CZ 3253 Computer Aided Drug design Lecture 10




- Slides: 46

CZ 3253: Computer Aided Drug design Lecture 10: Overview of Drug Testing Methods I: ADME Test Prof. Chen Yu Zong Tel: 6874 -6877 Email: csccyz@nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, National University of Singapore

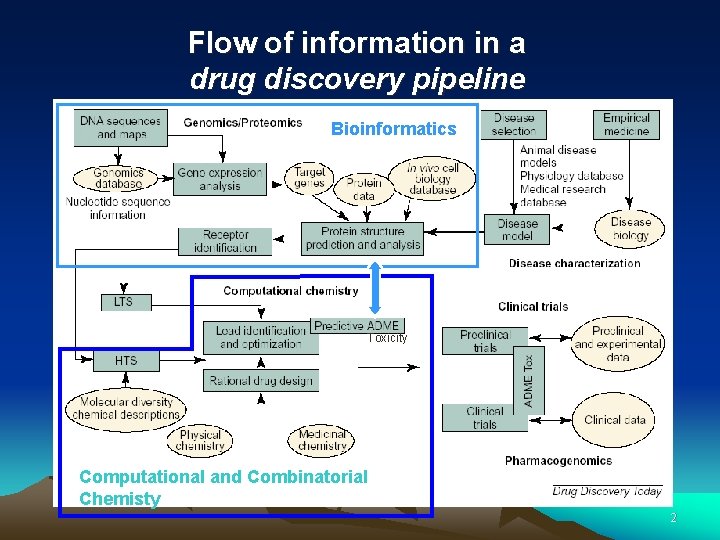
Flow of information in a drug discovery pipeline Bioinformatics Toxicity Computational and Combinatorial Chemisty 2

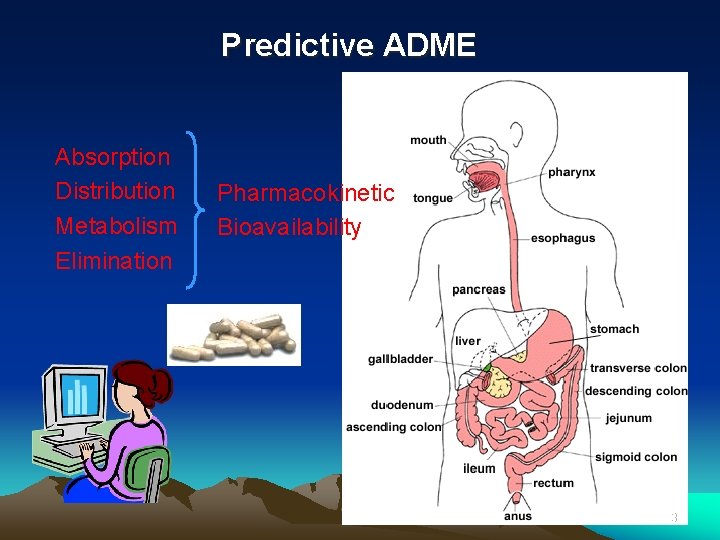
Predictive ADME Absorption Distribution Metabolism Elimination Pharmacokinetic Bioavailability 3

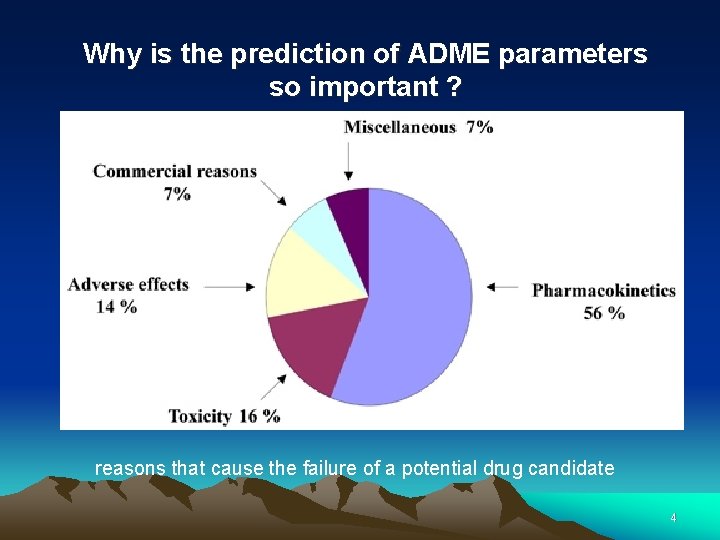
Why is the prediction of ADME parameters so important ? reasons that cause the failure of a potential drug candidate 4

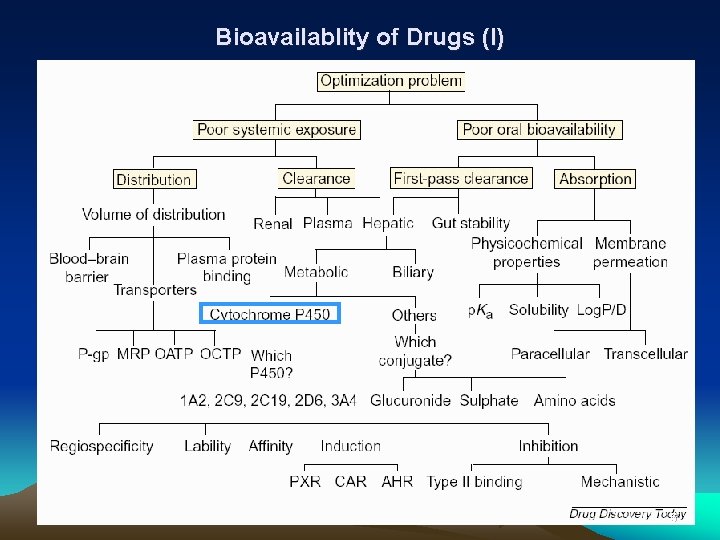
Bioavailablity of Drugs (I) 5

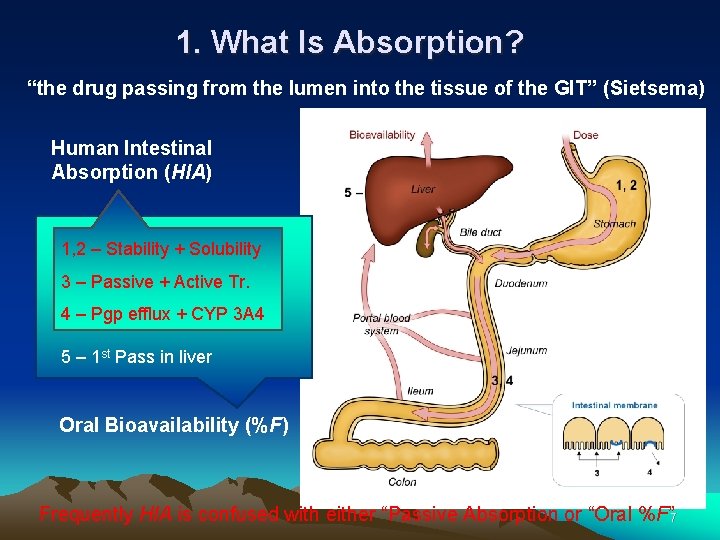
Bioavailability of Drugs (II) Uptake of orally administered drug proceeds after the stomach passage via the small intestine. In the liver, a series of metabolic transformation occurs. 6

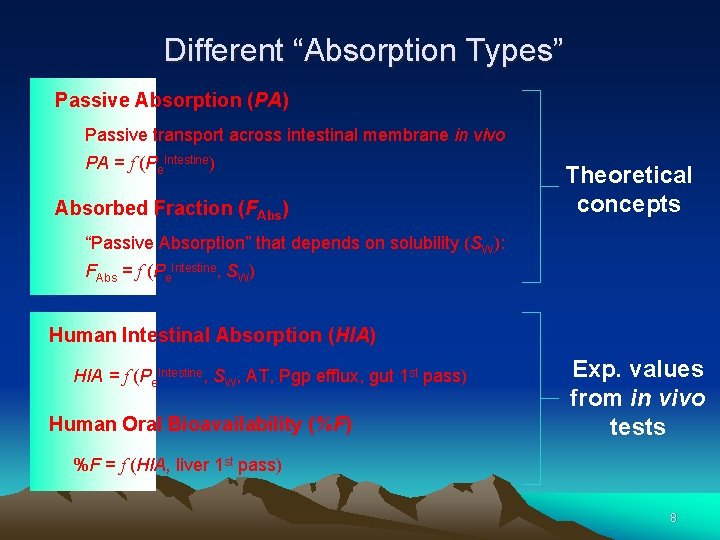
1. What Is Absorption? “the drug passing from the lumen into the tissue of the GIT” (Sietsema) Human Intestinal Absorption (HIA) 1, 2 – Stability + Solubility 3 – Passive + Active Tr. 4 – Pgp efflux + CYP 3 A 4 5 – 1 st Pass in liver Oral Bioavailability (%F) Frequently HIA is confused with either “Passive Absorption or “Oral %F” 7

