CZ 3253 Computer Aided Drug design Lecture 8








- Slides: 33

CZ 3253: Computer Aided Drug design Lecture 8: Drug Design Methods III Ligand-Protein Docking (Part I) Prof. Chen Yu Zong Tel: 6874 -6877 Email: csccyz@nus. edu. sg http: //xin. cz 3. nus. edu. sg Room 07 -24, level 7, SOC 1, National University of Singapore

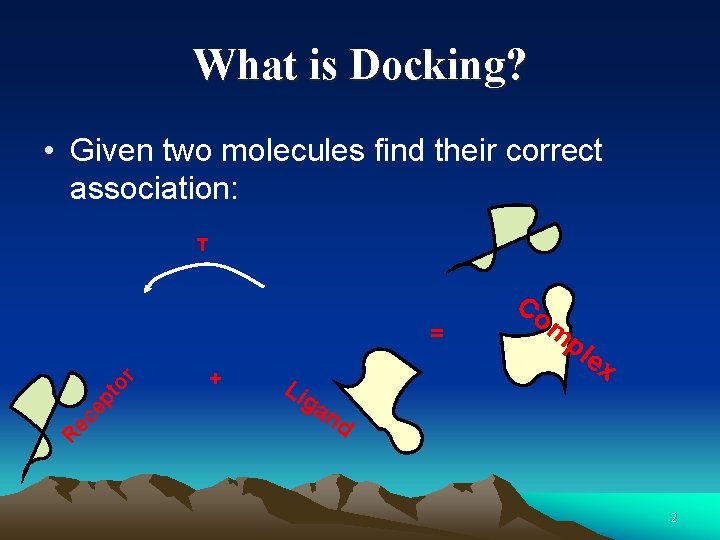
What is Docking? • Given two molecules find their correct association: T Re ce pt or = + Li Co m pl ex ga nd 2

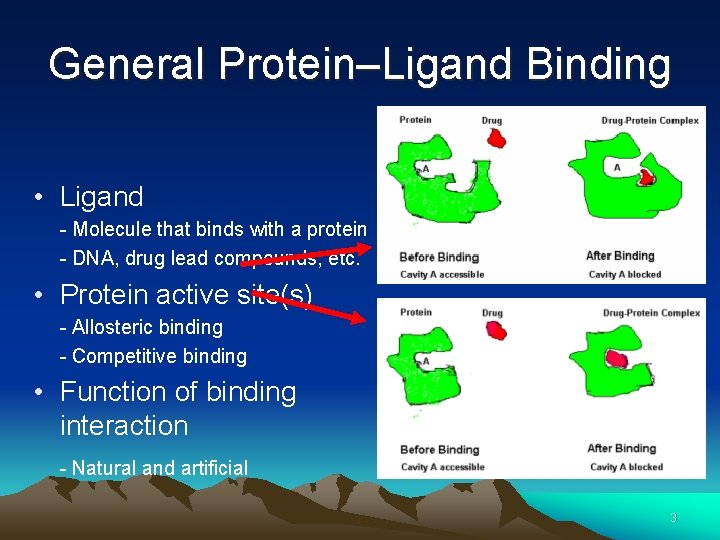
General Protein–Ligand Binding • Ligand - Molecule that binds with a protein - DNA, drug lead compounds, etc. • Protein active site(s) - Allosteric binding - Competitive binding • Function of binding interaction - Natural and artificial 3

What is Protein-Ligand Docking? • Definition: Computationally predict the structures of protein-ligand complexes from their conformations and orientations. The orientation that maximizes the interaction reveals the most accurate structure of the complex. • Importance of complexes - structure -> function 4

