CZ 3253 Computer Aided Drug design Lecture 7













- Slides: 40

CZ 3253: Computer Aided Drug design Lecture 7: Drug Design Methods II: SVM Prof. Chen Yu Zong Tel: 6874 -6877 Email: csccyz@nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, National University of Singapore

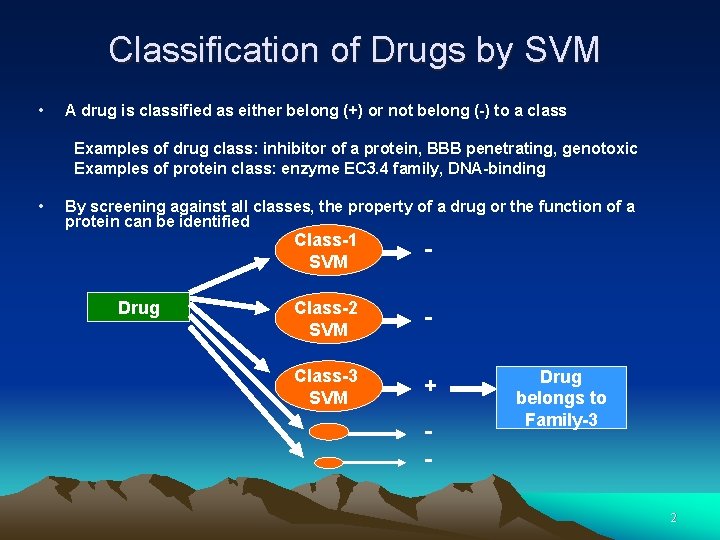
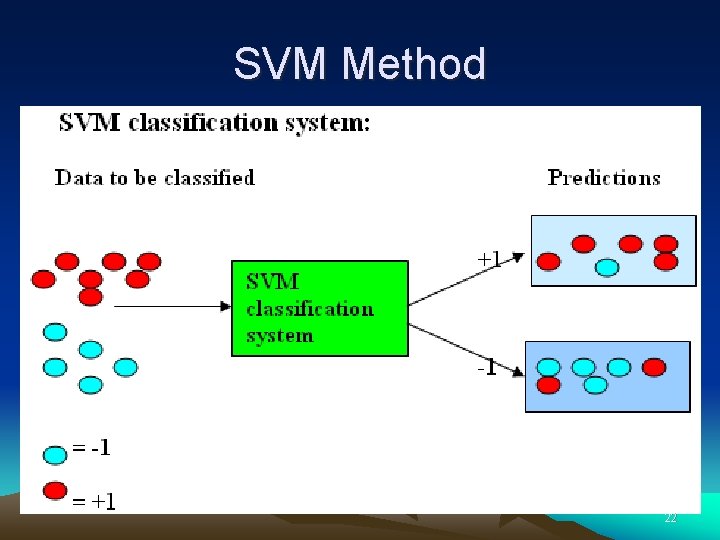
Classification of Drugs by SVM • A drug is classified as either belong (+) or not belong (-) to a class Examples of drug class: inhibitor of a protein, BBB penetrating, genotoxic Examples of protein class: enzyme EC 3. 4 family, DNA-binding • By screening against all classes, the property of a drug or the function of a protein can be identified Drug Class-1 SVM - Class-2 SVM - Class-3 SVM + - Drug belongs to Family-3 2

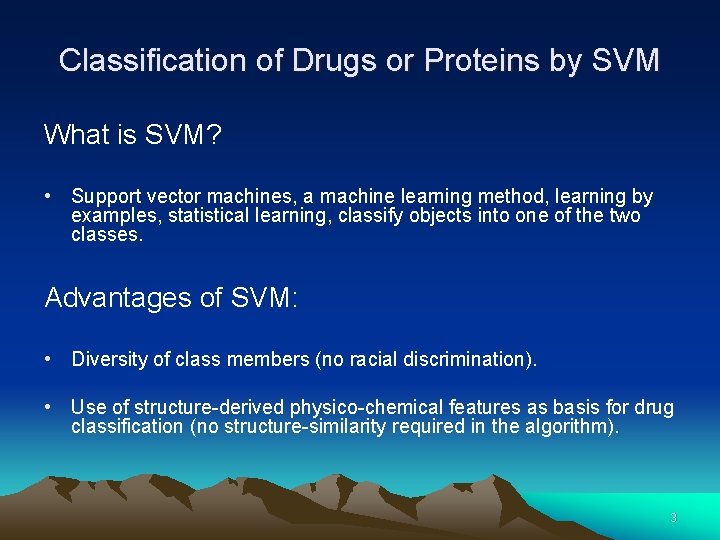
Classification of Drugs or Proteins by SVM What is SVM? • Support vector machines, a machine learning method, learning by examples, statistical learning, classify objects into one of the two classes. Advantages of SVM: • Diversity of class members (no racial discrimination). • Use of structure-derived physico-chemical features as basis for drug classification (no structure-similarity required in the algorithm). 3