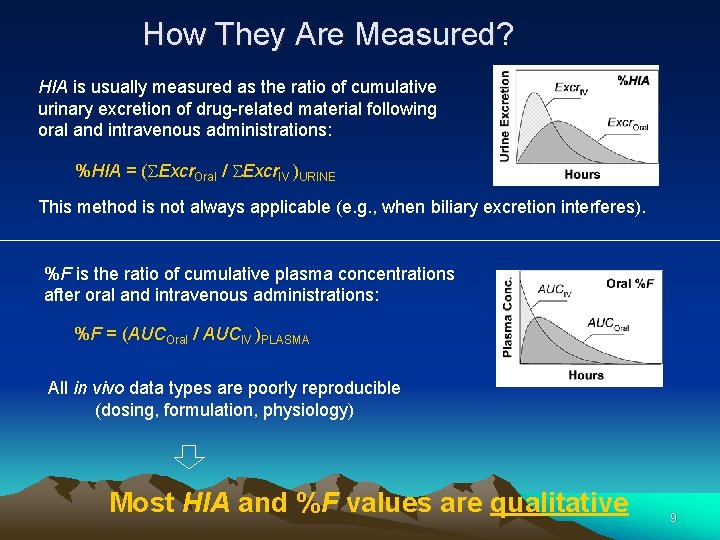
Different “Absorption Types” Passive Absorption (PA) Passive transport across intestinal membrane in vivo PA = f (Pe. Intestine) Absorbed Fraction (FAbs) Theoretical concepts “Passive Absorption” that depends on solubility (SW): FAbs = f (Pe. Intestine, SW) Human Intestinal Absorption (HIA) HIA = f (Pe. Intestine, SW, AT, Pgp efflux, gut 1 st pass) Human Oral Bioavailability (%F) Exp. values from in vivo tests %F = f (HIA, liver 1 st pass) 8

How They Are Measured? HIA is usually measured as the ratio of cumulative urinary excretion of drug-related material following oral and intravenous administrations: %HIA = ( Excr. Oral / Excr. IV )URINE This method is not always applicable (e. g. , when biliary excretion interferes). %F is the ratio of cumulative plasma concentrations after oral and intravenous administrations: %F = (AUCOral / AUCIV )PLASMA All in vivo data types are poorly reproducible (dosing, formulation, physiology) Most HIA and %F values are qualitative 9

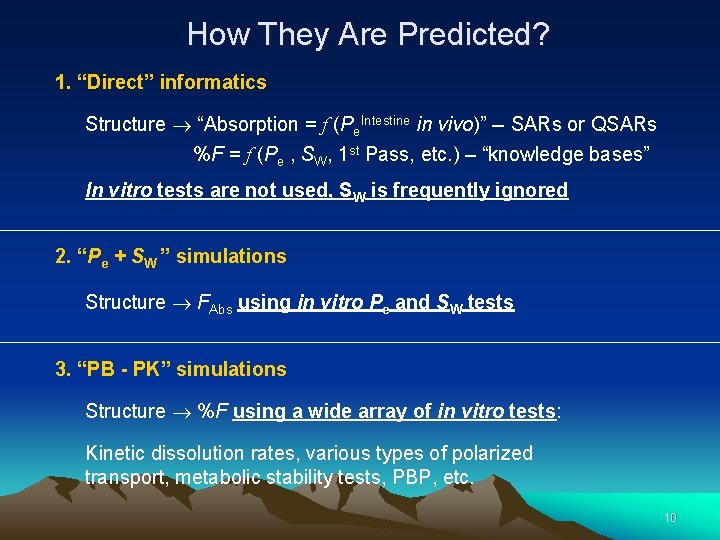
How They Are Predicted? 1. “Direct” informatics Structure “Absorption = f (Pe. Intestine in vivo)” -- SARs or QSARs %F = f (Pe , SW, 1 st Pass, etc. ) – “knowledge bases” In vitro tests are not used, SW is frequently ignored 2. “Pe + SW ” simulations Structure FAbs using in vitro Pe and SW tests 3. “PB - PK” simulations Structure %F using a wide array of in vitro tests: Kinetic dissolution rates, various types of polarized transport, metabolic stability tests, PBP, etc. 10

How They Are Predicted? 4. Statistical Learning Methods Structure Molecular Descriptors Training of Prediction System Trained using samples of absorption and non-absorption compounds Like any other statistical learning method, prediction accuracy dependent on the diversity and representativity of training data 11

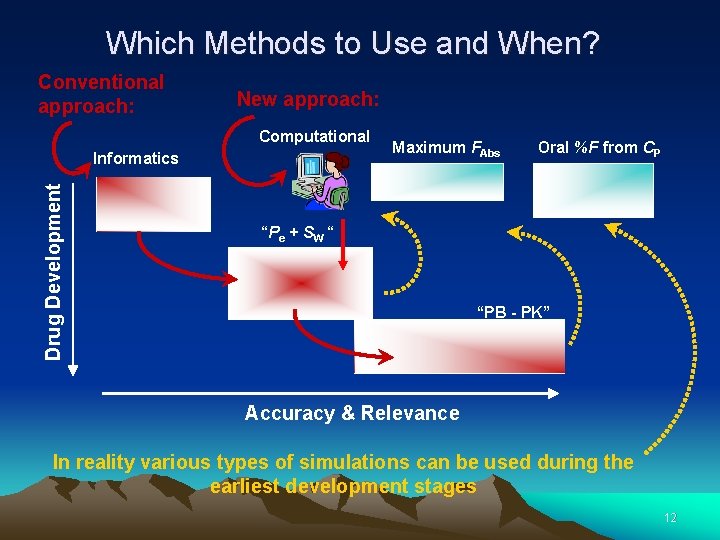
Which Methods to Use and When? Conventional approach: New approach: Computational Drug Development Informatics Maximum FAbs Oral %F from CP “Pe + SW “ “PB - PK” Accuracy & Relevance In reality various types of simulations can be used during the earliest development stages 12

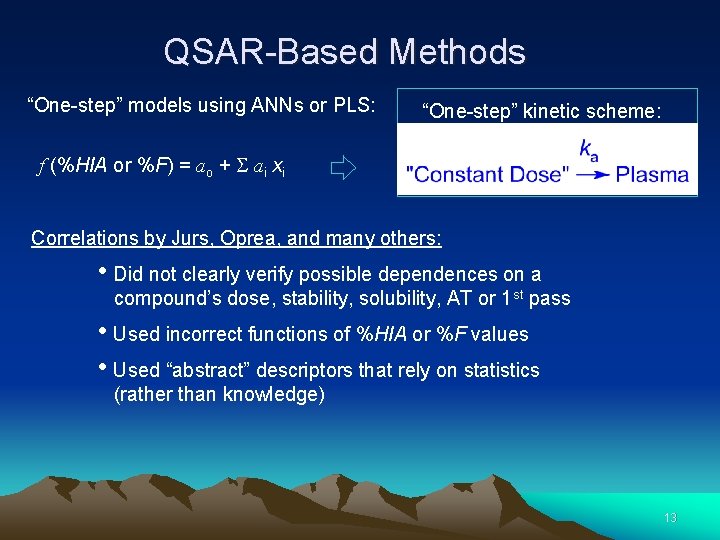
QSAR-Based Methods “One-step” models using ANNs or PLS: “One-step” kinetic scheme: f (%HIA or %F) = ao + ai xi Correlations by Jurs, Oprea, and many others: • Did not clearly verify possible dependences on a compound’s dose, stability, solubility, AT or 1 st pass • Used incorrect functions of %HIA or %F values • Used “abstract” descriptors that rely on statistics (rather than knowledge) 13

