CZ 3253 Computer Aided Drug design Lecture 4



- Slides: 49

CZ 3253: Computer Aided Drug design Lecture 4: Structural modeling of chemical molecules Prof. Chen Yu Zong Tel: 6874 -6877 Email: csccyz@nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, National University of Singapore

Formula for chemical compounds: Molecular formula: – Element(No. of atom). Element (No. of atom). . – Elements are placed in the order of C, N, H, O, and others. There can be exceptions to this rule. Examples: • Carbon dioxide CO 2 • Ammonia NH 3 • Octane C 8 H 18 2

Formula for chemical compounds: • Condensed formula – Formula reflects organization. – Members of the same functional group are placed together. • Example: 3

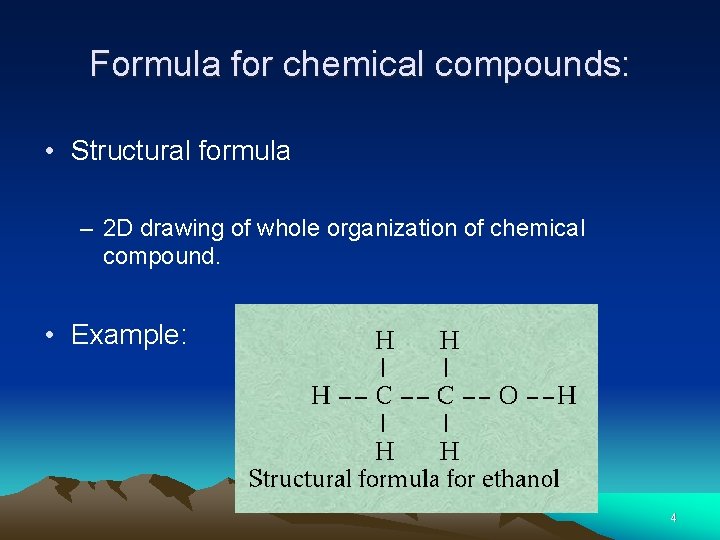
Formula for chemical compounds: • Structural formula – 2 D drawing of whole organization of chemical compound. • Example: 4

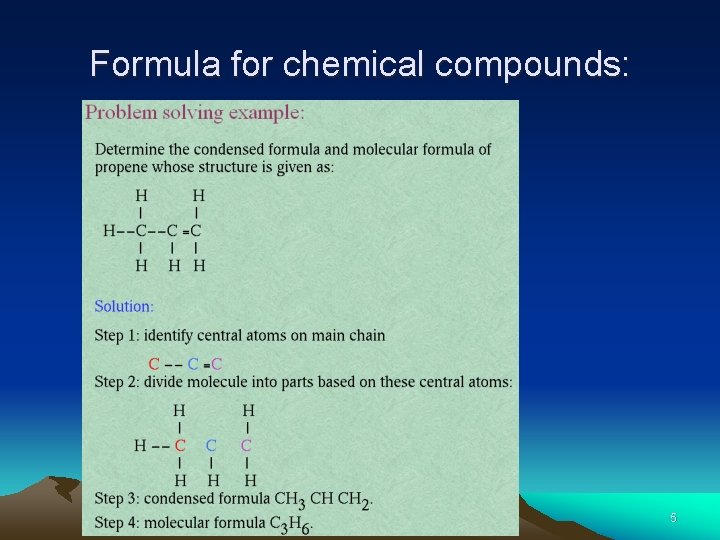
Formula for chemical compounds: 5

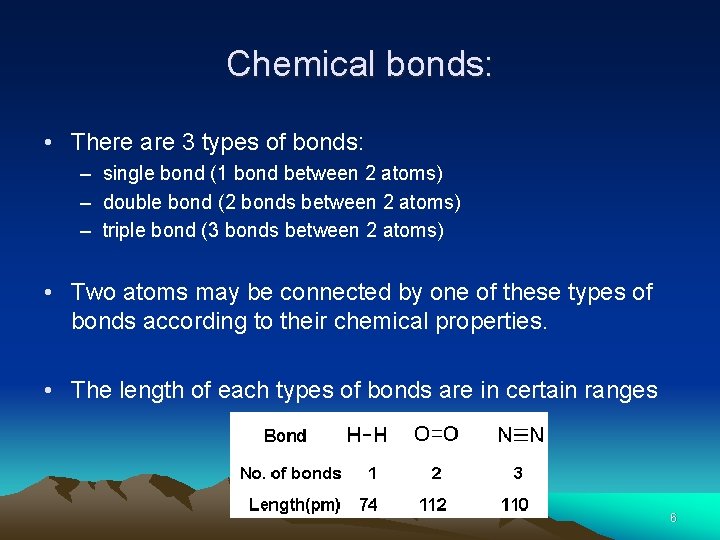
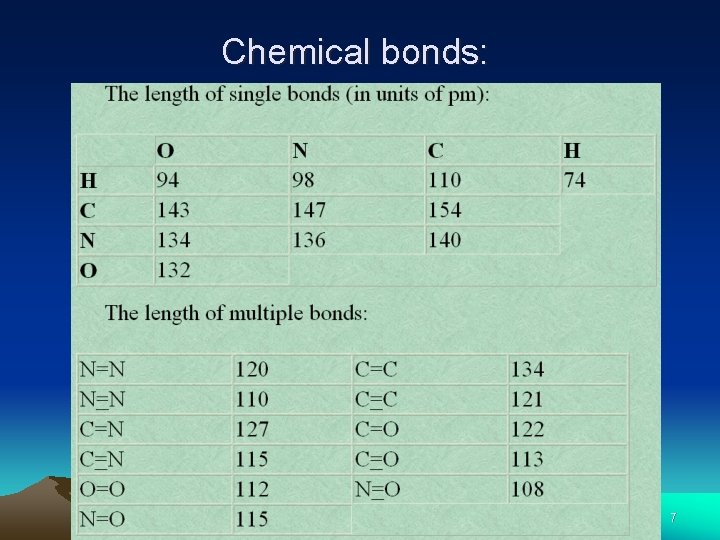
Chemical bonds: • There are 3 types of bonds: – single bond (1 bond between 2 atoms) – double bond (2 bonds between 2 atoms) – triple bond (3 bonds between 2 atoms) • Two atoms may be connected by one of these types of bonds according to their chemical properties. • The length of each types of bonds are in certain ranges 6

Chemical bonds: 7

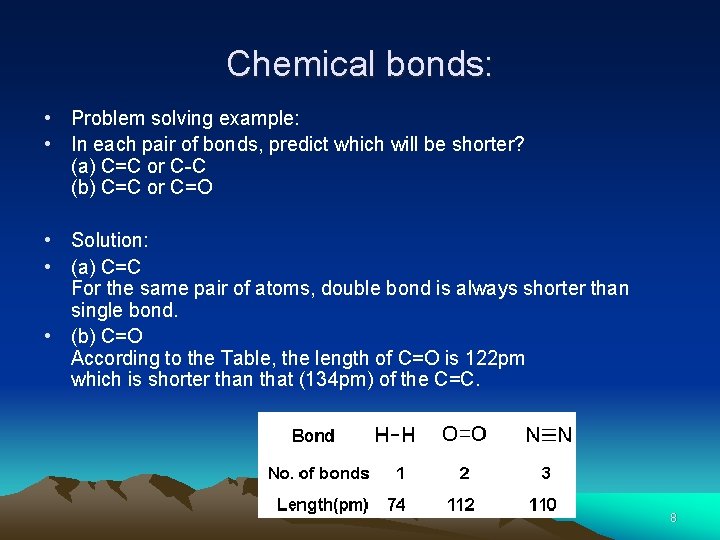
Chemical bonds: • Problem solving example: • In each pair of bonds, predict which will be shorter? (a) C=C or C-C (b) C=C or C=O • Solution: • (a) C=C For the same pair of atoms, double bond is always shorter than single bond. • (b) C=O According to the Table, the length of C=O is 122 pm which is shorter than that (134 pm) of the C=C. 8