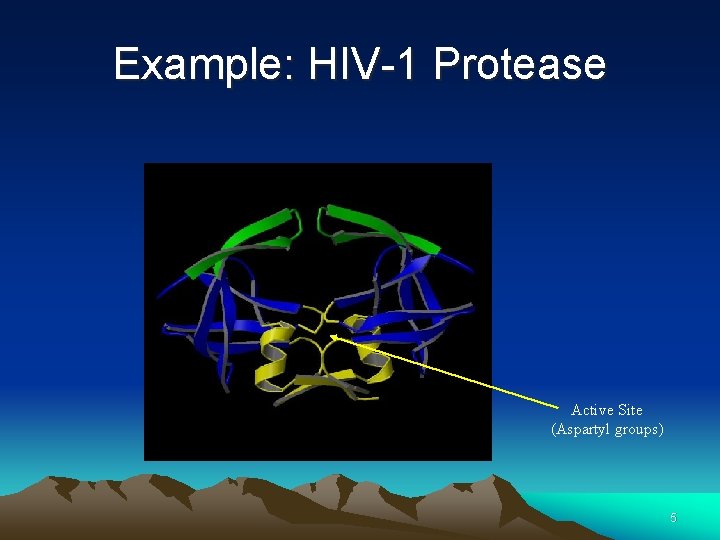
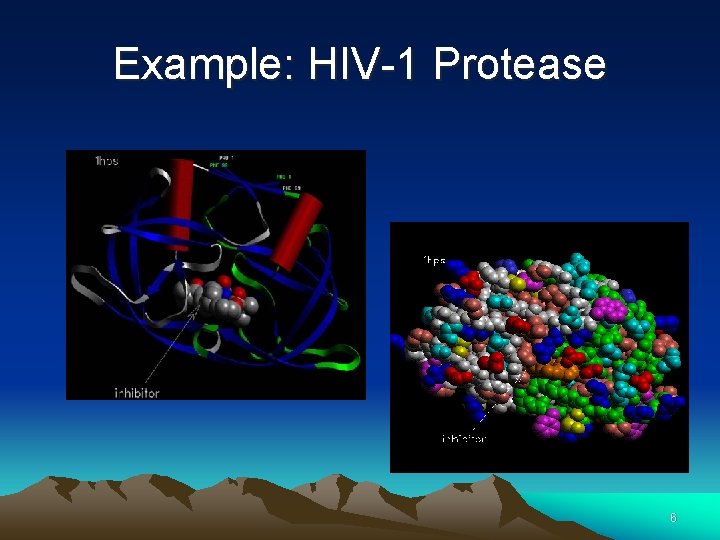
Example: HIV-1 Protease Active Site (Aspartyl groups) 5

Example: HIV-1 Protease 6

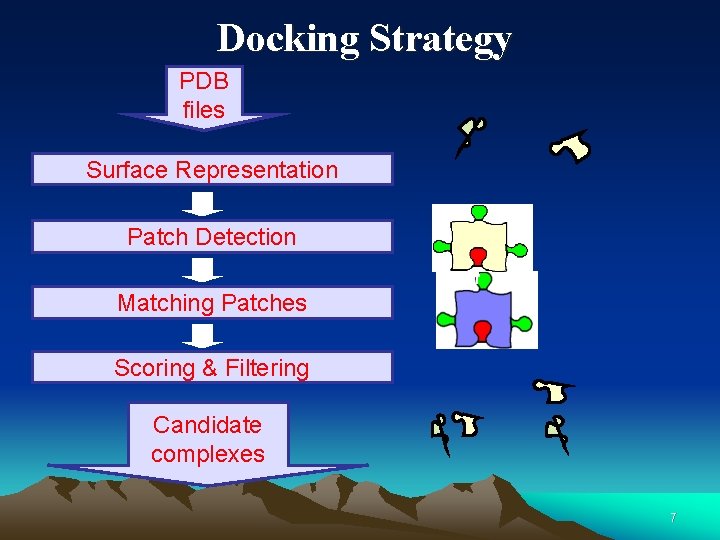
Docking Strategy PDB files Surface Representation Patch Detection Matching Patches Scoring & Filtering Candidate complexes 7