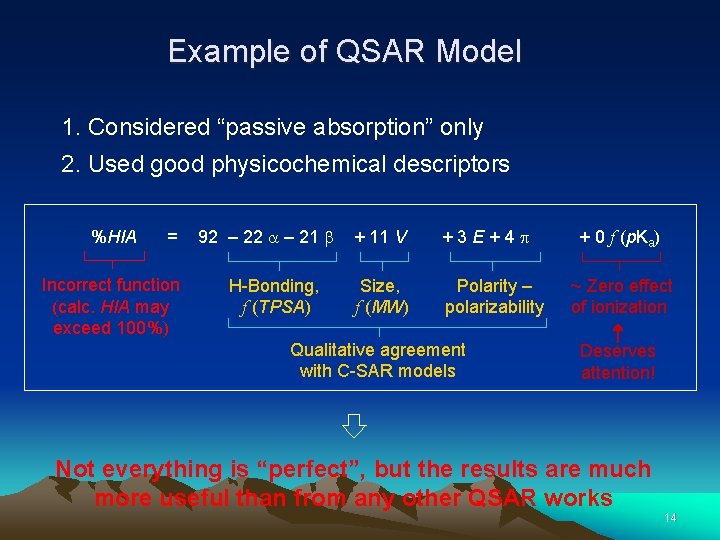
Example of QSAR Model 1. Considered “passive absorption” only 2. Used good physicochemical descriptors %HIA = Incorrect function (calc. HIA may exceed 100%) 92 – 22 – 21 H-Bonding, f (TPSA) + 11 V +3 E+4 Size, f (MW) Polarity – polarizability Qualitative agreement with C-SAR models + 0 f (p. Ka) ~ Zero effect of ionization Deserves attention! Not everything is “perfect”, but the results are much more useful than from any other QSAR works 14

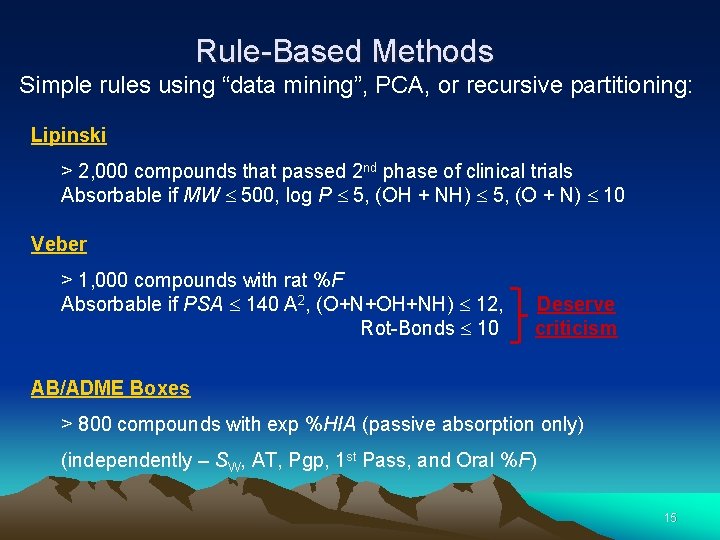
Rule-Based Methods Simple rules using “data mining”, PCA, or recursive partitioning: Lipinski > 2, 000 compounds that passed 2 nd phase of clinical trials Absorbable if MW 500, log P 5, (OH + NH) 5, (O + N) 10 Veber > 1, 000 compounds with rat %F Absorbable if PSA 140 A 2, (O+N+OH+NH) 12, Rot-Bonds 10 Deserve criticism AB/ADME Boxes > 800 compounds with exp %HIA (passive absorption only) (independently – SW, AT, Pgp, 1 st Pass, and Oral %F) 15

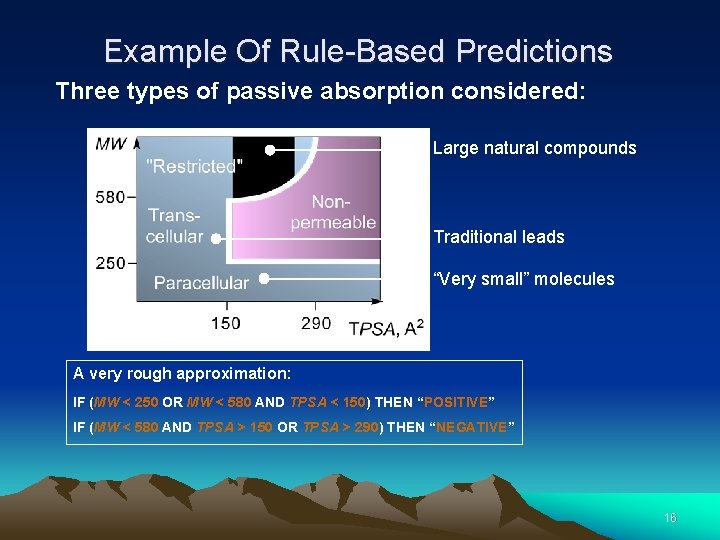
Example Of Rule-Based Predictions Three types of passive absorption considered: Large natural compounds Traditional leads “Very small” molecules A very rough approximation: IF (MW < 250 OR MW < 580 AND TPSA < 150) THEN “POSITIVE” IF (MW < 580 AND TPSA > 150 OR TPSA > 290) THEN “NEGATIVE” 16

Generalization of the Rules: The “Rule of Five” Formulation for Drug-Like Molecules Poor absorption or permeation are more likely when: • • There are more than 5 H-bond donors. The molecular weight is over 500. The Log. P is over 5. There are more than 10 H-bond acceptors. 17

Exception to the rule of five Compound classes that are substrates for biological transporters: • Antibiotics • Fungicides-Protozoacides antiseptics • Vitamins • Cardiac glycosides. 18

Computational calculations for new chemical entities • Applied to entities introduced between 1990 -1993 • Average values: – Mlog. P=1. 80 – H-bond donor sum=2. 53 – Molecular weight =408 – H-bond acceptor sum=6. 95 • Alerts for possible poor absorption-12% 19

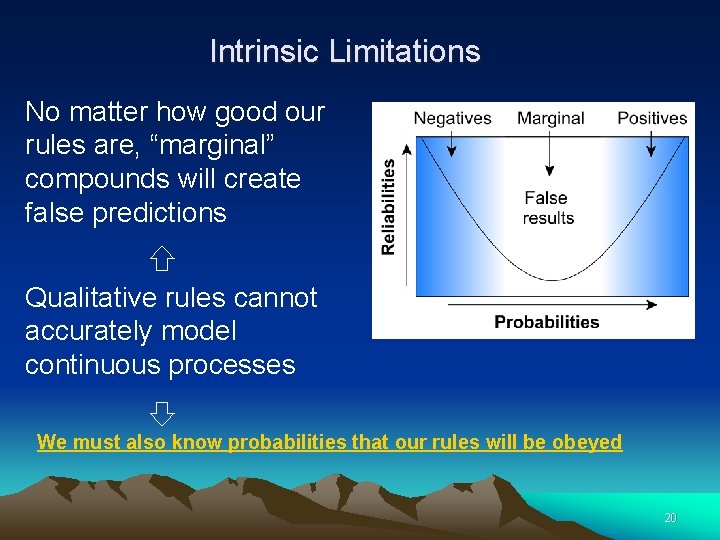
Intrinsic Limitations No matter how good our rules are, “marginal” compounds will create false predictions Qualitative rules cannot accurately model continuous processes We must also know probabilities that our rules will be obeyed 20