SVM References • C. Burges, "A tutorial on support vector machines for pattern recognition", Data Mining and Knowledge Discovery, Kluwer Academic Publishers, 1998 (on-line). • R. Duda, P. Hart, and D. Stork, Pattern Classification, John-Wiley, 2 nd edition, 2001 (section 5. 11, hard-copy). • S. Gong et al. Dynamic Vision: From Images to Face Recognition, Imperial College Pres, 2001 (sections 3. 6. 2, 3. 7. 2, hard copy). • Online lecture notes (http: //www. cs. unr. edu/~bebis/Math. Methods/SVM/lecture. pdf ) • Publications of SVM drug prediction: – J. Chem. Inf. Comput. Sci. 44, 1630 (2004) – J. Chem. Inf. Comput. Sci. 44, 1497 (2004) – Toxicol. Sci. 79, 170 (2004). 4

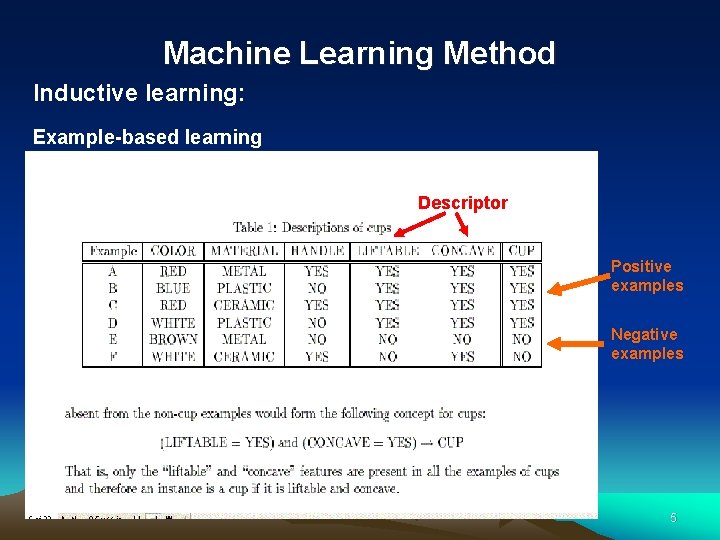
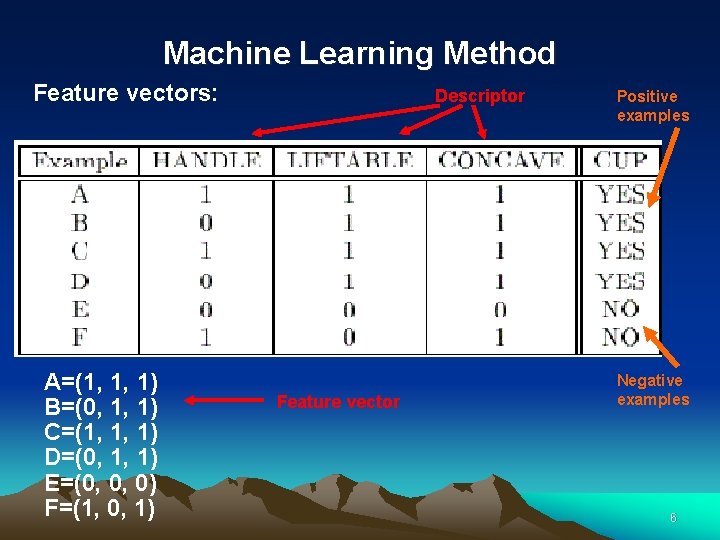
Machine Learning Method Inductive learning: Example-based learning Descriptor Positive examples Negative examples 5

Machine Learning Method Feature vectors: A=(1, 1, 1) B=(0, 1, 1) C=(1, 1, 1) D=(0, 1, 1) E=(0, 0, 0) F=(1, 0, 1) Descriptor Feature vector Positive examples Negative examples 6

SVM Method Feature vectors in input space: Z Input space Feature vector A=(1, 1, 1) B=(0, 1, 1) C=(1, 1, 1) D=(0, 1, 1) E=(0, 0, 0) F=(1, 0, 1) F E A B Y X 7

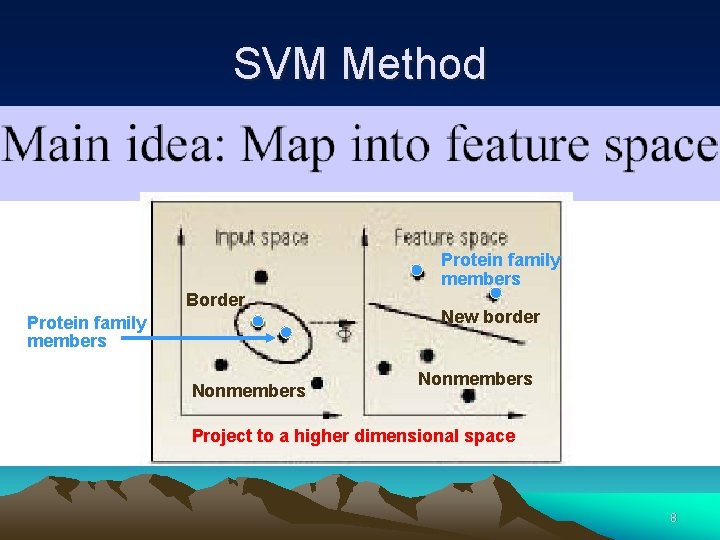
SVM Method Protein family members Border Protein family members Nonmembers New border Nonmembers Project to a higher dimensional space 8