Issues Involved in Docking • Protein Structure and Active Site - Assumed knowledge (PDBs, Homology modeling etc. ) - PROCAT database: 3 d enzyme active site templates • Ligand Structure - Pharmacophore (base fragment) in potential drug compound - well known groups • Rigid vs. Flexible - In solution or in vacum - Structure fixed, partly fixed, modeling of flexibility 8

Algorithmic Approaches to Docking • Qualitative – Geometric – Shape complementarity and fitting • Quantitative – Energy calculations – Determine global minimum energy – Free energy measure • Hybrid – Geometric and energy complementarity – 2 phase process: soft and hard docking 9

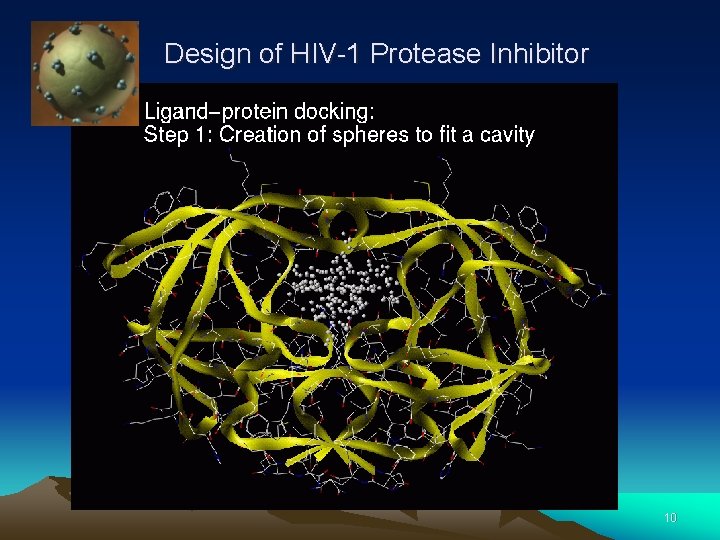
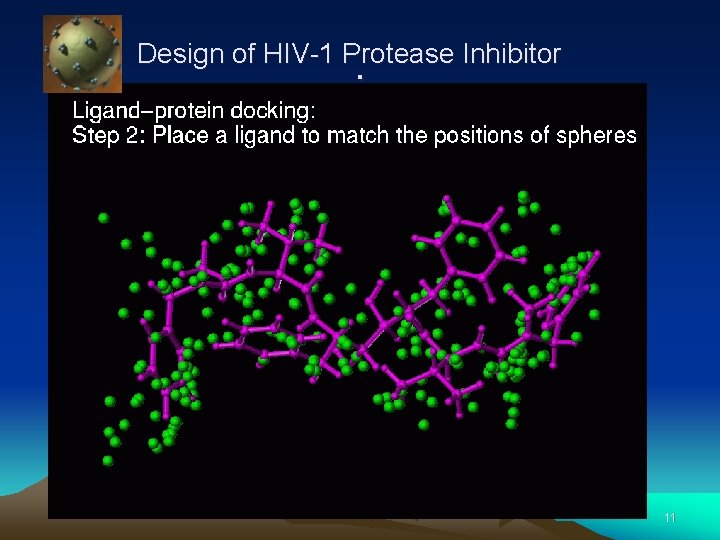
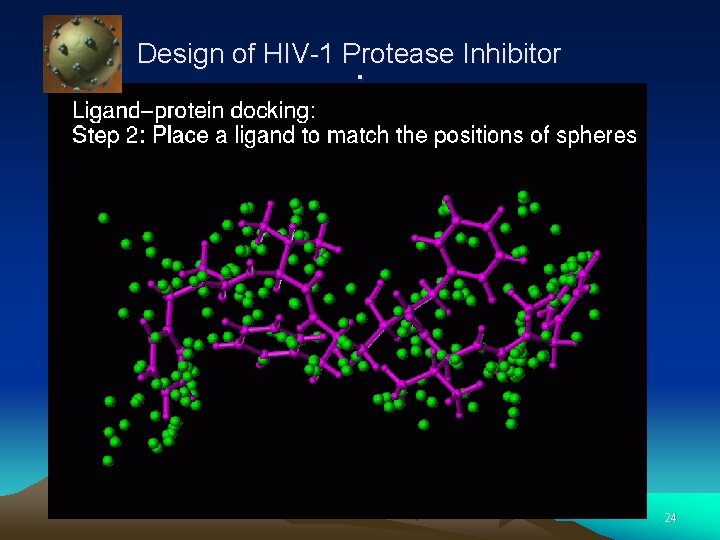
Design of HIV-1 Protease Inhibitor . 10