Chemical Compounds: Stereochemistry • Three features: – Configuration (atom organization). – Conformation (atom spatial arrangement). – Shape (Surface landscape, steric packing) 9

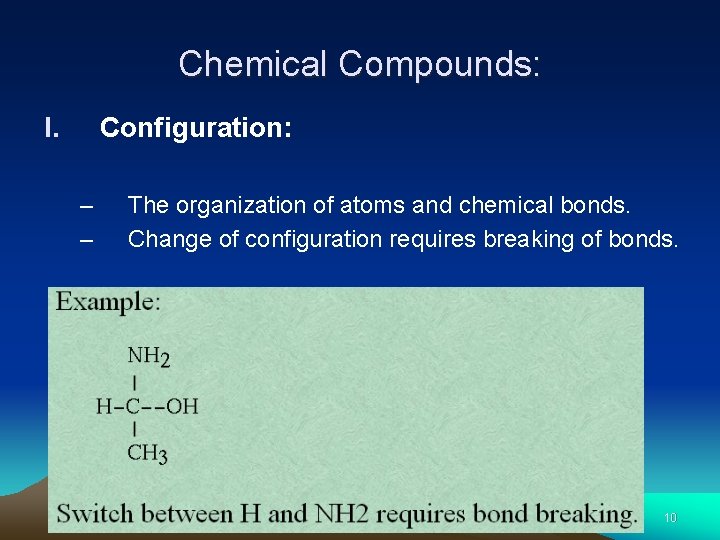
Chemical Compounds: I. Configuration: – – The organization of atoms and chemical bonds. Change of configuration requires breaking of bonds. 10

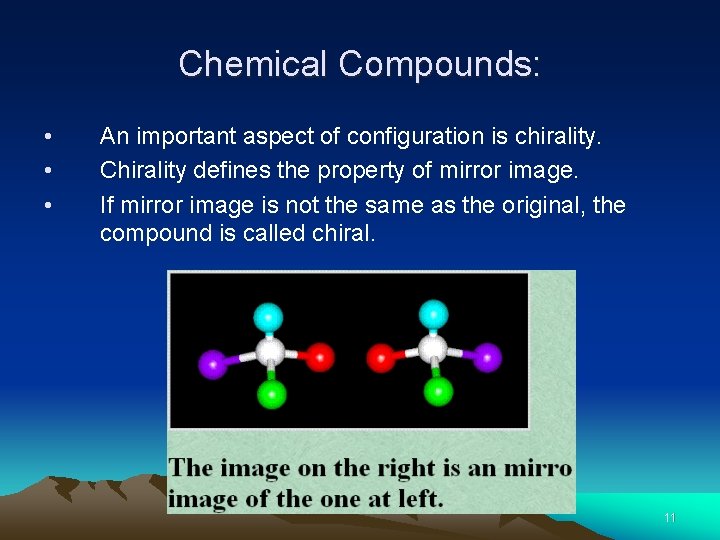
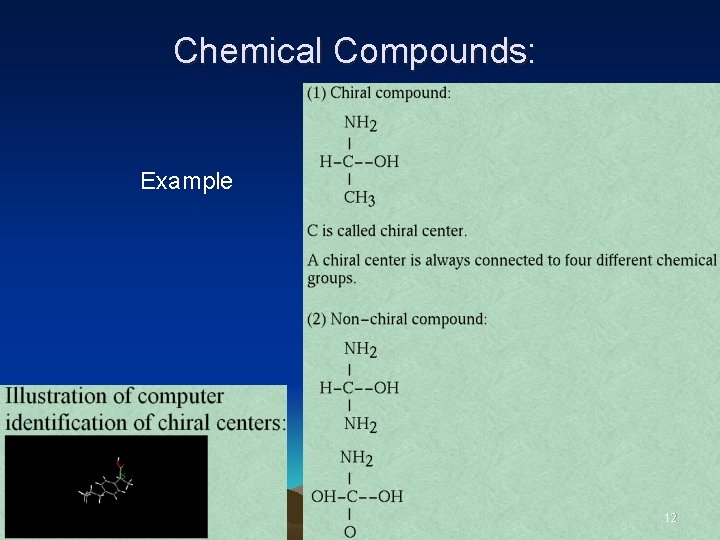
• • • Chemical Compounds: An important aspect of configuration is chirality. Chirality defines the property of mirror image. If mirror image is not the same as the original, the compound is called chiral. 11

Chemical Compounds: Example 12

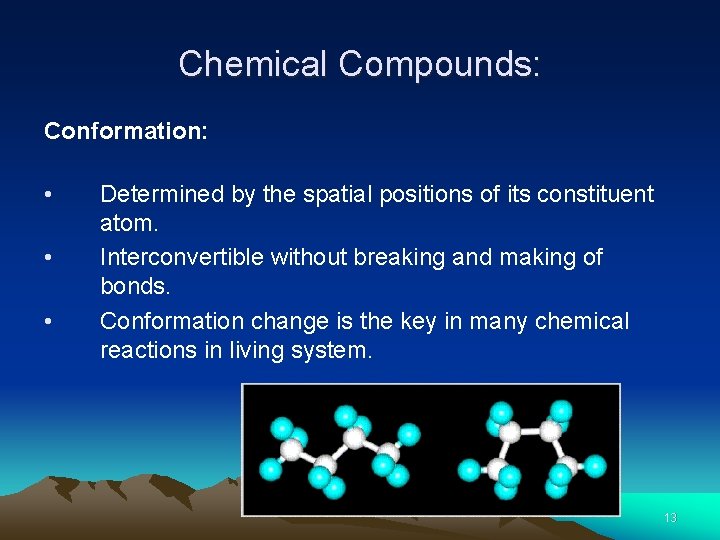
Chemical Compounds: Conformation: • Determined by the spatial positions of its constituent atom. • Interconvertible without breaking and making of bonds. • Conformation change is the key in many chemical reactions in living system. 13

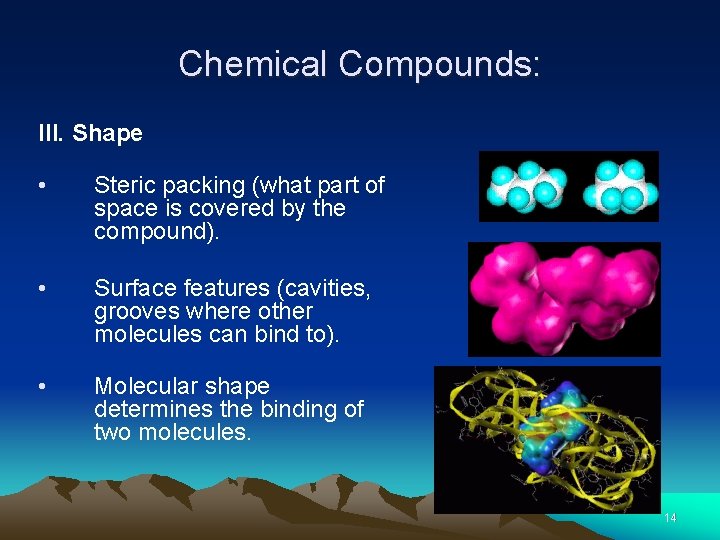
Chemical Compounds: III. Shape • Steric packing (what part of space is covered by the compound). • • Surface features (cavities, grooves where other molecules can bind to). Molecular shape determines the binding of two molecules. 14