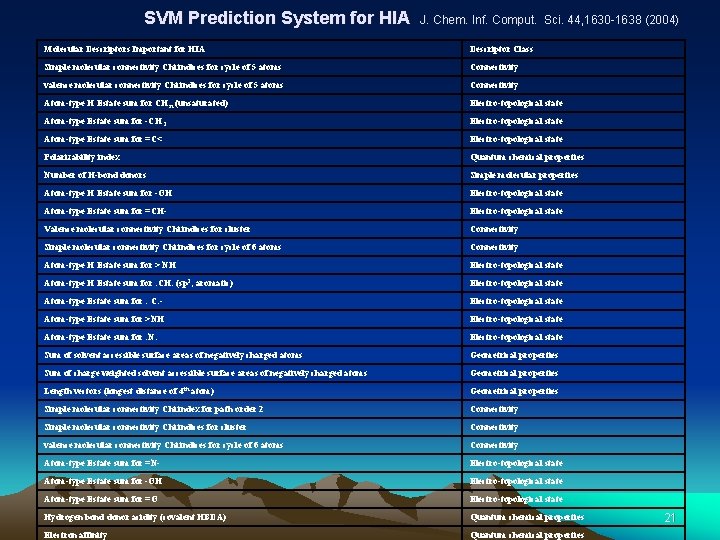
SVM Prediction System for HIA J. Chem. Inf. Comput. Sci. 44, 1630 -1638 (2004) Molecular Descriptors Important for HIA Descriptor Class Simple molecular connectivity Chi indices for cycle of 5 atoms Connectivity valence molecular connectivity Chi indices for cycle of 5 atoms Connectivity Atom-type H Estate sum for CH n (unsaturated) Electro-topological state Atom-type Estate sum for -CH 3 Electro-topological state Atom-type Estate sum for =C< Electro-topological state Polarizability index Quantum chemical properties Number of H-bond donors Simple molecular properties Atom-type H Estate sum for -OH Electro-topological state Atom-type Estate sum for =CH- Electro-topological state Valence molecular connectivity Chi indices for cluster Connectivity Simple molecular connectivity Chi indices for cycle of 6 atoms Connectivity Atom-type H Estate sum for > NH Electro-topological state Atom-type H Estate sum for : CH: (sp 2, aromatic) Electro-topological state Atom-type Estate sum for : C: - Electro-topological state Atom-type Estate sum for >NH Electro-topological state Atom-type Estate sum for : N: Electro-topological state Sum of solvent accessible surface areas of negatively charged atoms Geometrical properties Sum of charge weighted solvent accessible surface areas of negatively charged atoms Geometrical properties Length vectors (longest distance of 4 th atom) Geometrical properties Simple molecular connectivity Chi index for path order 2 Connectivity Simple molecular connectivity Chi indices for cluster Connectivity valence molecular connectivity Chi indices for cycle of 6 atoms Connectivity Atom-type Estate sum for =N- Electro-topological state Atom-type Estate sum for -OH Electro-topological state Atom-type Estate sum for =O Electro-topological state Hydrogen bond donor acidity (covalent HBDA) Quantum chemical properties Electron affinity Quantum chemical properties 21

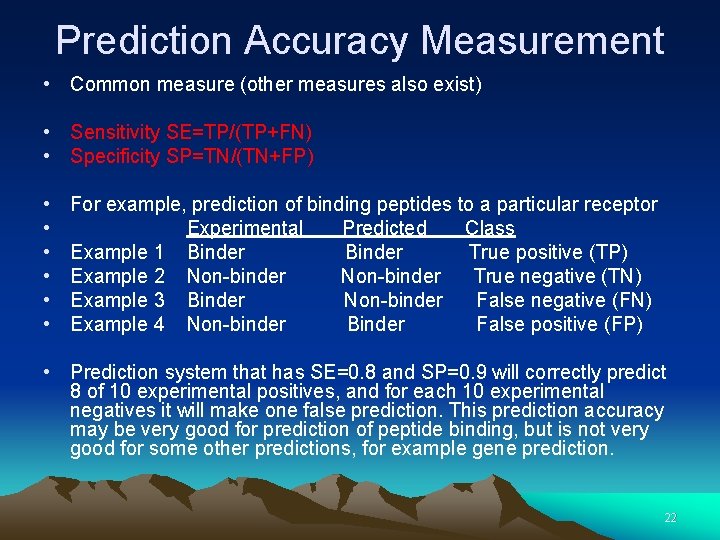
Prediction Accuracy Measurement • Common measure (other measures also exist) • Sensitivity SE=TP/(TP+FN) • Specificity SP=TN/(TN+FP) • • • For example, prediction of binding peptides to a particular receptor Experimental Predicted Class Example 1 Binder True positive (TP) Example 2 Non-binder True negative (TN) Example 3 Binder Non-binder False negative (FN) Example 4 Non-binder Binder False positive (FP) • Prediction system that has SE=0. 8 and SP=0. 9 will correctly predict 8 of 10 experimental positives, and for each 10 experimental negatives it will make one false prediction. This prediction accuracy may be very good for prediction of peptide binding, but is not very good for some other predictions, for example gene prediction. 22

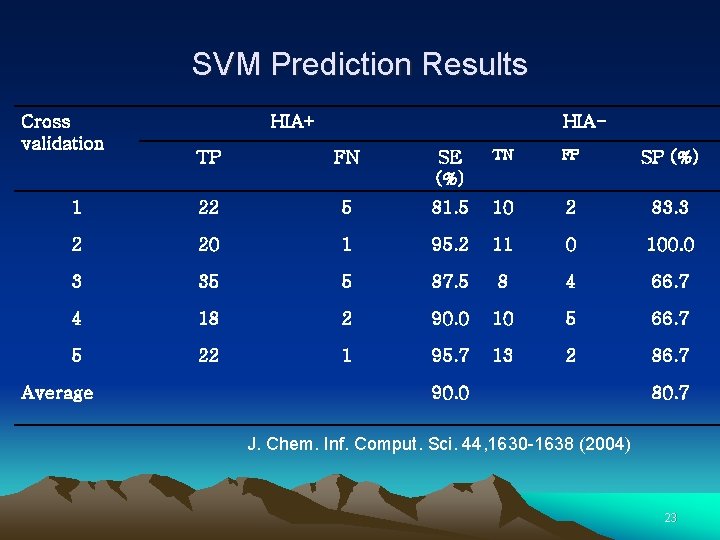
SVM Prediction Results Cross validation HIA+ HIA- TP FN SE (%) TN FP SP (%) 1 22 5 81. 5 10 2 83. 3 2 20 1 95. 2 11 0 100. 0 3 35 5 87. 5 8 4 66. 7 4 18 2 90. 0 10 5 66. 7 5 22 1 95. 7 13 2 86. 7 Average 90. 0 80. 7 J. Chem. Inf. Comput. Sci. 44, 1630 -1638 (2004) 23

Cytochrome P 450 The super-family of cytochrome P 450 enzymes has a crucial role in the metabolism of drugs. Almost every drug is processed by some of these enzymes. This causes a reduced bioavailability. Cytochrome P 450 enzymes show extensive structural polymorphism (differences in the coding region). 24

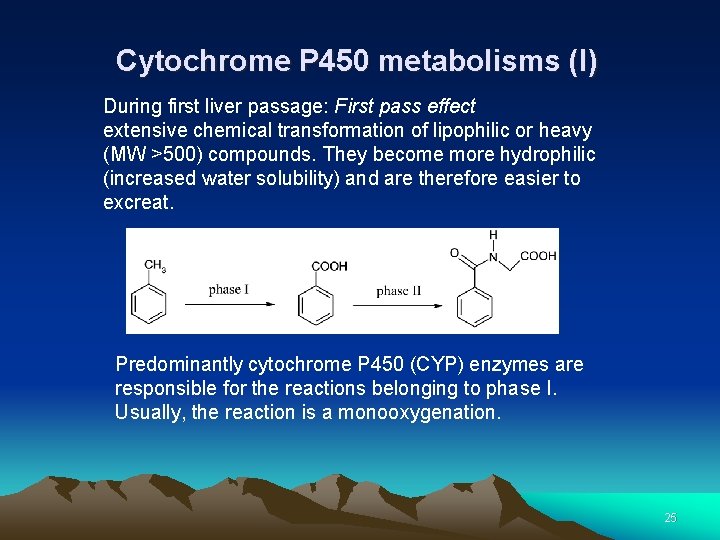
Cytochrome P 450 metabolisms (I) During first liver passage: First pass effect extensive chemical transformation of lipophilic or heavy (MW >500) compounds. They become more hydrophilic (increased water solubility) and are therefore easier to excreat. Predominantly cytochrome P 450 (CYP) enzymes are responsible for the reactions belonging to phase I. Usually, the reaction is a monooxygenation. 25