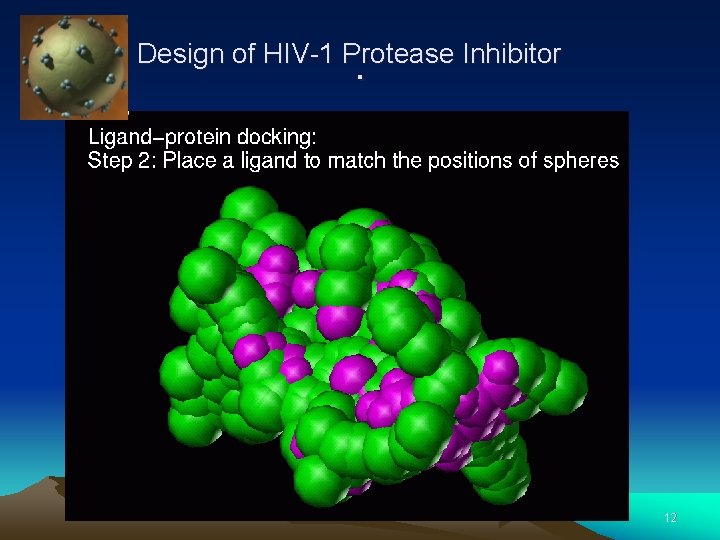
Design of HIV-1 Protease Inhibitor . 11

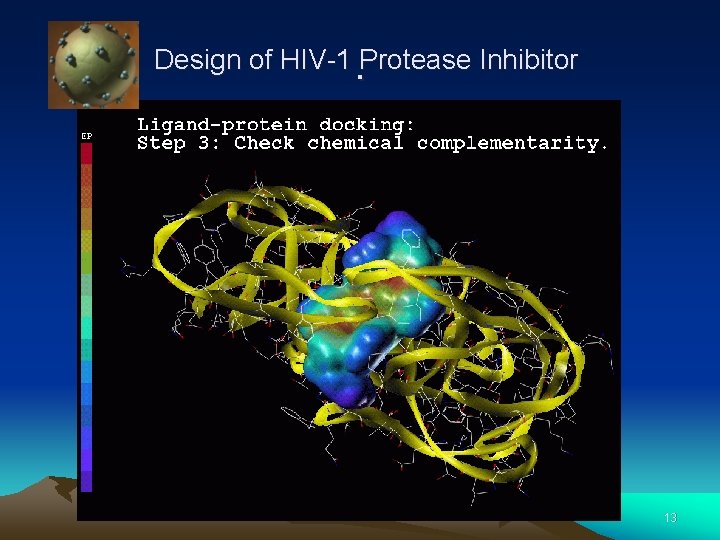
Design of HIV-1 Protease Inhibitor . 12

Design of HIV-1. Protease Inhibitor 13

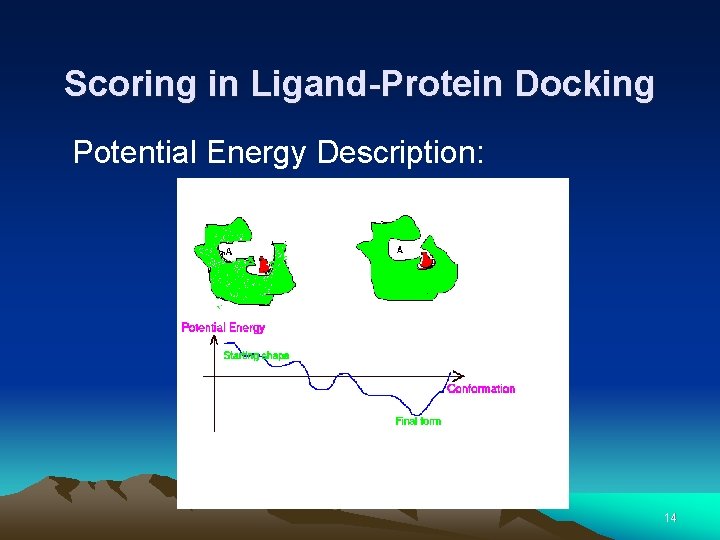
Scoring in Ligand-Protein Docking Potential Energy Description: 14