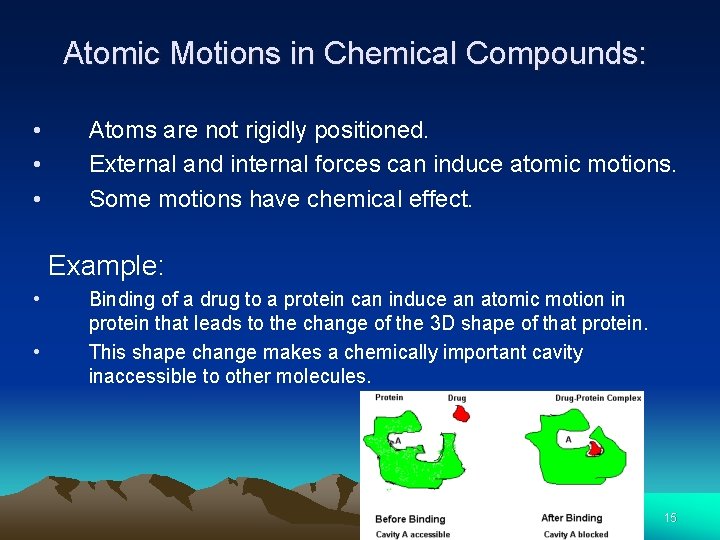
Atomic Motions in Chemical Compounds: • • • Atoms are not rigidly positioned. External and internal forces can induce atomic motions. Some motions have chemical effect. Example: • • Binding of a drug to a protein can induce an atomic motion in protein that leads to the change of the 3 D shape of that protein. This shape change makes a chemically important cavity inaccessible to other molecules. 15

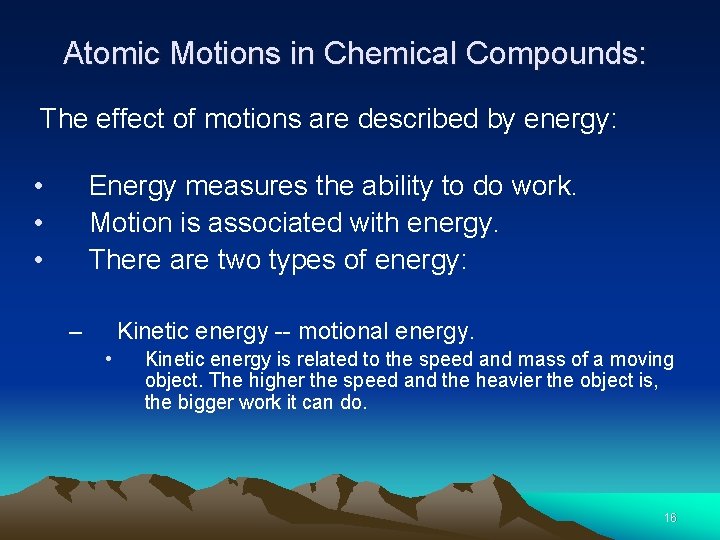
Atomic Motions in Chemical Compounds: The effect of motions are described by energy: • Energy measures the ability to do work. • Motion is associated with energy. • There are two types of energy: – Kinetic energy -- motional energy. • Kinetic energy is related to the speed and mass of a moving object. The higher the speed and the heavier the object is, the bigger work it can do. 16

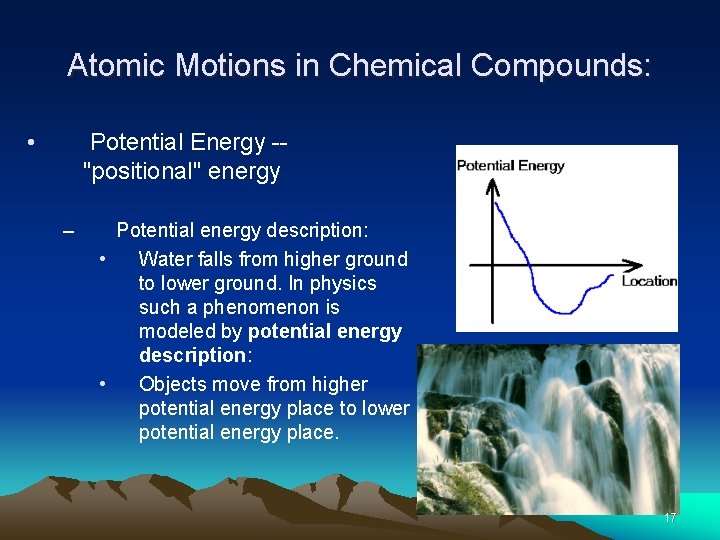
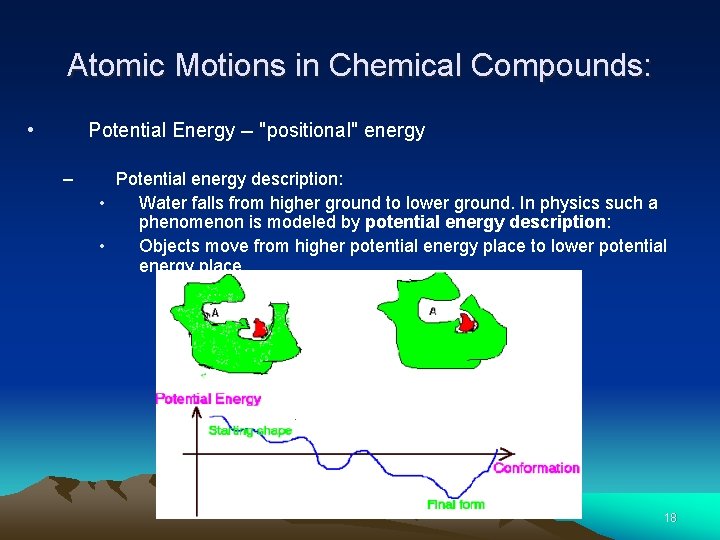
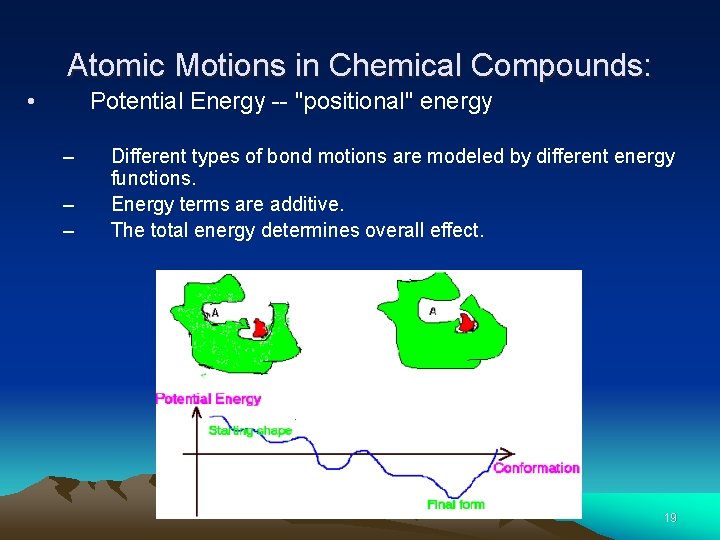
Atomic Motions in Chemical Compounds: • Potential Energy -- "positional" energy – Potential energy description: • Water falls from higher ground to lower ground. In physics such a phenomenon is modeled by potential energy description: • Objects move from higher potential energy place to lower potential energy place. 17

Atomic Motions in Chemical Compounds: • Potential Energy -- "positional" energy – Potential energy description: • Water falls from higher ground to lower ground. In physics such a phenomenon is modeled by potential energy description: • Objects move from higher potential energy place to lower potential energy place. 18

Atomic Motions in Chemical Compounds: • Potential Energy -- "positional" energy – – – Different types of bond motions are modeled by different energy functions. Energy terms are additive. The total energy determines overall effect. 19