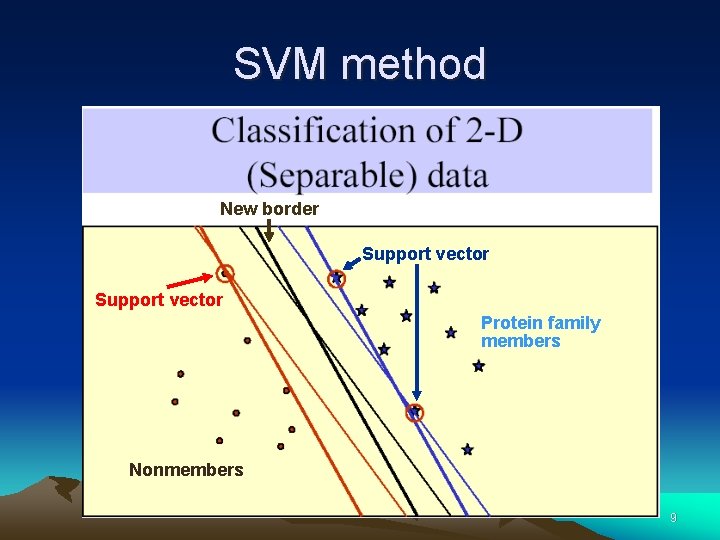
SVM method New border Support vector Protein family members Nonmembers 9

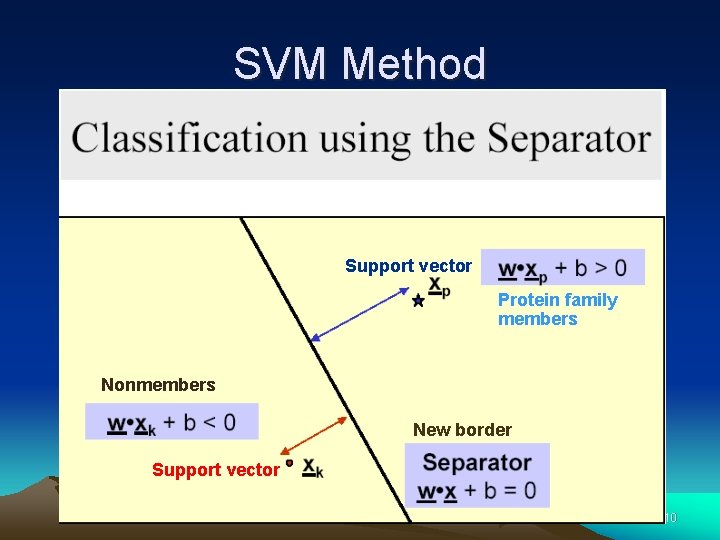
SVM Method Support vector Protein family members Nonmembers New border Support vector 10

Best Linear Separator? 11

Best Linear Separator? 12

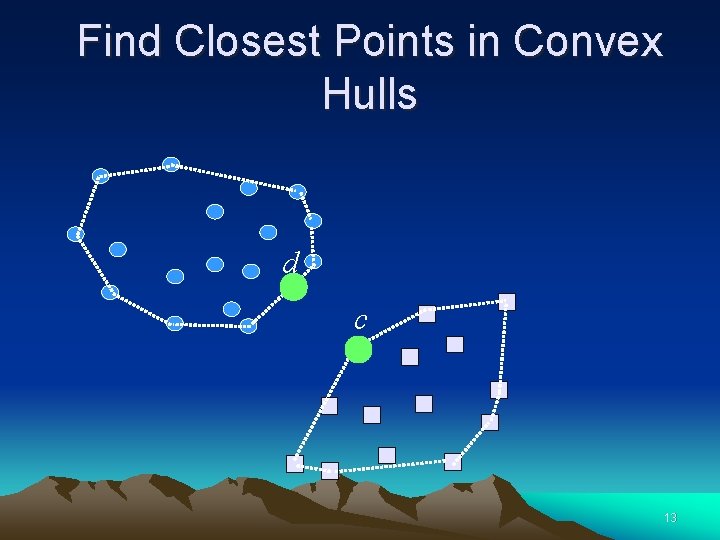
Find Closest Points in Convex Hulls d c 13