Preprocessing • Determine internal representation - Convert coordinates of both molecules from PDB files - e. g. Michael Connolly’s MS program (www. biohedron. com) - dot surface - Auto. Grid - 3 d grid (array) with discrete values - often used in rigid docking 15

Some techniques • Surface representation, that efficiently represents the docking surface and identifies the regions of interest (cavities and protrusions) • • Connolly surface Lenhoff technique Kuntz et al. Clustered-Spheres Alpha shapes • Surface matching that matches surfaces to optimize a binding score: • Geometric Hashing 16

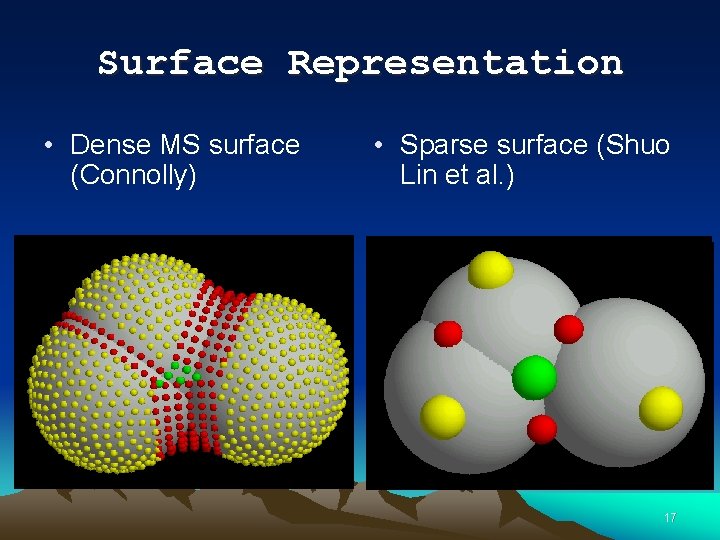
Surface Representation • Dense MS surface (Connolly) • Sparse surface (Shuo Lin et al. ) 17

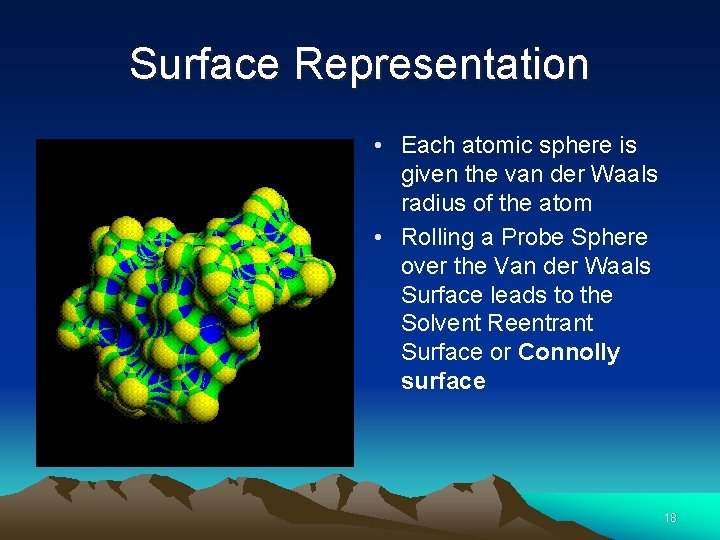
Surface Representation • Each atomic sphere is given the van der Waals radius of the atom • Rolling a Probe Sphere over the Van der Waals Surface leads to the Solvent Reentrant Surface or Connolly surface 18