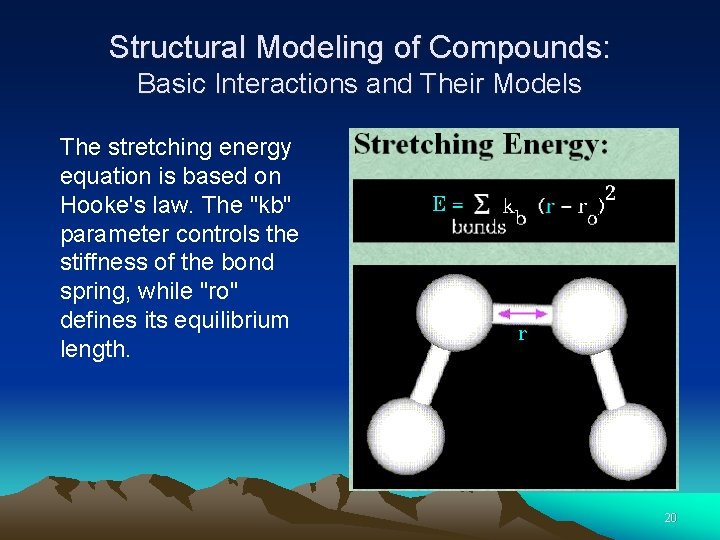
Structural Modeling of Compounds: Basic Interactions and Their Models The stretching energy equation is based on Hooke's law. The "kb" parameter controls the stiffness of the bond spring, while "ro" defines its equilibrium length. 20

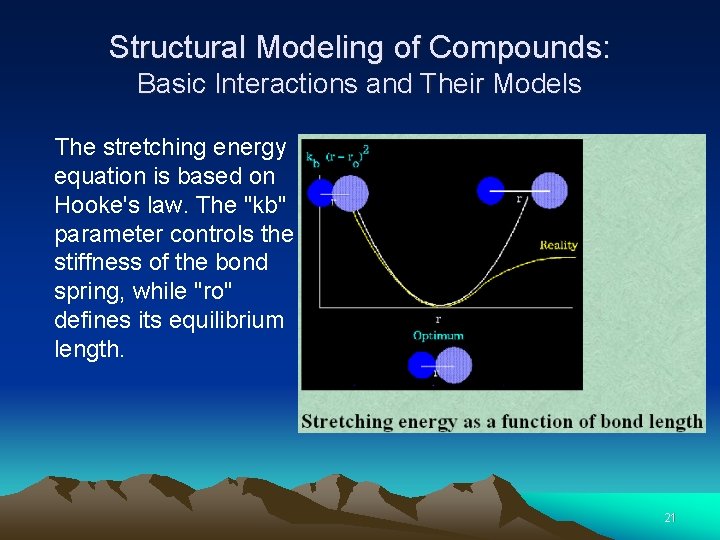
Structural Modeling of Compounds: Basic Interactions and Their Models The stretching energy equation is based on Hooke's law. The "kb" parameter controls the stiffness of the bond spring, while "ro" defines its equilibrium length. 21

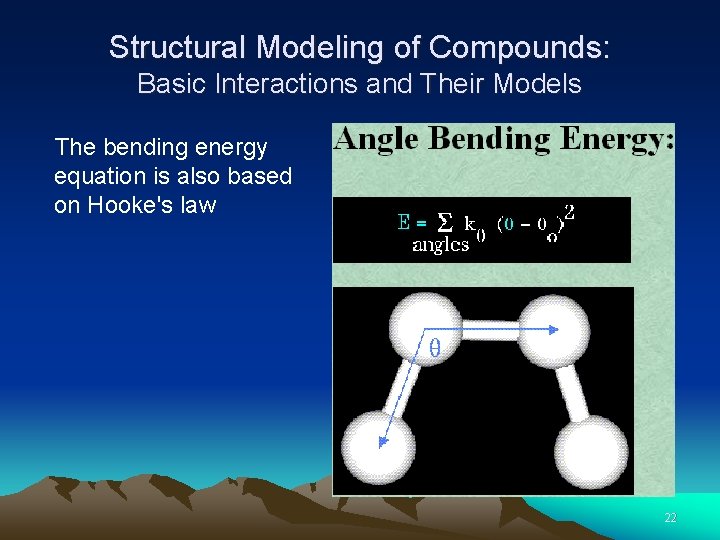
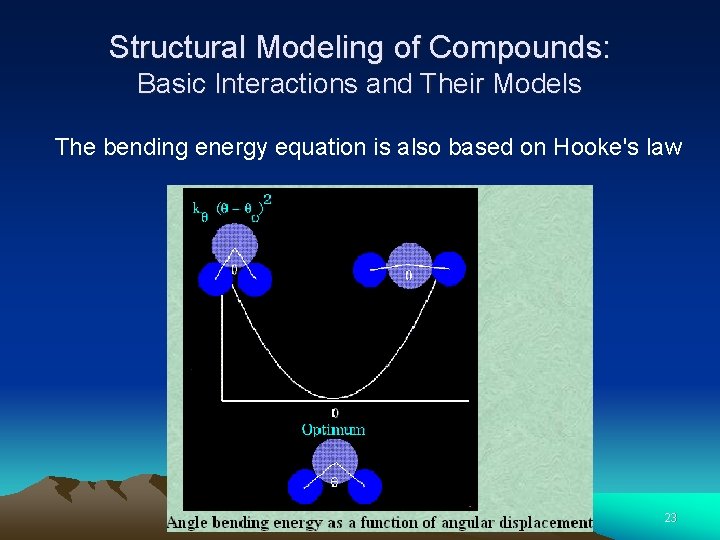
Structural Modeling of Compounds: Basic Interactions and Their Models The bending energy equation is also based on Hooke's law 22

Structural Modeling of Compounds: Basic Interactions and Their Models The bending energy equation is also based on Hooke's law 23

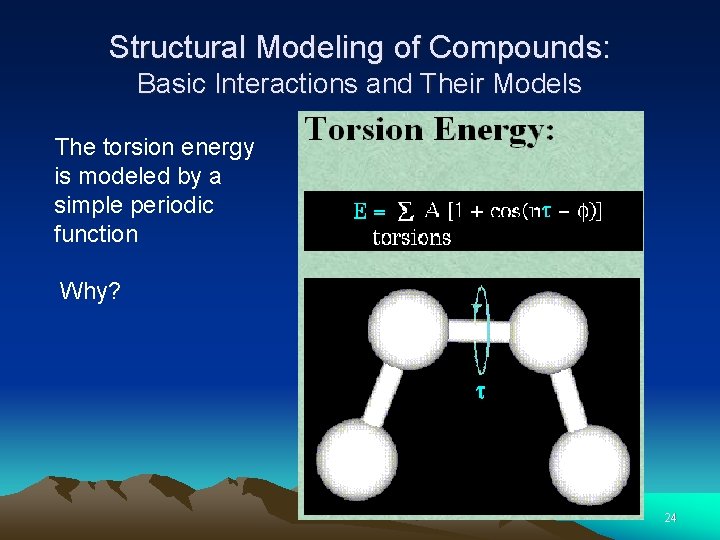
Structural Modeling of Compounds: Basic Interactions and Their Models The torsion energy is modeled by a simple periodic function Why? 24

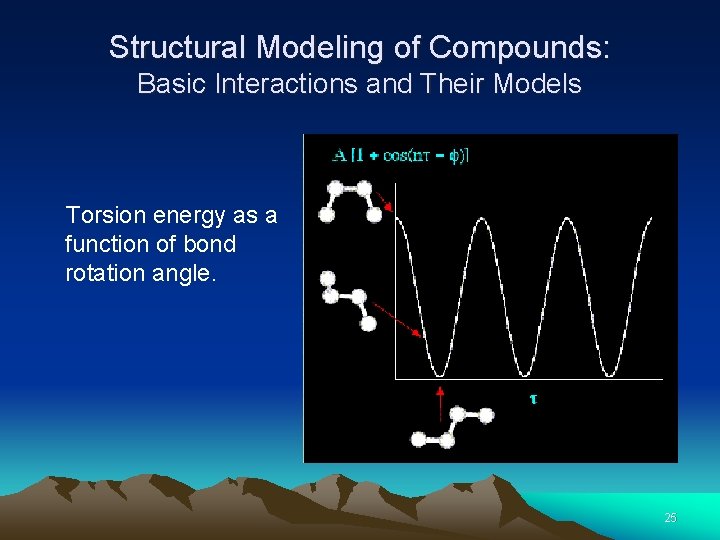
Structural Modeling of Compounds: Basic Interactions and Their Models Torsion energy as a function of bond rotation angle. 25