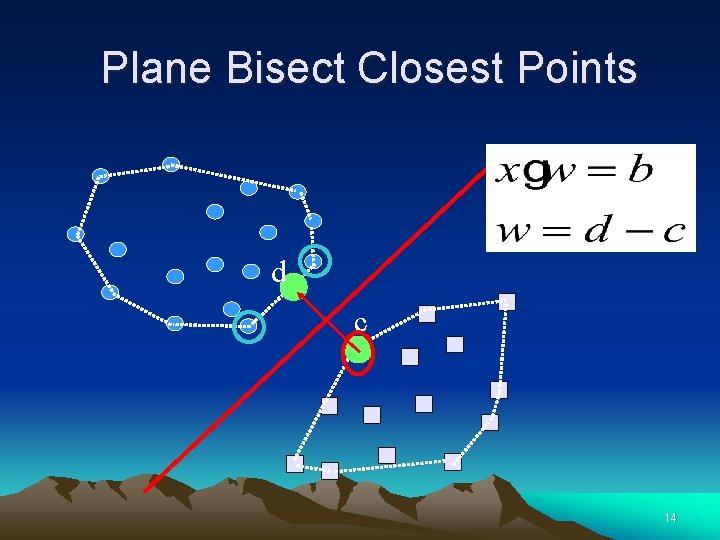
Plane Bisect Closest Points d c 14

Find using quadratic program Many existing and new solvers. 15

Best Linear Separator: Supporting Plane Method Maximize distance Between two paral supporting planes Distance = “Margin” = 16

Best Linear Separator? 17

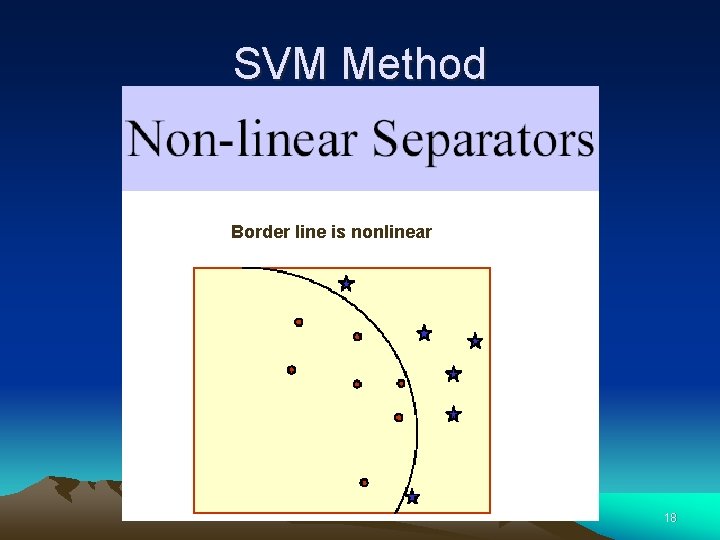
SVM Method Border line is nonlinear 18

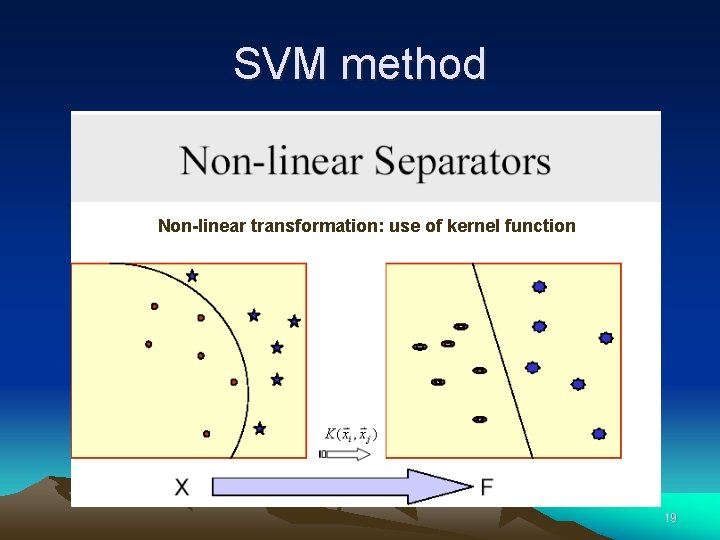
SVM method Non-linear transformation: use of kernel function 19