Lenhoff technique • Computes a “complementary” surface for the receptor instead of the Connolly surface, i. e. computes possible positions for the atom centers of the ligand Atom centers of the ligand van der Waals surface 19

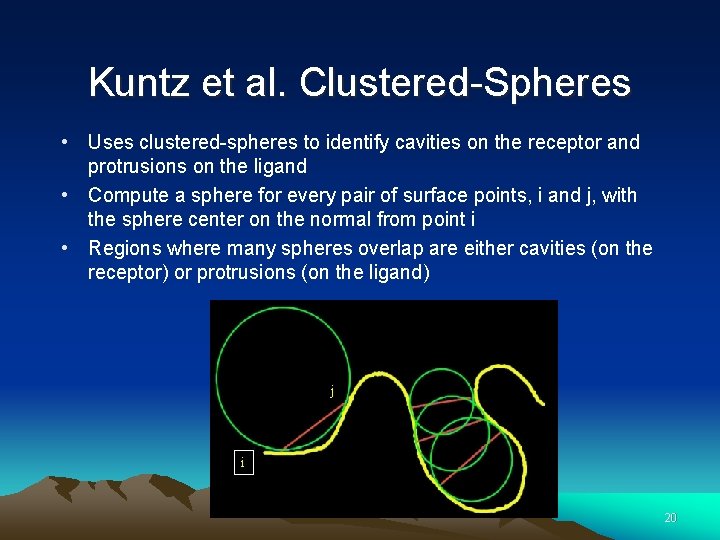
Kuntz et al. Clustered-Spheres • Uses clustered-spheres to identify cavities on the receptor and protrusions on the ligand • Compute a sphere for every pair of surface points, i and j, with the sphere center on the normal from point i • Regions where many spheres overlap are either cavities (on the receptor) or protrusions (on the ligand) j i 20

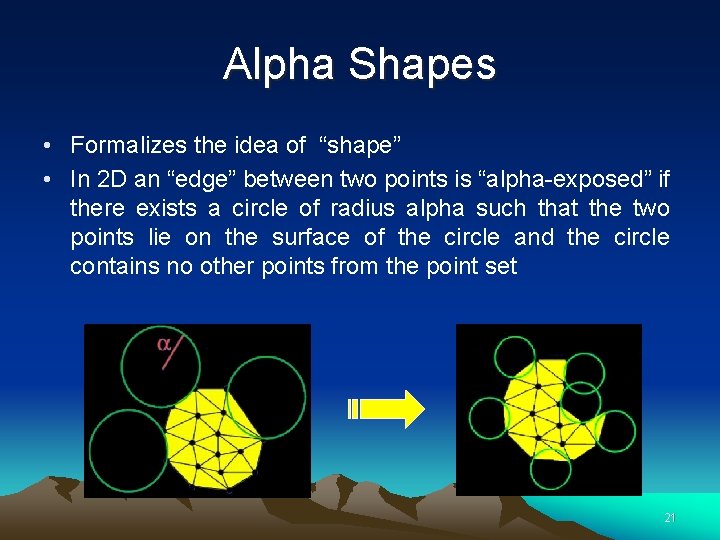
Alpha Shapes • Formalizes the idea of “shape” • In 2 D an “edge” between two points is “alpha-exposed” if there exists a circle of radius alpha such that the two points lie on the surface of the circle and the circle contains no other points from the point set 21