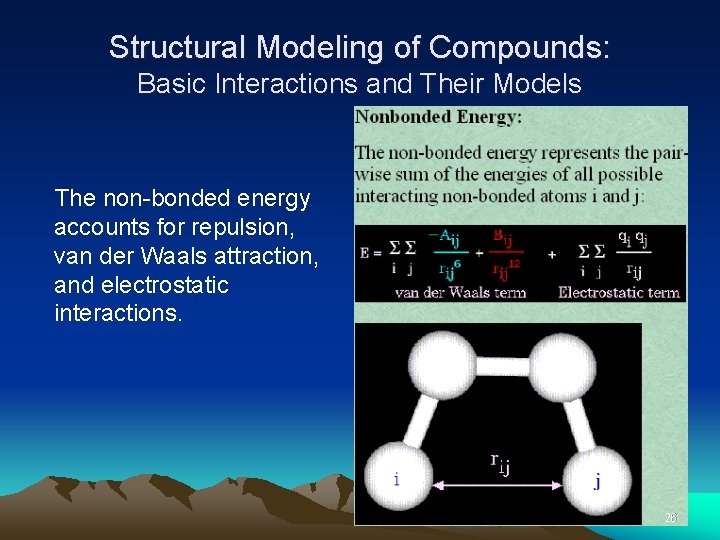
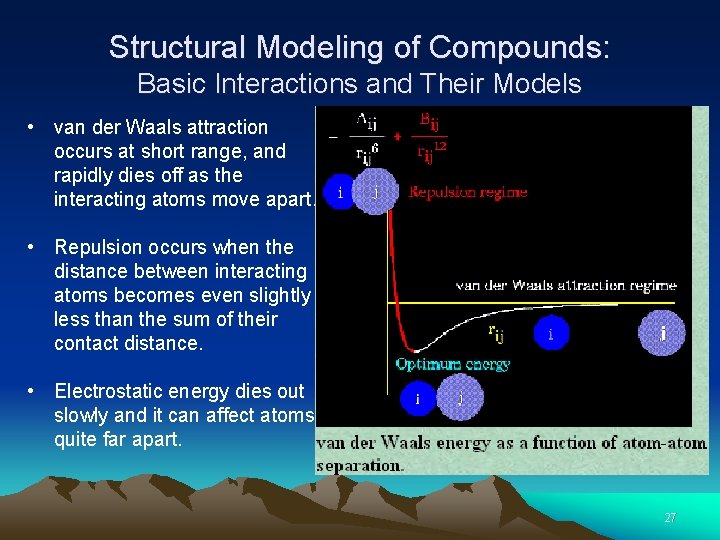
Structural Modeling of Compounds: Basic Interactions and Their Models The non-bonded energy accounts for repulsion, van der Waals attraction, and electrostatic interactions. 26

Structural Modeling of Compounds: Basic Interactions and Their Models • van der Waals attraction occurs at short range, and rapidly dies off as the interacting atoms move apart. • Repulsion occurs when the distance between interacting atoms becomes even slightly less than the sum of their contact distance. • Electrostatic energy dies out slowly and it can affect atoms quite far apart. 27

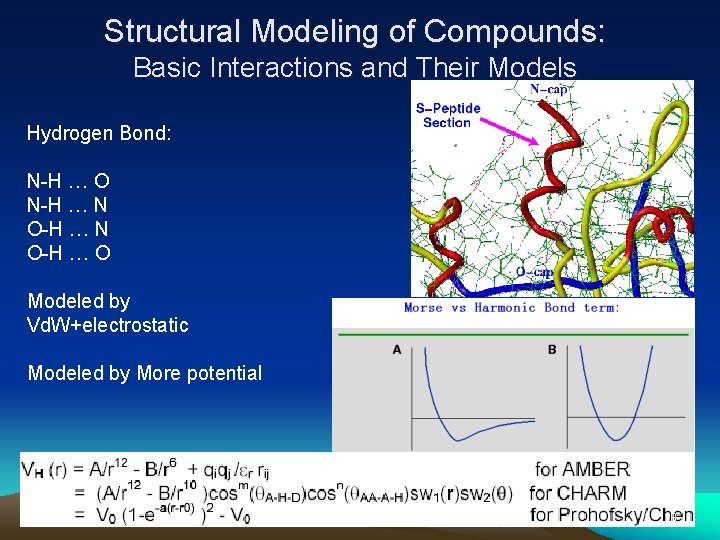
Structural Modeling of Compounds: Basic Interactions and Their Models Hydrogen Bond: N-H … O N-H … N O-H … O Modeled by Vd. W+electrostatic Modeled by More potential 28

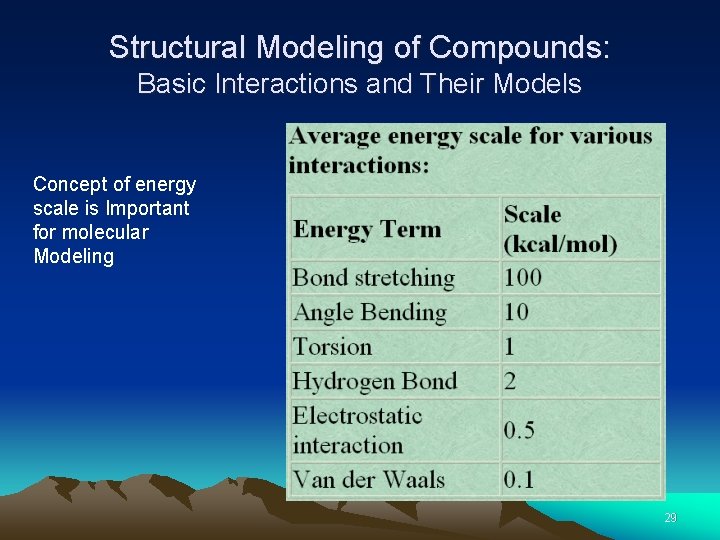
Structural Modeling of Compounds: Basic Interactions and Their Models Concept of energy scale is Important for molecular Modeling 29

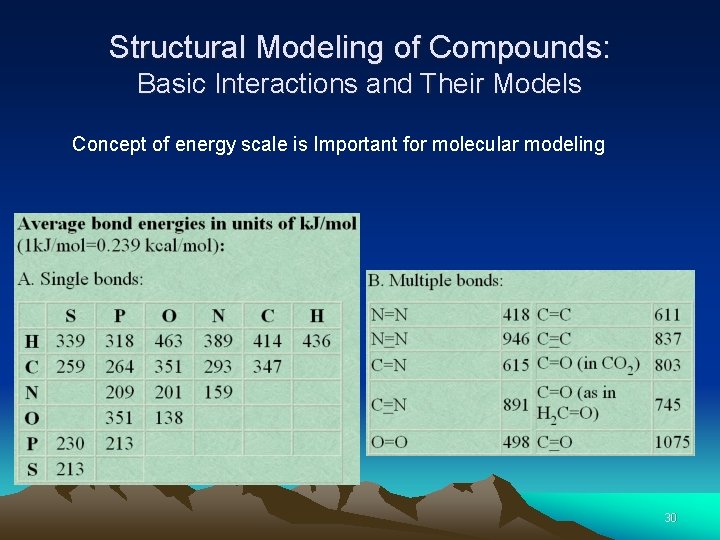
Structural Modeling of Compounds: Basic Interactions and Their Models Concept of energy scale is Important for molecular modeling 30

Structural Modeling of Compounds: Basic Interactions and Their Models Sources of force parameters: Bonds, Vd. W, Electrostatic (for amino acids, nucleotides only): • AMBER: J. Am. Chem. Soc. 117, 5179 -5197 • CHARMM: J. Comp. Chem. 4, 187 -217 H-bonds (Morse potential): • Nucleic Acids Res. 20, 415 -419. • Biophys. J. 66, 820 -826 Electrostatic parameters of organic molecules need to be computed individually by using special software (such as Gaussian) 31