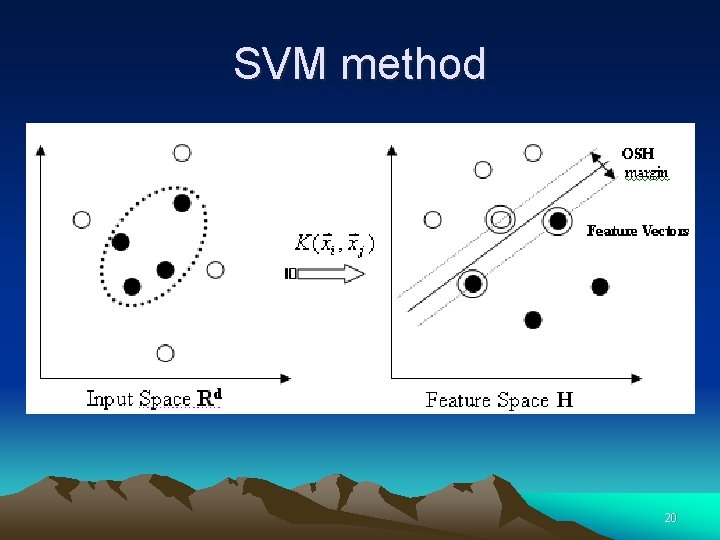
SVM method Non-linear transformation 20

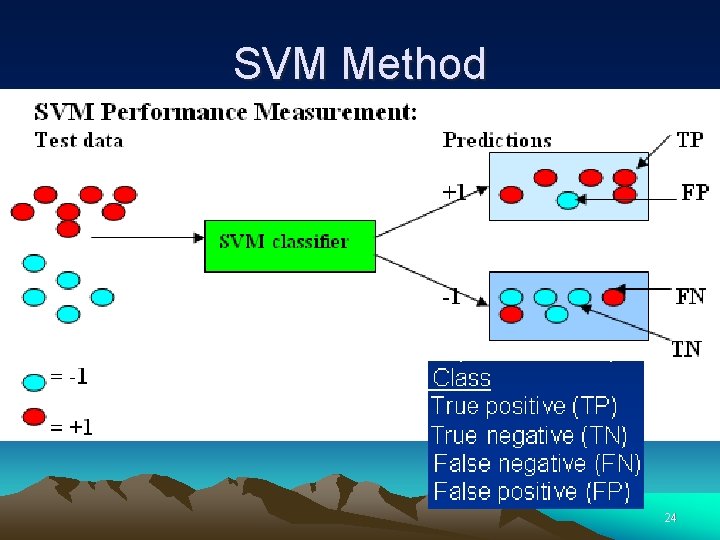
SVM Method 21

SVM Method 22

SVM Method 23

SVM Method 24

SVM for Classification of Drugs How to represent a drug? • Each structure represented by specific feature vector assembled from structural, physico-chemical properties: – Simple molecular properties (molecular weight, no. of rotatable bonds etc. 18 in total) – Molecular Connectivity and shape (28 in total) – Electro-topological state polarity (84 in total) – Quantum chemical properties (electric charge, polaritability etc. 13 in total) – Geometrical properties (molecular size vector, van der Waals volume, molecular surface etc. 16 in total) J. Chem. Inf. Comput. Sci. 44, 1630 (2004) J. Chem. Inf. Comput. Sci. 44, 1497 (2004) Toxicol. Sci. 79, 170 (2004). 25

SVM Feature Selection CACO 2 - 718 descriptors Average of 10 Models Q 2 is MSE scaled by variance: = (mean square error) / Test Q 2 =. 7073 (true variance) 26

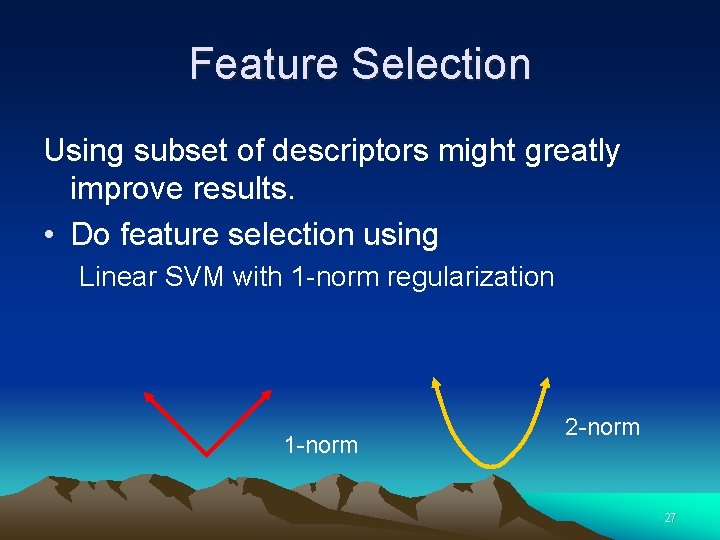
Feature Selection Using subset of descriptors might greatly improve results. • Do feature selection using Linear SVM with 1 -norm regularization 1 -norm 27