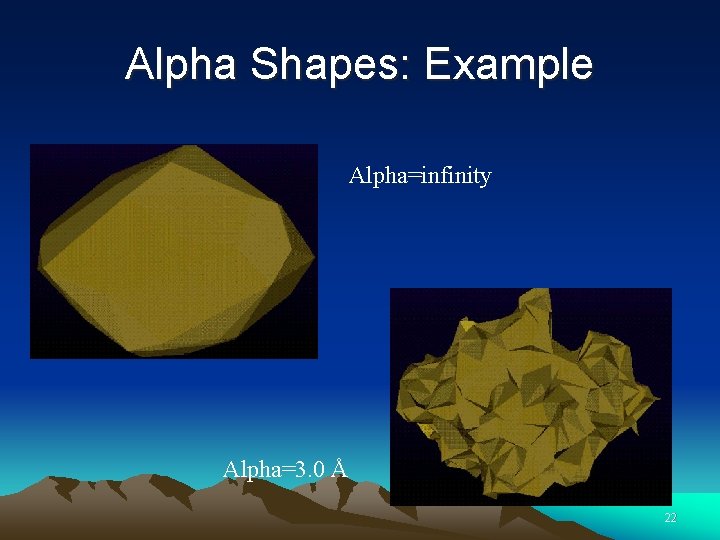
Alpha Shapes: Example Alpha=infinity Alpha=3. 0 Å 22

Surface Matching • Find the transformation (rotation + translation) that will maximize the number of matching surface points from the receptor and the ligand First satisfy steric constraints… • Find the best fit of the receptor and ligand using only geometrical constraints … then use energy calculations to refine the docking • Select the fit that has the minimum energy 23

Design of HIV-1 Protease Inhibitor . 24

Docking Programs More information in: http: //www. bmm. icnet. uk/~smithgr/soft. html The programs are: • DOCK (I. D. Kuntz, UCSF) • Auto. DOCK (Arthur Olson, The Scripps Research Institute) • Rosetta. DOCK (Baker, Washington Univ. , Gray, Johns Hopkins Univ. ) • INVDOCK (Y. Z. Chen, NUS) 25

DOCK as an Example DOCK works in 5 steps: • Step 1 Start with 3 D coordinates of target receptor • Step 2 Generate molecular surface for receptor • Step 3 Generate spheres to fill the active site of the receptor: The spheres become potential locations for ligand atoms • Step 4 Matching: Sphere centers are then matched to the ligand atoms, to determine possible orientations for the ligand • Step 5 Scoring: Find the top scoring orientation 26

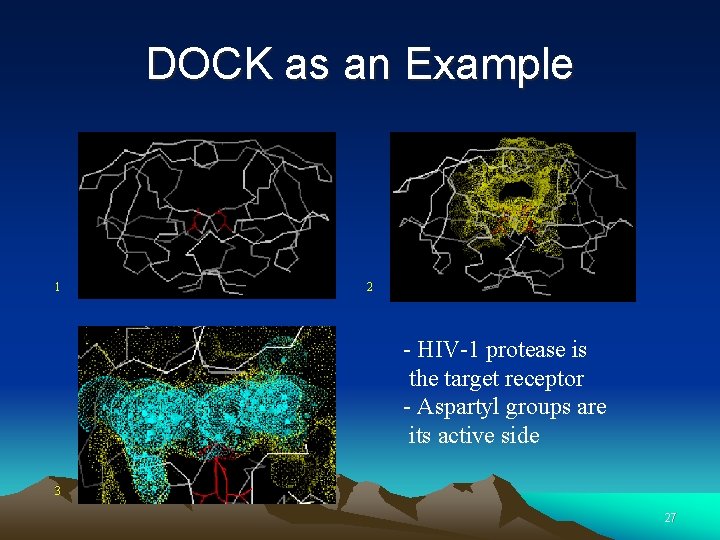
DOCK as an Example 1 2 - HIV-1 protease is the target receptor - Aspartyl groups are its active side 3 27