Representation of Chemical Compounds How to represent a compound? • Each structure represented by digital form of structural, physicochemical properties: – Simple molecular properties (molecular weight, no. of rotatable bonds etc. ) – Molecular Connectivity and shape – Electro-topological state polarity – Quantum chemical properties (electric charge, polaritability etc. 13 in total) – Geometrical properties (molecular size vectors, van der Waals volume, molecular surface etc. 16 in total) J. Chem. Inf. Comput. Sci. 44, 1630 (2004) J. Chem. Inf. Comput. Sci. 44, 1497 (2004) Toxicol. Sci. 79, 170 (2004). 32

Molecular Descriptors • Constitutional – MW, N atoms, • Topological – Connectivity, Weiner index • Electrostatic – Polarity, polarizability, partial charges • Geometrical Descriptors – Length, width, Molecular volume • Quantum Chemical – HOMO and LUMO energies – Vibrational frequencies – Bond orders – Total energy 33

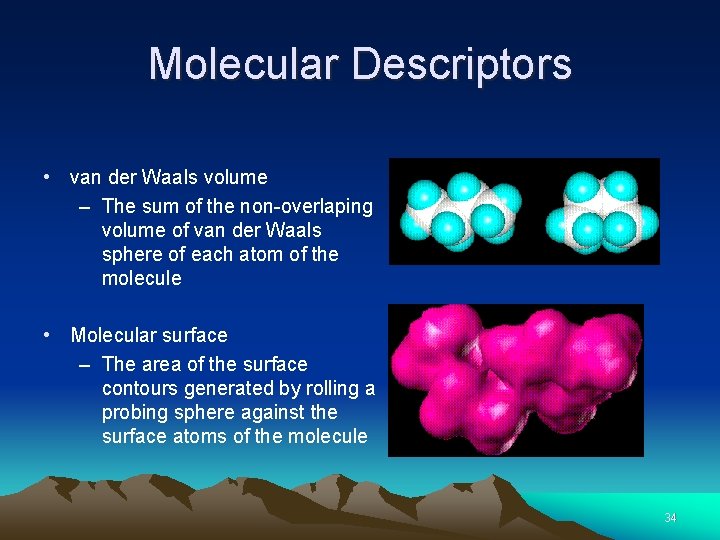
Molecular Descriptors • van der Waals volume – The sum of the non-overlaping volume of van der Waals sphere of each atom of the molecule • Molecular surface – The area of the surface contours generated by rolling a probing sphere against the surface atoms of the molecule 34

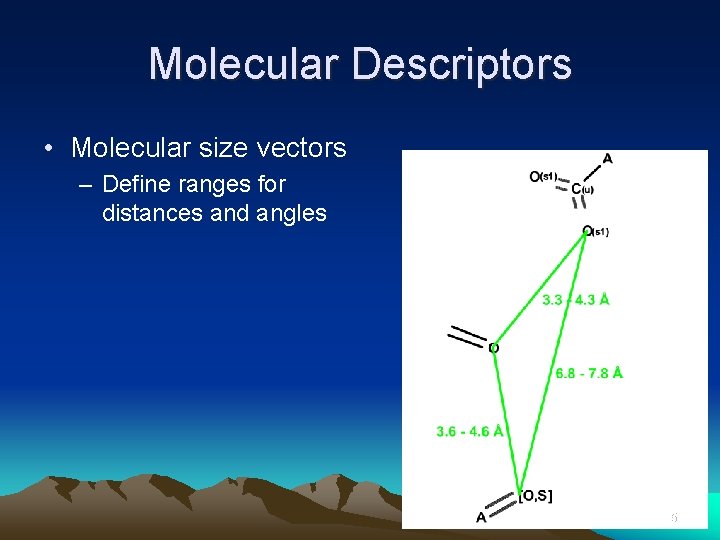
Molecular Descriptors • Molecular size vectors – Define ranges for distances and angles 35

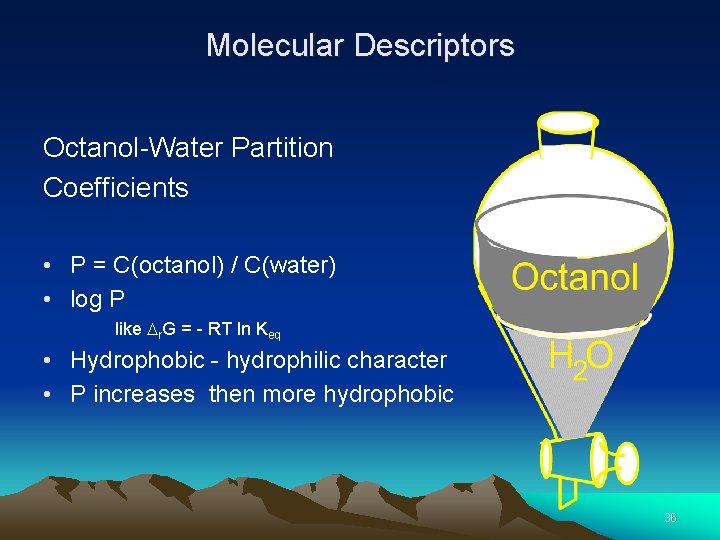
Molecular Descriptors Octanol-Water Partition Coefficients • P = C(octanol) / C(water) • log P like r. G = - RT ln Keq • Hydrophobic - hydrophilic character • P increases then more hydrophobic 36

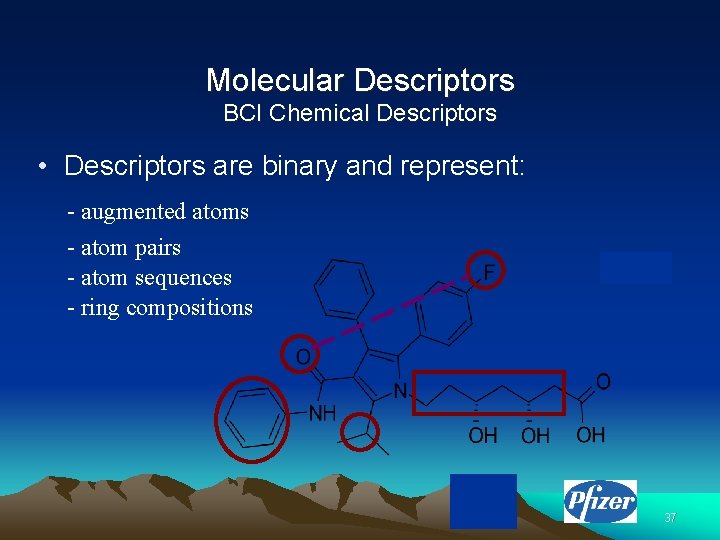
Molecular Descriptors BCI Chemical Descriptors • Descriptors are binary and represent: - augmented atoms - atom pairs - atom sequences - ring compositions 37

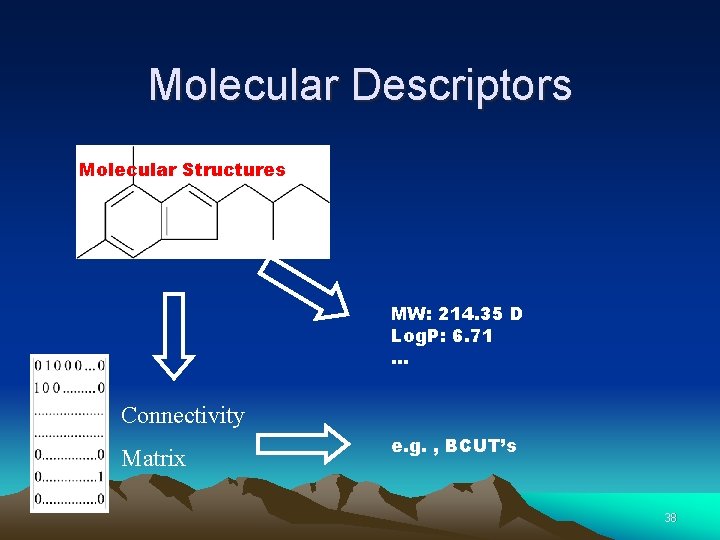
Molecular Descriptors Molecular Structures MW: 214. 35 D Log. P: 6. 71 … Connectivity Matrix e. g. , BCUT’s 38