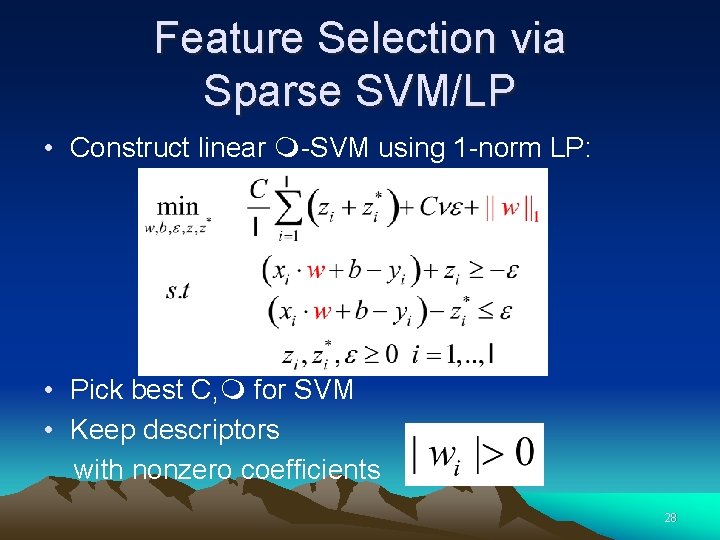
Feature Selection via Sparse SVM/LP • Construct linear -SVM using 1 -norm LP: • Pick best C, for SVM • Keep descriptors with nonzero coefficients 28

Bagged Feature Selection Partition Training Data Training Set Validation Set Linear SVM Algorithm For Feature Selection Repeat B times Random Variable - r A Linear Regression Model Bag B Models and Obtain Subset of Features 29

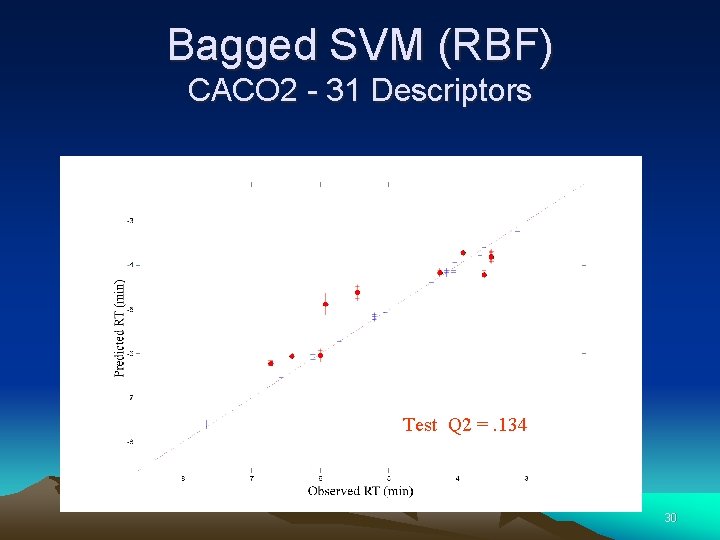
Bagged SVM (RBF) CACO 2 - 31 Descriptors Test Q 2 =. 134 30

Starplot Caco 2 - 31 Descriptors ABSDRN 6 a. don KB 54 SMR. VSA 2 BNP 8 DRNB 10 DRNB 00 KB 11 PEOE. VSA. 4 PEOE. VSA. FPPOS ANGLEB 45 PIPB 53 Slog. P. VSA 6 apol ABSFUKMIN PIPB 04 PEOE. VSA. FPOL PIPMAX PEOE. VSA. FHYD PEOE. VSA. PPOS EP 2 PEOE. VSA. FNEG Slog. P. VSA 0 BNPB 31 FUKB 14 BNPB 50 Slog. P. VSA 9 pmi. Z BNPB 21 ABSKMIN SIKIA 31

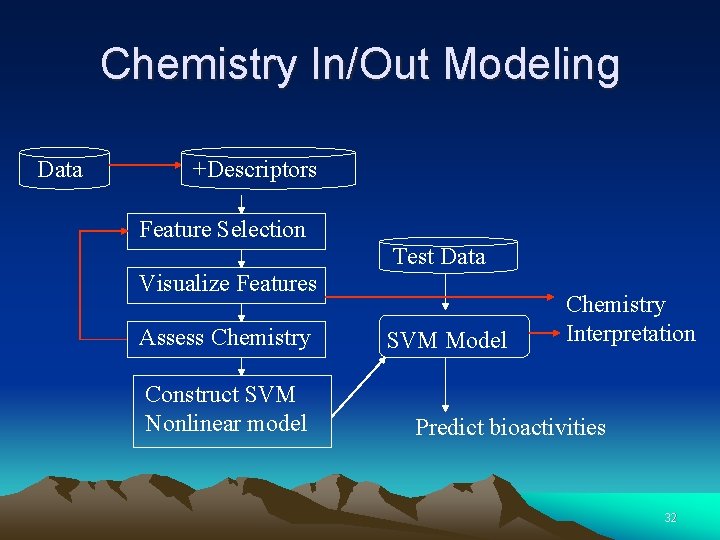
Chemistry In/Out Modeling Data +Descriptors Feature Selection Test Data Visualize Features Assess Chemistry Construct SVM Nonlinear model SVM Model Chemistry Interpretation Predict bioactivities 32