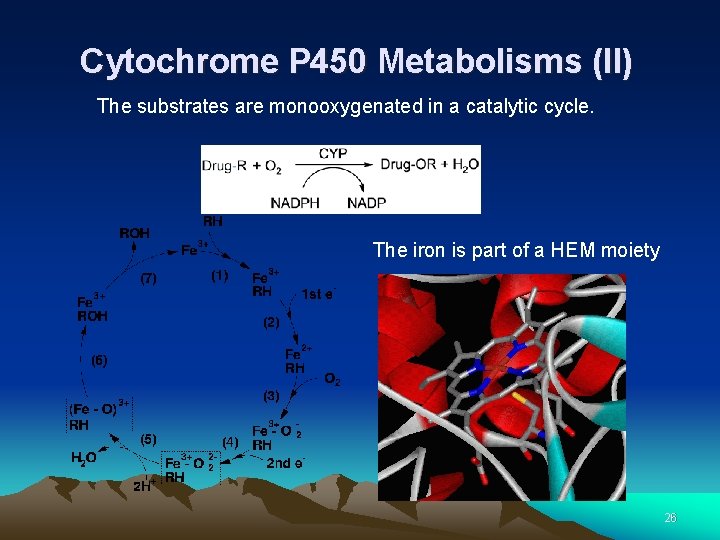
Cytochrome P 450 Metabolisms (II) The substrates are monooxygenated in a catalytic cycle. The iron is part of a HEM moiety 26

Cytochrome P 450 Metabolisms (III) The cytochromes involved in the metabolism are mainly monooxygenases that evolved from the steroid and fatty acid biosynthesis. So far, 17 families of CYPs with about 50 isoforms have been characterized in the human genome. classification: CYP 3 A 4 *15 A-B family isoenzyme allel >40% sequencesub-family homology >55% sequencehomology 27

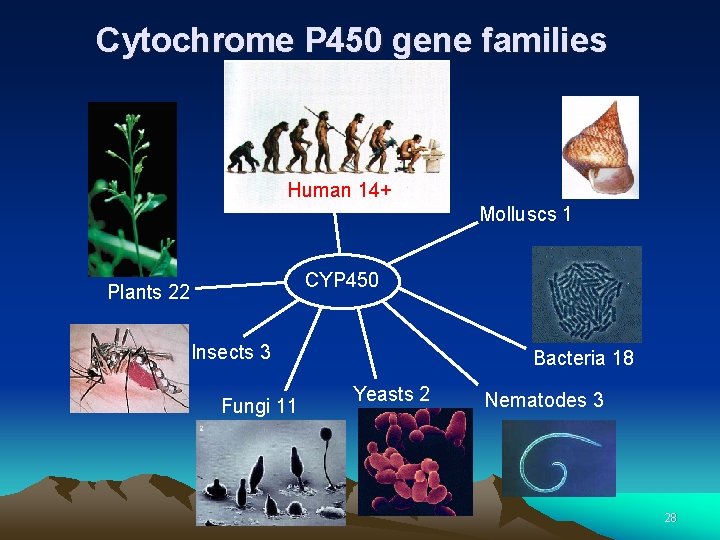
Cytochrome P 450 gene families Human 14+ Molluscs 1 CYP 450 Plants 22 Insects 3 Fungi 11 Bacteria 18 Yeasts 2 Nematodes 3 28

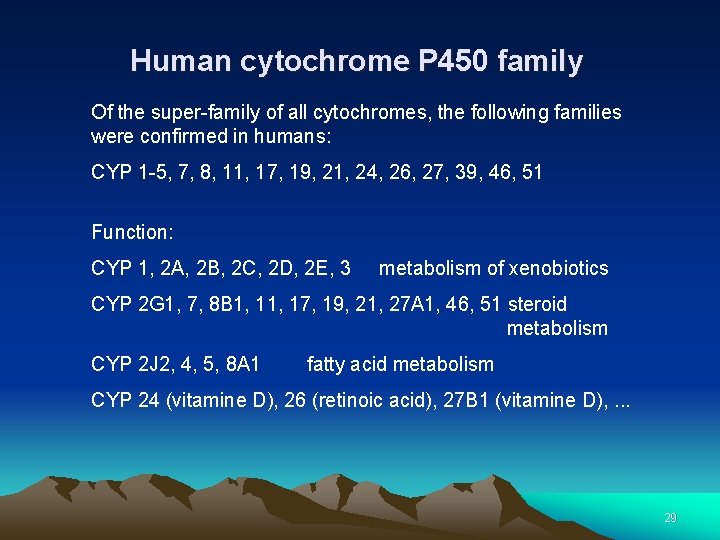
Human cytochrome P 450 family Of the super-family of all cytochromes, the following families were confirmed in humans: CYP 1 -5, 7, 8, 11, 17, 19, 21, 24, 26, 27, 39, 46, 51 Function: CYP 1, 2 A, 2 B, 2 C, 2 D, 2 E, 3 metabolism of xenobiotics CYP 2 G 1, 7, 8 B 1, 17, 19, 21, 27 A 1, 46, 51 steroid metabolism CYP 2 J 2, 4, 5, 8 A 1 fatty acid metabolism CYP 24 (vitamine D), 26 (retinoic acid), 27 B 1 (vitamine D), . . . 29

Cytochrome P 450 enzymes (I) Flavin Monooxygenase Isoenzyme Alkohol Dehydrogenase Aldehyd Oxidase Monoamin Dehydrogenase (MAO) The redox activity is mediated by an iron porphyrin in the active center 30

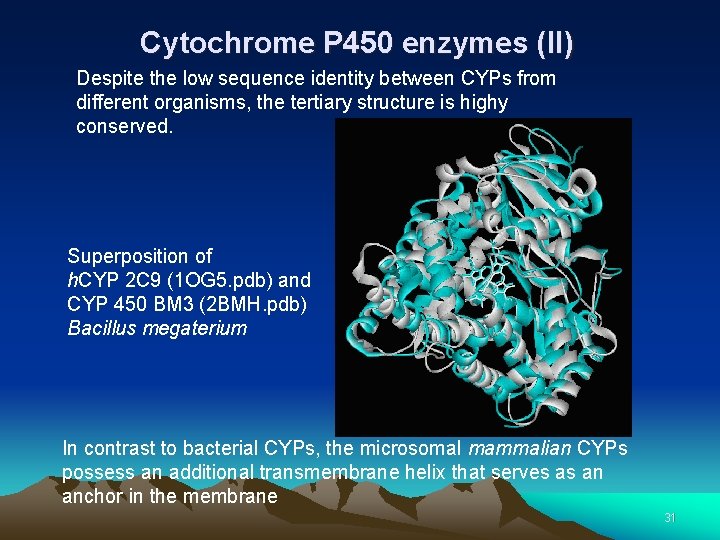
Cytochrome P 450 enzymes (II) Despite the low sequence identity between CYPs from different organisms, the tertiary structure is highy conserved. Superposition of h. CYP 2 C 9 (1 OG 5. pdb) and CYP 450 BM 3 (2 BMH. pdb) Bacillus megaterium In contrast to bacterial CYPs, the microsomal mammalian CYPs possess an additional transmembrane helix that serves as an anchor in the membrane 31