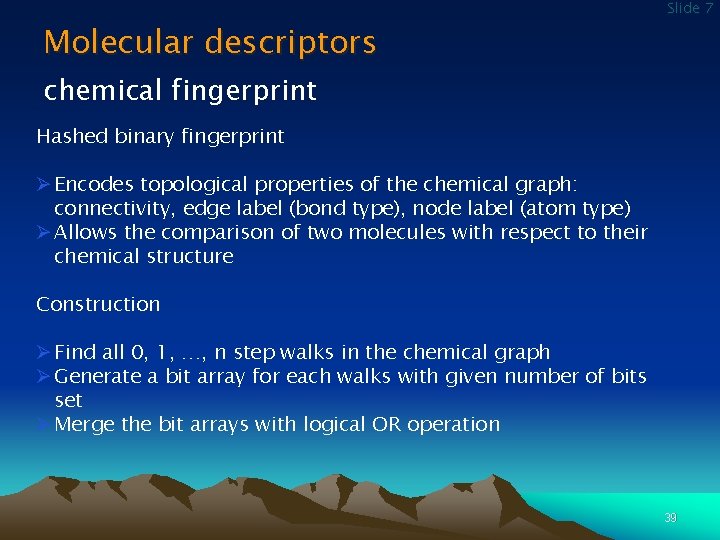
Molecular descriptors Slide 7 chemical fingerprint Hashed binary fingerprint Ø Encodes topological properties of the chemical graph: connectivity, edge label (bond type), node label (atom type) Ø Allows the comparison of two molecules with respect to their chemical structure Construction Ø Find all 0, 1, …, n step walks in the chemical graph Ø Generate a bit array for each walks with given number of bits set Ø Merge the bit arrays with logical OR operation 39

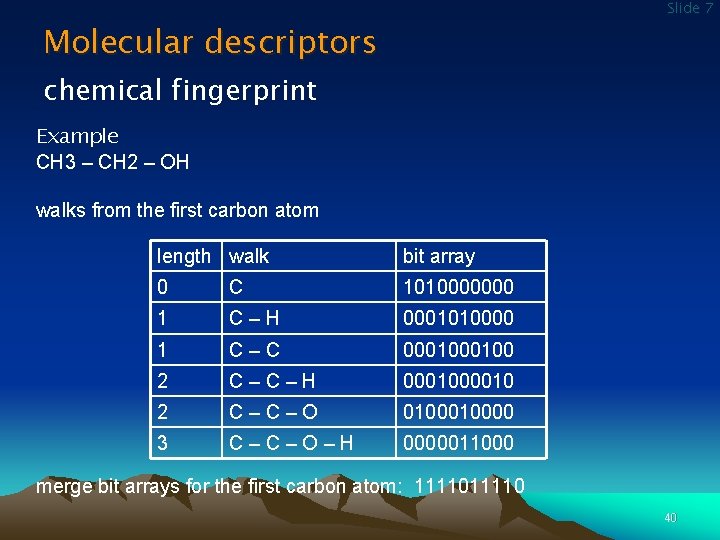
Slide 7 Molecular descriptors chemical fingerprint Example CH 3 – CH 2 – OH walks from the first carbon atom length walk bit array 0 C 1010000000 1 C – H 0001010000 1 C – C 000100 2 C – H 00010 2 C – O 010000 3 C – O – H 0000011000 merge bit arrays for the first carbon atom: 11110 40

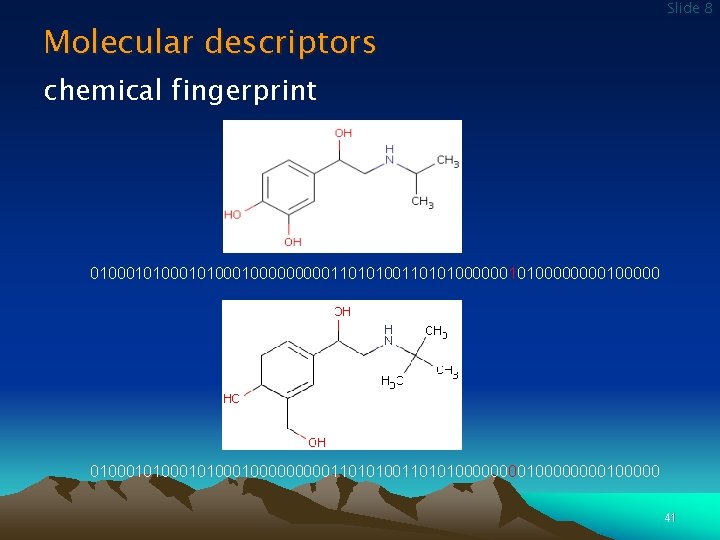
Molecular descriptors Slide 8 chemical fingerprint 010001010000000001101010000001010000100000 0100010100000000011010100000000100000 41

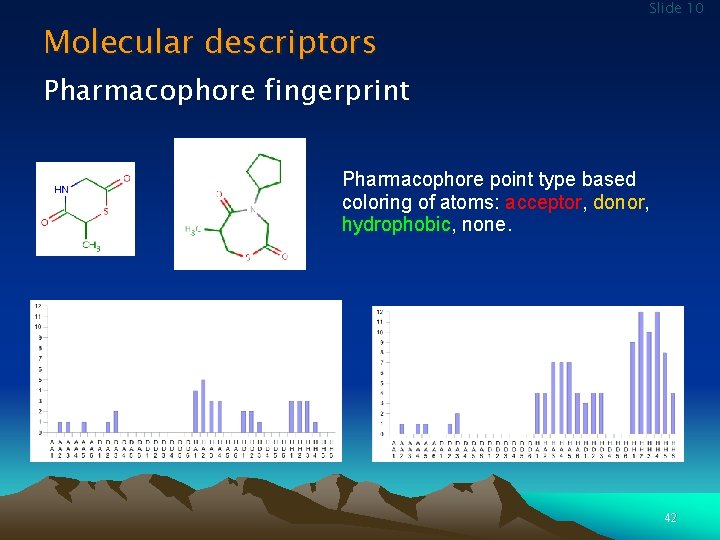
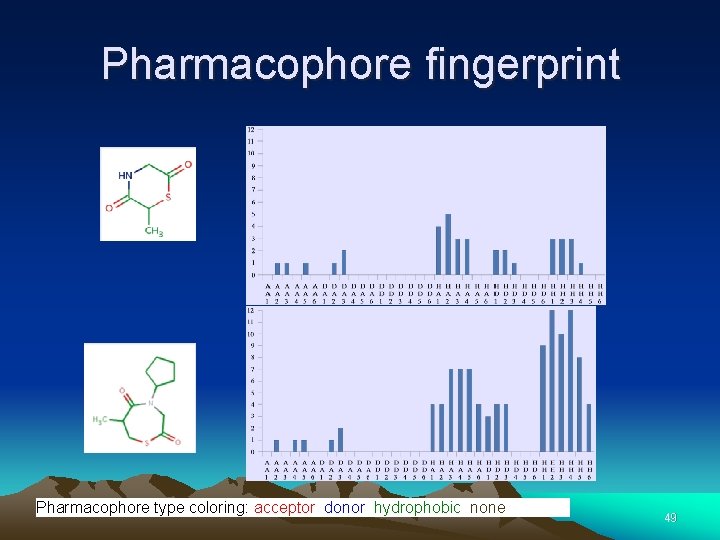
Molecular descriptors Slide 10 Pharmacophore fingerprint Pharmacophore point type based coloring of atoms: acceptor, donor, hydrophobic, none. 42