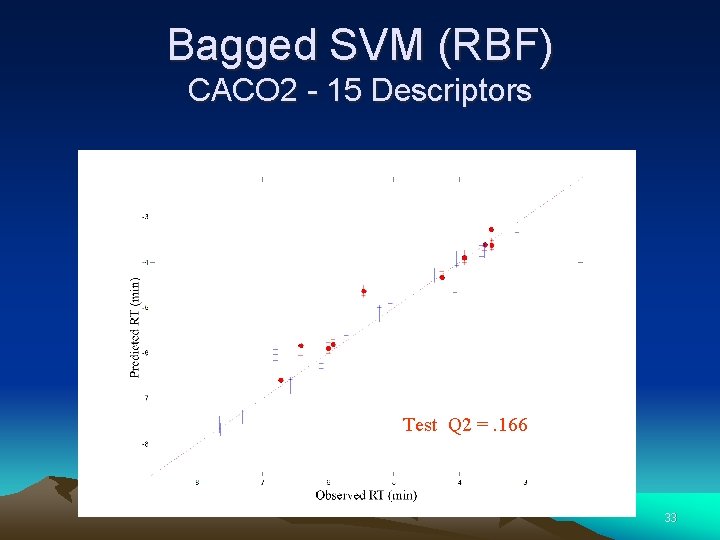
Bagged SVM (RBF) CACO 2 - 15 Descriptors Test Q 2 =. 166 33

CACO 2 – 15 Variables a. don DRNB 10 PEOE. VSA. FNEG BNPB 31 KB 54 ABSDRN 6 ABSKMIN FUKB 14 SMR. VSA 2 PEOE. VSA. FPPOS SIKIA Slog. P. VSA 0 ANGLEB 45 DRNB 00 pmi. Z 34

Chemical Insights • Hydrophobicity - a. don • SIZE and Shape ABSDRN 6, SMR. VSA 2, ANGLEB 45, Pmi. Z Large is bad. Flat is bad. Globular is good. • Polarity – PEOE. VSA. FPPOS, PEOE. VSA. FNEG: negative partial charge good. Correspond to conventional wisdom – rule of 5. 35

Hybrid TAE/SHAPE • Shape important overall factor – DRNB 10, DRNB 00: del rho dot N – BNP 31: bare nuclear potential – KB 54: kinetic energy descriptors very large lipophilic molecules don’t work – FUKB 14: Fukui Surface • • Interpretations difficult Point to chemistry challenges/hypotheses 36

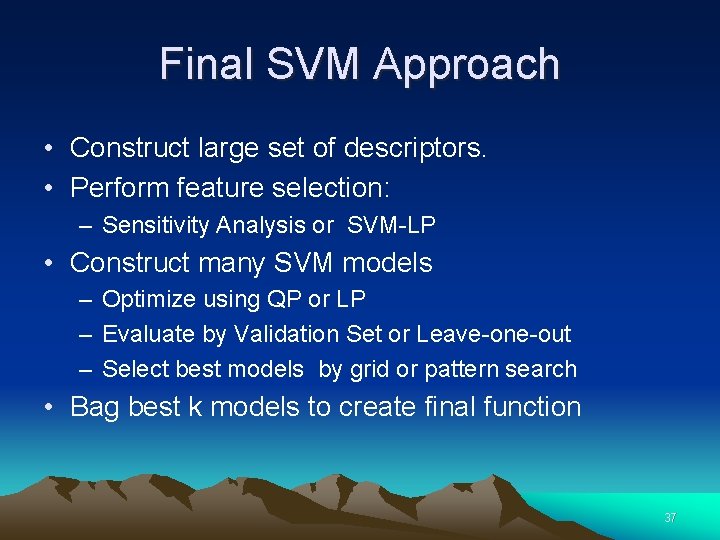
Final SVM Approach • Construct large set of descriptors. • Perform feature selection: – Sensitivity Analysis or SVM-LP • Construct many SVM models – Optimize using QP or LP – Evaluate by Validation Set or Leave-one-out – Select best models by grid or pattern search • Bag best k models to create final function 37

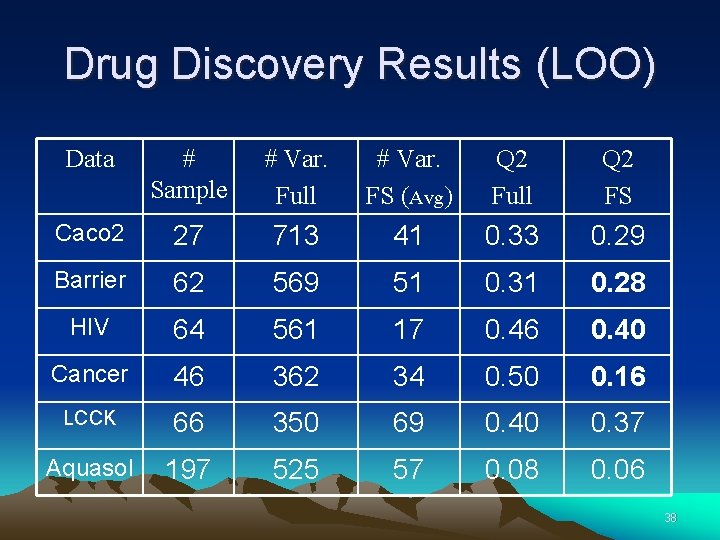
Drug Discovery Results (LOO) Data # Sample # Var. Full # Var. FS (Avg) Q 2 Full Q 2 FS Caco 2 27 713 41 0. 33 0. 29 Barrier 62 569 51 0. 31 0. 28 HIV 64 561 17 0. 46 0. 40 Cancer 46 362 34 0. 50 0. 16 LCCK 66 350 69 0. 40 0. 37 Aquasol 197 525 57 0. 08 0. 06 38