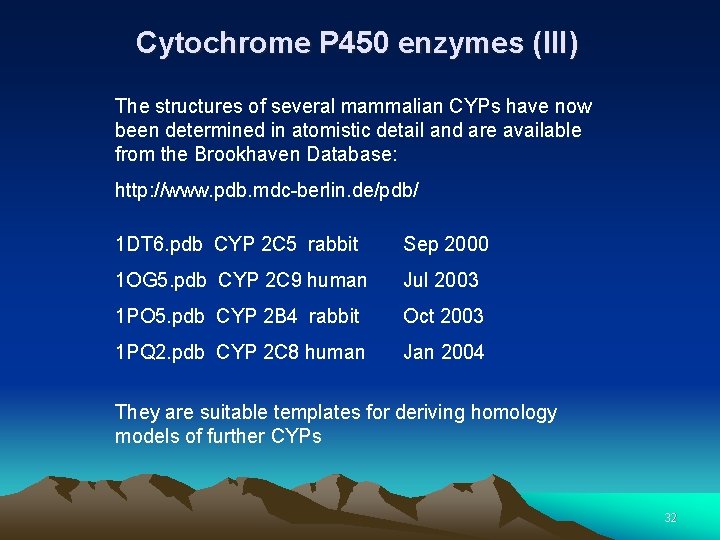
Cytochrome P 450 enzymes (III) The structures of several mammalian CYPs have now been determined in atomistic detail and are available from the Brookhaven Database: http: //www. pdb. mdc-berlin. de/pdb/ 1 DT 6. pdb CYP 2 C 5 rabbit Sep 2000 1 OG 5. pdb CYP 2 C 9 human Jul 2003 1 PO 5. pdb CYP 2 B 4 rabbit Oct 2003 1 PQ 2. pdb CYP 2 C 8 human Jan 2004 They are suitable templates for deriving homology models of further CYPs 32

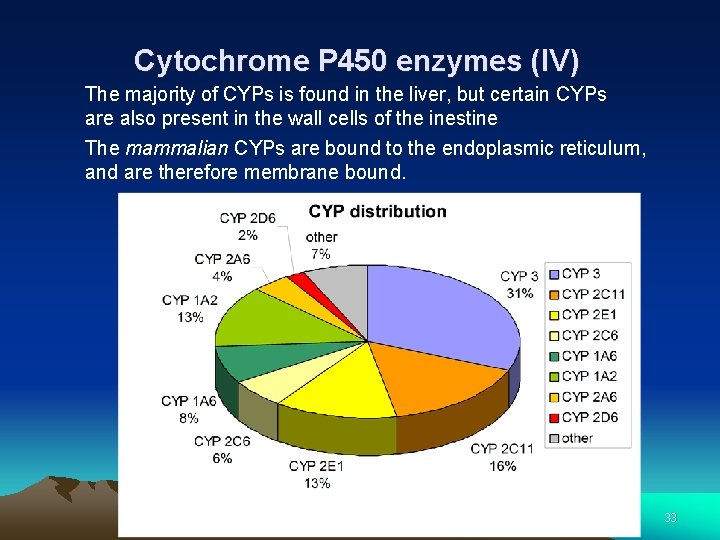
Cytochrome P 450 enzymes (IV) The majority of CYPs is found in the liver, but certain CYPs are also present in the wall cells of the inestine The mammalian CYPs are bound to the endoplasmic reticulum, and are therefore membrane bound. 33

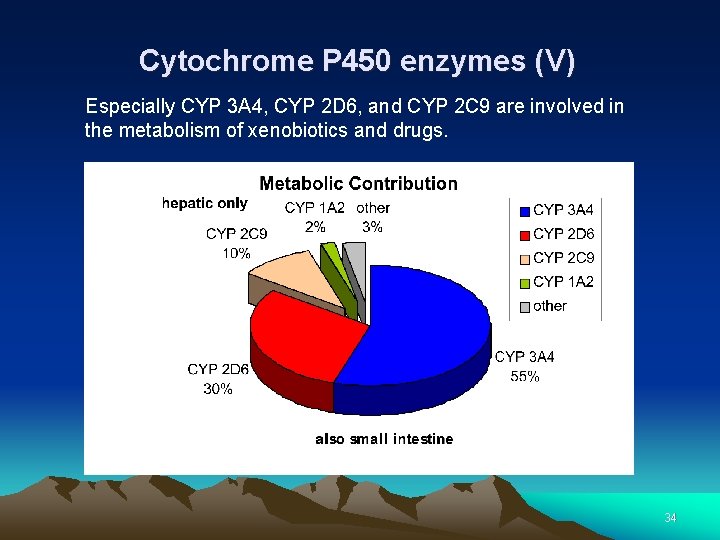
Cytochrome P 450 enzymes (V) Especially CYP 3 A 4, CYP 2 D 6, and CYP 2 C 9 are involved in the metabolism of xenobiotics and drugs. 34

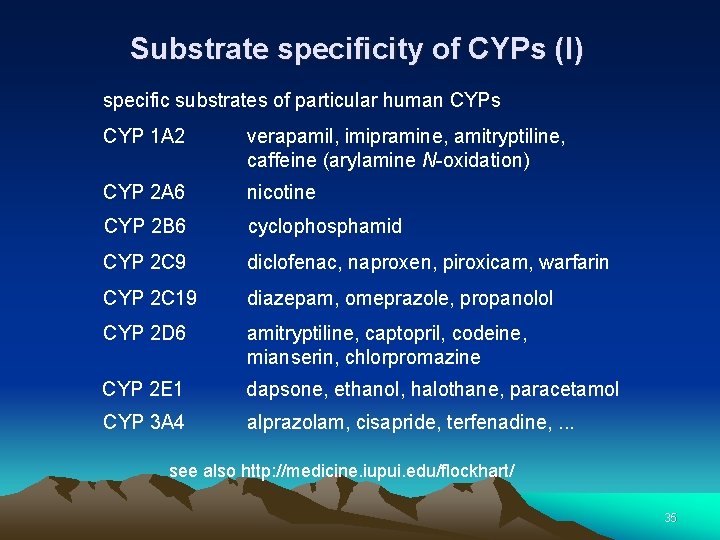
Substrate specificity of CYPs (I) specific substrates of particular human CYPs CYP 1 A 2 verapamil, imipramine, amitryptiline, caffeine (arylamine N-oxidation) CYP 2 A 6 nicotine CYP 2 B 6 cyclophosphamid CYP 2 C 9 diclofenac, naproxen, piroxicam, warfarin CYP 2 C 19 diazepam, omeprazole, propanolol CYP 2 D 6 amitryptiline, captopril, codeine, mianserin, chlorpromazine CYP 2 E 1 dapsone, ethanol, halothane, paracetamol CYP 3 A 4 alprazolam, cisapride, terfenadine, . . . see also http: //medicine. iupui. edu/flockhart/ 35

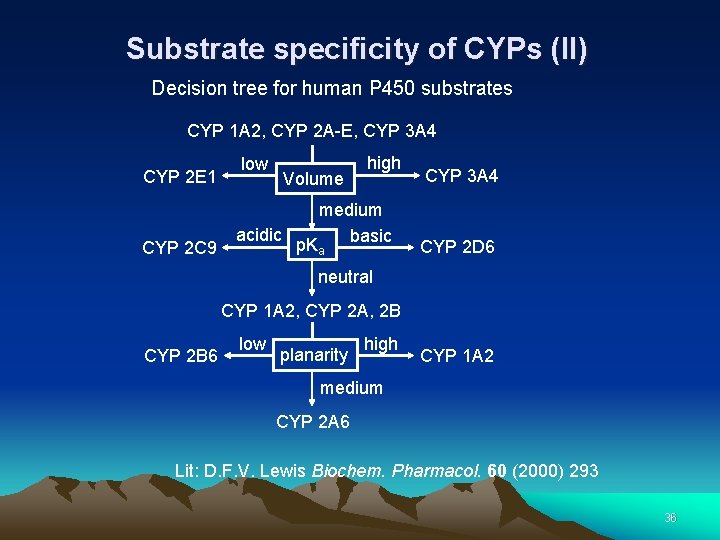
Substrate specificity of CYPs (II) Decision tree for human P 450 substrates CYP 1 A 2, CYP 2 A-E, CYP 3 A 4 CYP 2 E 1 CYP 2 C 9 low Volume high medium acidic basic p. K a CYP 3 A 4 CYP 2 D 6 neutral CYP 1 A 2, CYP 2 A, 2 B CYP 2 B 6 low planarity high CYP 1 A 2 medium CYP 2 A 6 Lit: D. F. V. Lewis Biochem. Pharmacol. 60 (2000) 293 36