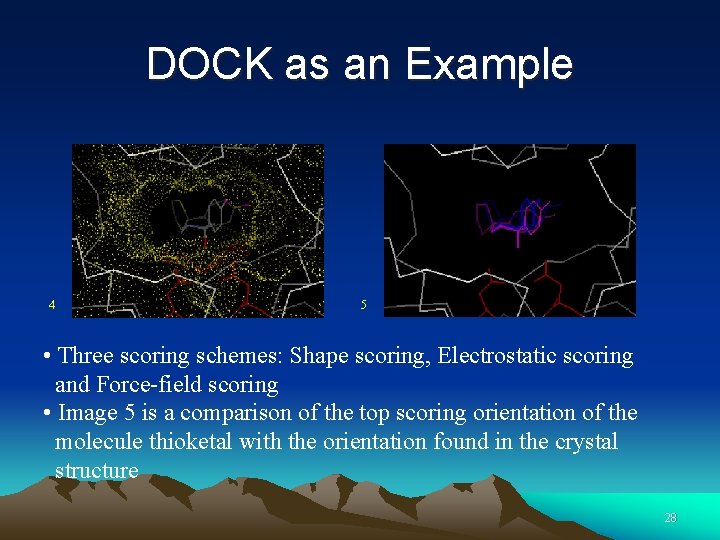
DOCK as an Example 4 5 • Three scoring schemes: Shape scoring, Electrostatic scoring and Force-field scoring • Image 5 is a comparison of the top scoring orientation of the molecule thioketal with the orientation found in the crystal structure 28

The DOCK Algorithm Two steps in rigid ligand mode: Orienting the putative ligand in the site Guided by matching distances, between predefined site points on the target to interatomic distances of the ligand. The RT matrix is used for the transform of the ligand. Scoring the resulting orientation Each orientation is scored for each quality fit. The process is repeated a user-defined number of orientations or maximum orientations 29

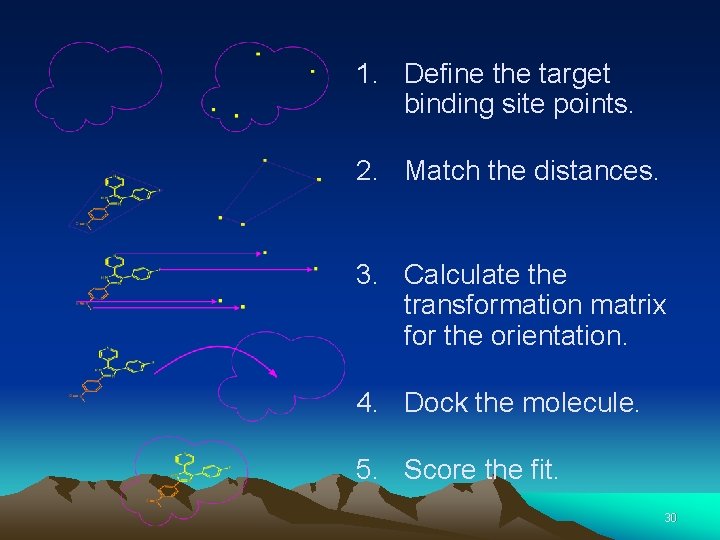
1. Define the target binding site points. 2. Match the distances. 3. Calculate the transformation matrix for the orientation. 4. Dock the molecule. 5. Score the fit. 30

Site Points Generation in DOCK • Program SPHGEN identifies the active site, and other sites of interest. • Each invagination is characterized by a set of overlapping spheres. • For receptors, a negative image of the surface invaginations is created; • For a ligand, the program creates a positive image of the entire molecule. 31

The Matching Can be directed by 2 additional features: • Chemical matching - labeling the site points such that only particular atom types are allowed to be matched to them. • Critical cluster - subsets of interest can be defined as critical clusters, so that at least one member of them will be part of any accepted ligand “match”. Increase in efficiency and speed due to elimination of potentially less promising orientations! 32

Other Docking programs Auto. Dock – Auto. Dock was designed to dock flexible ligands into receptor binding sites – The strongest feature of Auto. Dock is the range of powerful optimization algorithms available Rosetta. DOCK – It models physical forces and creates a very large number of decoys – It uses degeneracy after clustering as a final criterion in decoy selection INVDOCK – Docking strategy and algorithm similar to DOCK, but with the capability of finding the receptors to which a molecule can bind to. 33