Topological pharmacophore fingreprint • Encodes pharmacophore properties of molecules as frequency counts of pharmacophore point pairs at given topological distance • Allows the comparison of two molecules with respect to their pharmacophore Construction 1. 2. 3. 4. Perceive pharmacophoric features Map pharmacophore point type to atoms Calculate length of shortest path between each pair of atoms Assign a histogram to every pharmacophore point pairs and count the frequency of the pair with respect to its distance 43

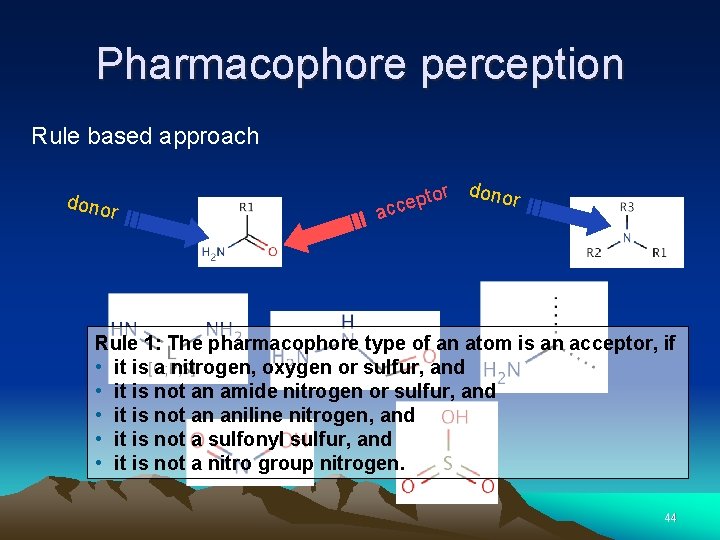
Pharmacophore perception Rule based approach dono r r donor to p e c ac Rule 1: The pharmacophore type of an atom is an acceptor, if • it is a nitrogen, oxygen or sulfur, and • it is not an amide nitrogen or sulfur, and • it is not an aniline nitrogen, and • it is not a sulfonyl sulfur, and • it is not a nitro group nitrogen. 44

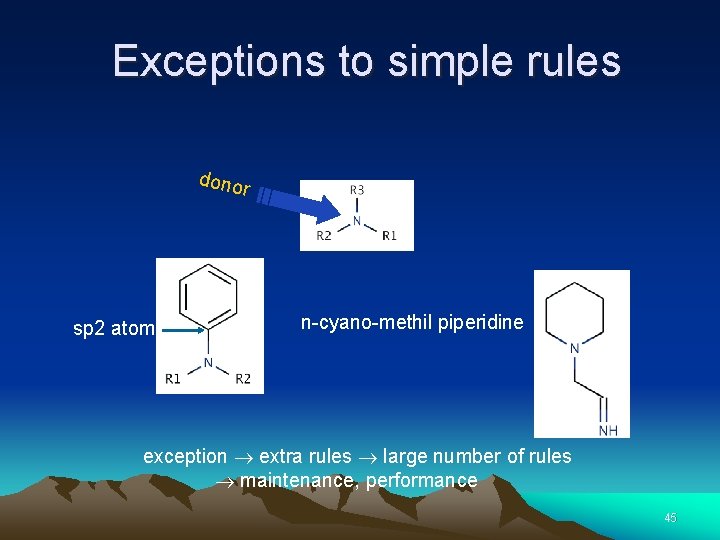
Exceptions to simple rules dono r sp 2 atom n-cyano-methil piperidine exception extra rules large number of rules maintenance, performance 45

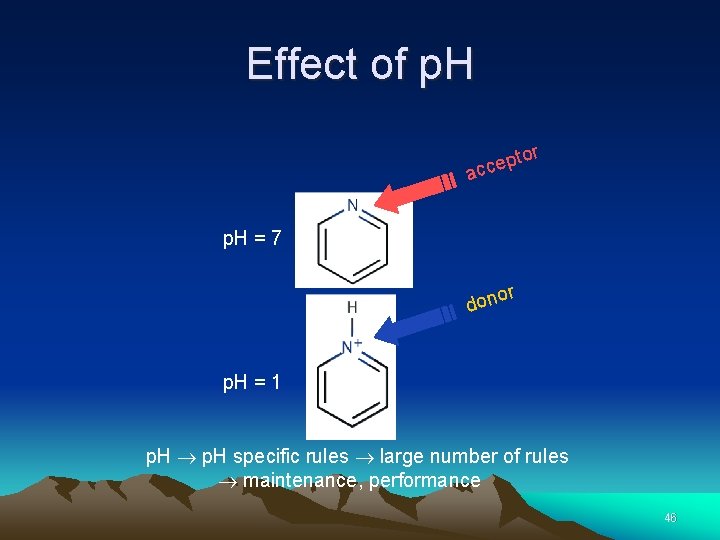
Effect of p. H to p e c ac r p. H = 7 or n o d p. H = 1 p. H specific rules large number of rules maintenance, performance 46

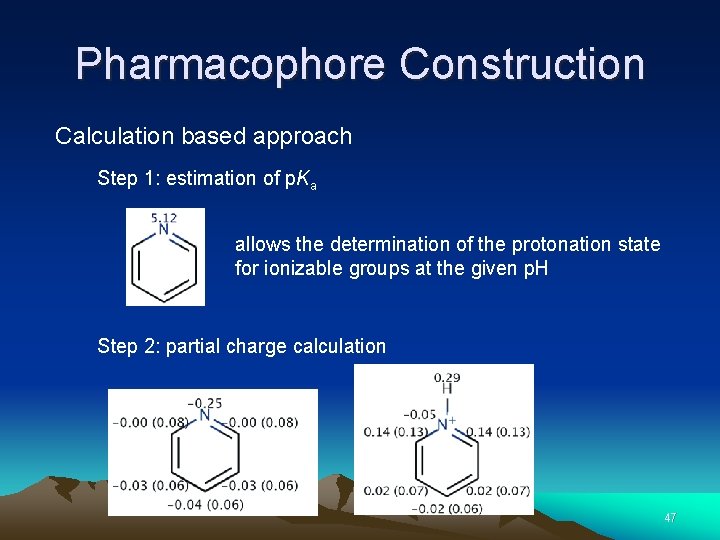
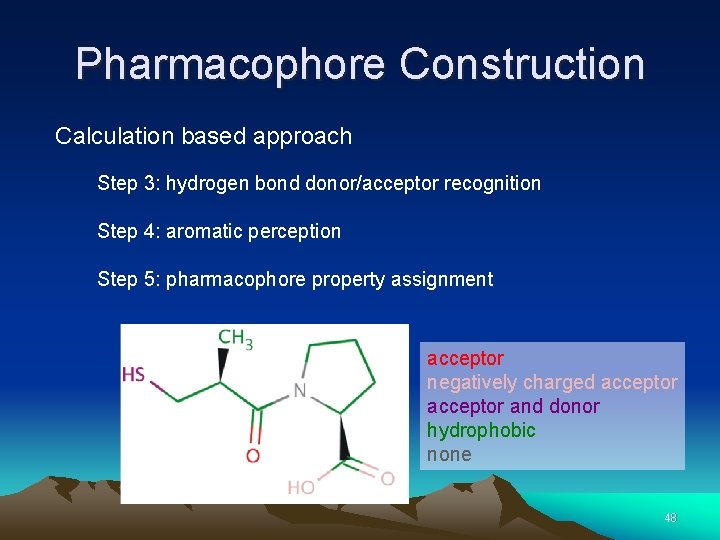
Pharmacophore Construction Calculation based approach Step 1: estimation of p. Ka allows the determination of the protonation state for ionizable groups at the given p. H Step 2: partial charge calculation 47

Pharmacophore Construction Calculation based approach Step 3: hydrogen bond donor/acceptor recognition Step 4: aromatic perception Step 5: pharmacophore property assignment acceptor negatively charged acceptor and donor hydrophobic none 48

Pharmacophore fingerprint Pharmacophore type coloring: acceptor, donor, hydrophobic, none. 49