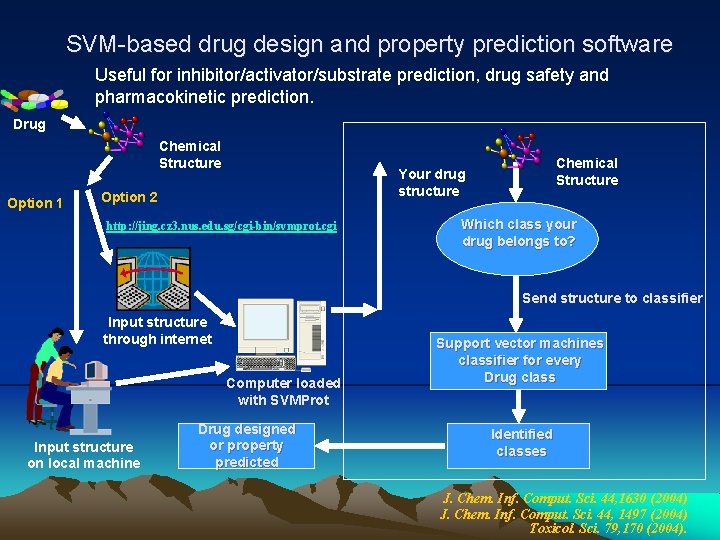
SVM-based drug design and property prediction software Useful for inhibitor/activator/substrate prediction, drug safety and pharmacokinetic prediction. Drug Chemical Structure Option 1 Chemical Structure Your drug structure Option 2 http: //jing. cz 3. nus. edu. sg/cgi-bin/svmprot. cgi Which class your drug belongs to? Send structure to classifier Input structure through internet Computer loaded with SVMProt Input structure on local machine Drug designed or property predicted Support vector machines classifier for every Drug class Identified classes J. Chem. Inf. Comput. Sci. 44, 1630 (2004) J. Chem. Inf. Comput. Sci. 44, 1497 (2004) Toxicol. Sci. 79, 170 (2004).

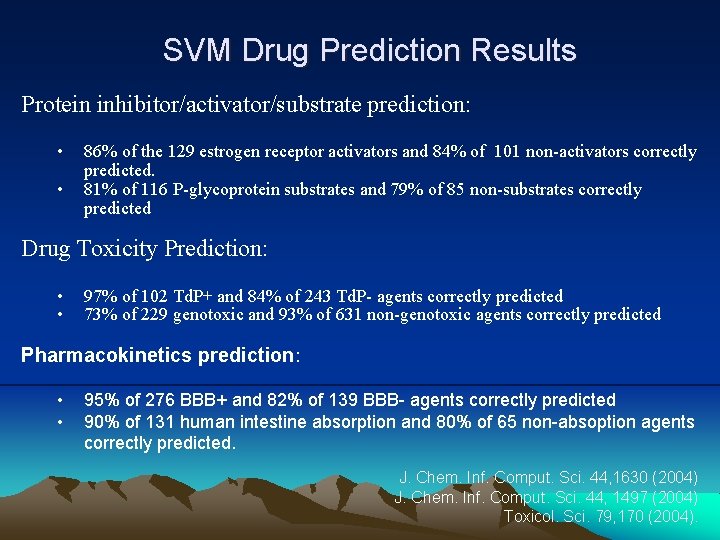
SVM Drug Prediction Results Protein inhibitor/activator/substrate prediction: • • 86% of the 129 estrogen receptor activators and 84% of 101 non-activators correctly predicted. 81% of 116 P-glycoprotein substrates and 79% of 85 non-substrates correctly predicted Drug Toxicity Prediction: • • 97% of 102 Td. P+ and 84% of 243 Td. P- agents correctly predicted 73% of 229 genotoxic and 93% of 631 non-genotoxic agents correctly predicted Pharmacokinetics prediction: • • 95% of 276 BBB+ and 82% of 139 BBB- agents correctly predicted 90% of 131 human intestine absorption and 80% of 65 non-absoption agents correctly predicted. J. Chem. Inf. Comput. Sci. 44, 1630 (2004) J. Chem. Inf. Comput. Sci. 44, 1497 (2004) Toxicol. Sci. 79, 170 (2004).