Cytochrome P 450 polymorphisms „Every human differs (more or less) “ The phenotype can be distinguished by the actual activity or the amount of the expressed CYP enzyme. The genotype, however, is determined by the individual DNA sequence. Human: two sets of chromosomes That means: The same genotype enables different phenotypes Depending on the metabolic activity, three major catagories of metabolizers are separated: extensive metabolizer (normal), poor metabolizer, and ultra-rapid metabolizer (increased metabolism of xenobiotics) Lit: K. Nagata et al. Drug Metabol. Pharmacokin 3 (2002) 167 37

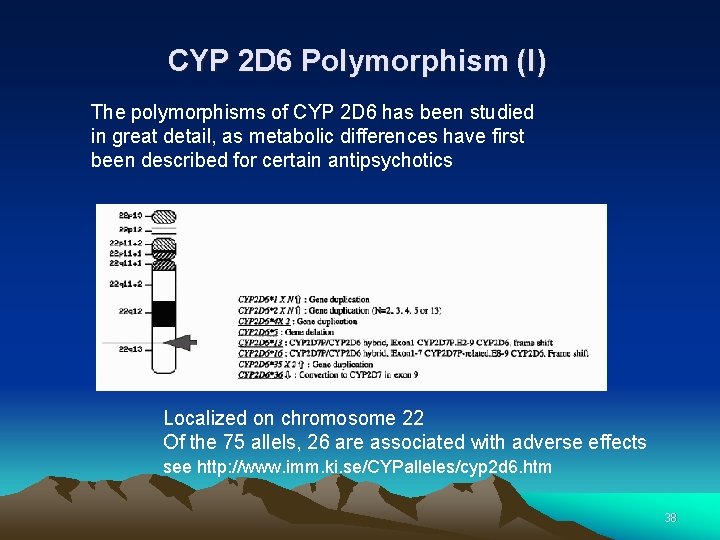
CYP 2 D 6 Polymorphism (I) The polymorphisms of CYP 2 D 6 has been studied in great detail, as metabolic differences have first been described for certain antipsychotics Localized on chromosome 22 Of the 75 allels, 26 are associated with adverse effects see http: //www. imm. ki. se/CYPalleles/cyp 2 d 6. htm 38

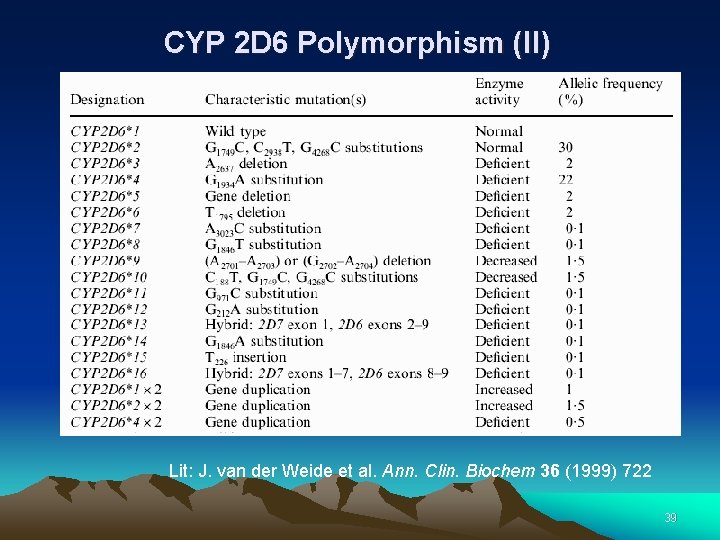
CYP 2 D 6 Polymorphism (II) Lit: J. van der Weide et al. Ann. Clin. Biochem 36 (1999) 722 39

CYP 2 D 6 Polymorphism (III) MGLEALVPLAVIVAIFLLLVDLMHRRQRWAARYPPGPLPLPGLGNLLHVDFQNTPYCFDQ poor debrisoquine metabolism S R impaired mechanism of sparteine LRRRFGDVFSLQLAWTPVVVLNGLAAVREALVTHGEDTADRPPVPITQILGFGPRSQGVF poor debrisoquine metabolism I LARYGPAWREQRRFSVSTLRNLGLGKKSLEQWVTEEAACLCAAFANHSGRPFRPNGLLDK poor debrisoquine metabolism R AVSNVIASLTCGRRFEYDDPRFLRLLDLAQEGLKEESGFLREVLNAVPVLLHIPALAGKV LRFQKAFLTQLDELLTEHRMTWDPAQPPRDLTEAFLAEMEKAKGNPESSFNDENLRIVVA missing in CYP 2 D 6*9 allele DLFSAGMVTTSTTLAWGLLLMILHPDVQRRVQQEIDDVIGQVRRPEMGDQAHMPYTTAVI P loss of activity in CYP 2 D 6*7 HEVQRFGDIVPLGMTHMTSRDIEVQGFRIPKGTTLITNLSSVLKDEAVWEKPFRFHPEHF LDAQGHFVKPEAFLPFSAGRRACLGEPLARMELFLFFTSLLQHFSFSVPTGQPRPSHHGV FAFLVSPSPYELCAVPR T impaired metabolism of sparteine in alleles 2, 10, 12, 14 and 17 of CYP 2 D 6 see http: //www. expasy. org/cgi-bin/niceprot. pl? P 10635 40

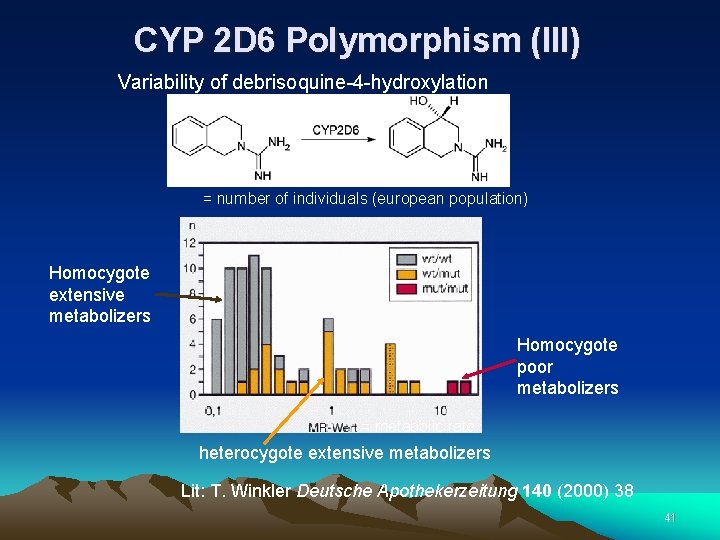
CYP 2 D 6 Polymorphism (III) Variability of debrisoquine-4 -hydroxylation = number of individuals (european population) Homocygote extensive metabolizers Homocygote poor metabolizers = metabolic rate heterocygote extensive metabolizers Lit: T. Winkler Deutsche Apothekerzeitung 140 (2000) 38 41

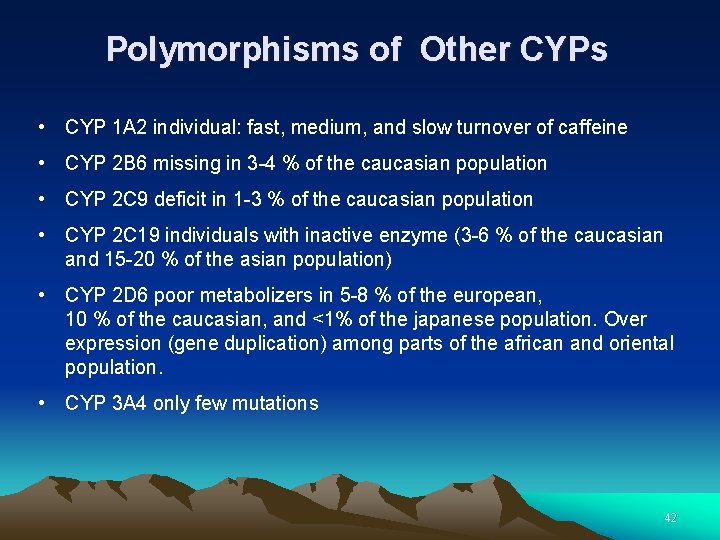
Polymorphisms of Other CYPs • CYP 1 A 2 individual: fast, medium, and slow turnover of caffeine • CYP 2 B 6 missing in 3 -4 % of the caucasian population • CYP 2 C 9 deficit in 1 -3 % of the caucasian population • CYP 2 C 19 individuals with inactive enzyme (3 -6 % of the caucasian and 15 -20 % of the asian population) • CYP 2 D 6 poor metabolizers in 5 -8 % of the european, 10 % of the caucasian, and <1% of the japanese population. Over expression (gene duplication) among parts of the african and oriental population. • CYP 3 A 4 only few mutations 42

