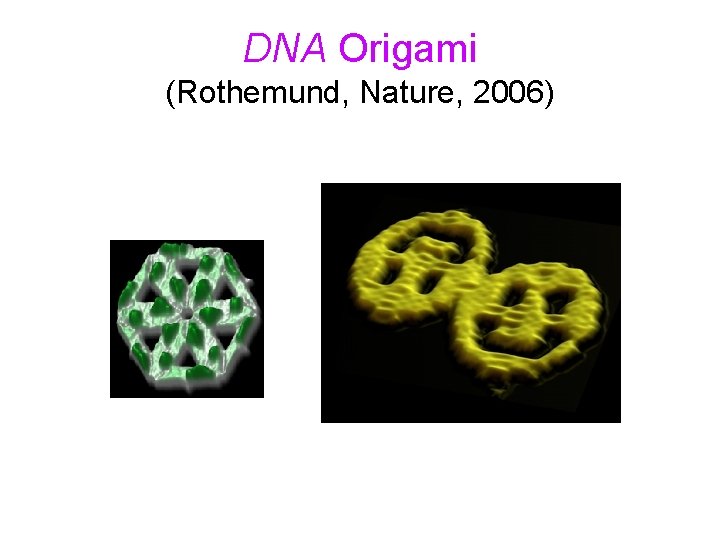
DNA Origami (Rothemund, Nature, 2006)

From DNA to TCS • • The genetic code Splicing systems Optimal encodings for DNA Computing Sticker systems Watson-Crick automata Combinatorics on DNA words Cellular computing DNA computation by self-assembly

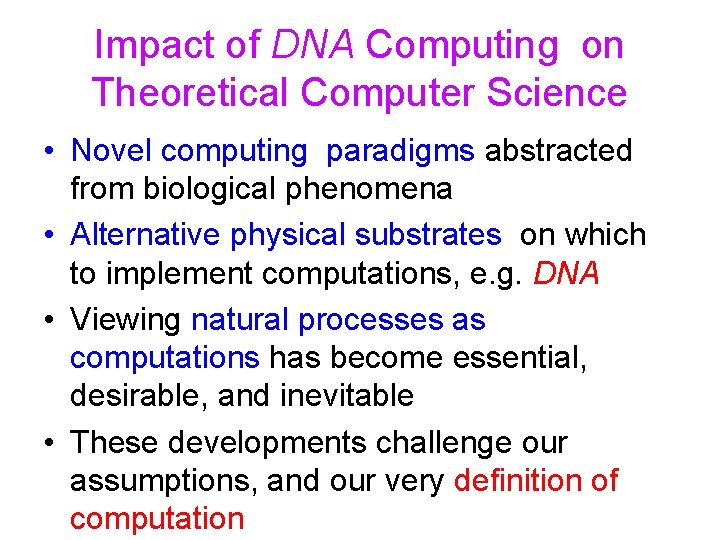
Impact of DNA Computing on Theoretical Computer Science • Novel computing paradigms abstracted from biological phenomena • Alternative physical substrates on which to implement computations, e. g. DNA • Viewing natural processes as computations has become essential, desirable, and inevitable • These developments challenge our assumptions, and our very definition of computation

Our Challenge • Discover a new, broader notion of computation • Understand the world around us in terms of information processing • “Biology and Computer Science – life and computation – are related. I am confident that at their interface great discoveries await whose who seek them. ” (Adleman’ 98)