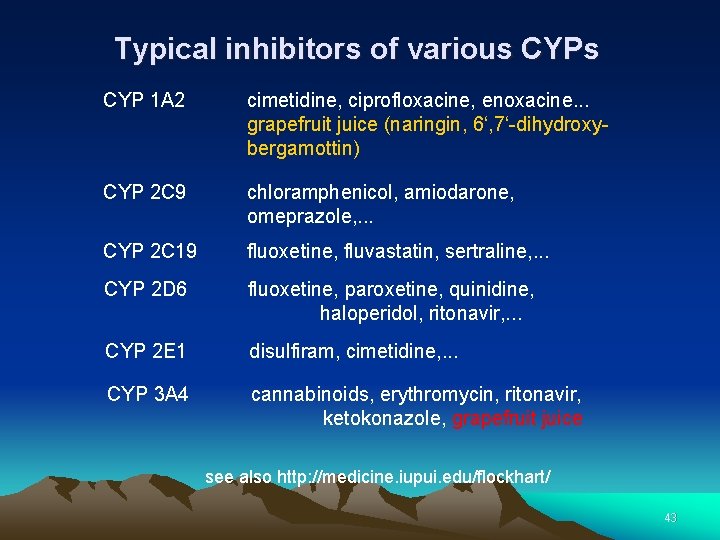
Typical inhibitors of various CYP 1 A 2 cimetidine, ciprofloxacine, enoxacine. . . grapefruit juice (naringin, 6‘, 7‘-dihydroxybergamottin) CYP 2 C 9 chloramphenicol, amiodarone, omeprazole, . . . CYP 2 C 19 fluoxetine, fluvastatin, sertraline, . . . CYP 2 D 6 fluoxetine, paroxetine, quinidine, haloperidol, ritonavir, . . . CYP 2 E 1 disulfiram, cimetidine, . . . CYP 3 A 4 cannabinoids, erythromycin, ritonavir, ketokonazole, grapefruit juice see also http: //medicine. iupui. edu/flockhart/ 43

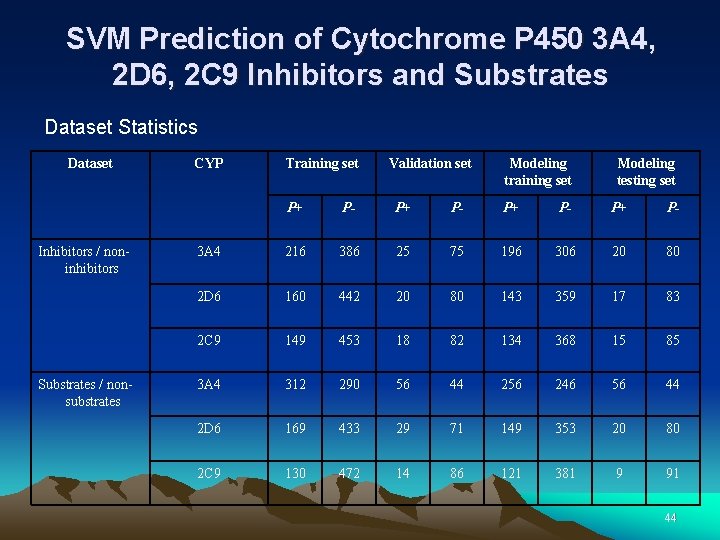
SVM Prediction of Cytochrome P 450 3 A 4, 2 D 6, 2 C 9 Inhibitors and Substrates Dataset Statistics Dataset Inhibitors / noninhibitors Substrates / nonsubstrates CYP Training set Validation set Modeling training set Modeling testing set P+ P- 3 A 4 216 386 25 75 196 306 20 80 2 D 6 160 442 20 80 143 359 17 83 2 C 9 149 453 18 82 134 368 15 85 3 A 4 312 290 56 44 256 246 56 44 2 D 6 169 433 29 71 149 353 20 80 2 C 9 130 472 14 86 121 381 9 91 44

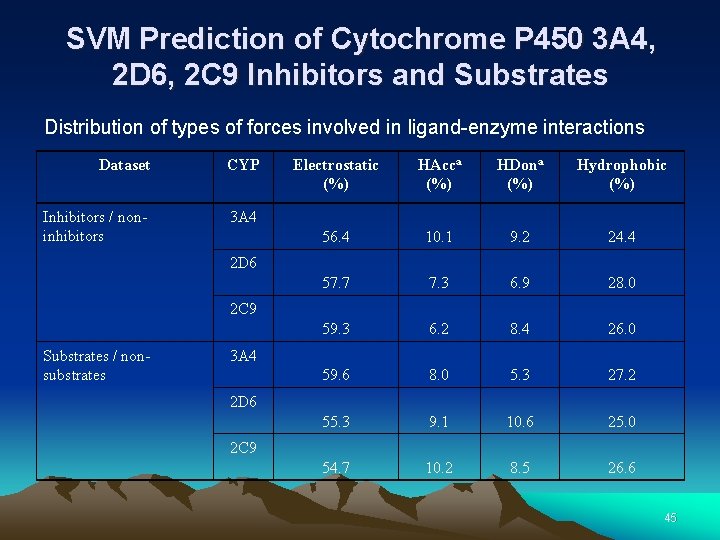
SVM Prediction of Cytochrome P 450 3 A 4, 2 D 6, 2 C 9 Inhibitors and Substrates Distribution of types of forces involved in ligand-enzyme interactions Dataset CYP Inhibitors / noninhibitors 3 A 4 Electrostatic (%) HAcca (%) HDona (%) Hydrophobic (%) 56. 4 10. 1 9. 2 24. 4 57. 7 7. 3 6. 9 28. 0 59. 3 6. 2 8. 4 26. 0 59. 6 8. 0 5. 3 27. 2 55. 3 9. 1 10. 6 25. 0 54. 7 10. 2 8. 5 26. 6 2 D 6 2 C 9 Substrates / nonsubstrates 3 A 4 2 D 6 2 C 9 45

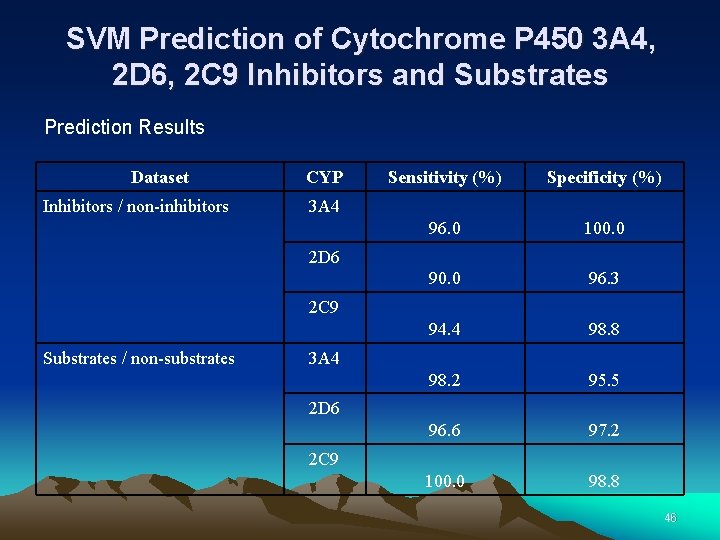
SVM Prediction of Cytochrome P 450 3 A 4, 2 D 6, 2 C 9 Inhibitors and Substrates Prediction Results Dataset Inhibitors / non-inhibitors CYP Sensitivity (%) Specificity (%) 96. 0 100. 0 96. 3 94. 4 98. 8 98. 2 95. 5 96. 6 97. 2 100. 0 98. 8 3 A 4 2 D 6 2 C 9 Substrates / non-substrates 3 A 4 2 D 6 2 C 9 46