Classical Molecular Dynamics CEC Inha University ChiOk Hwang

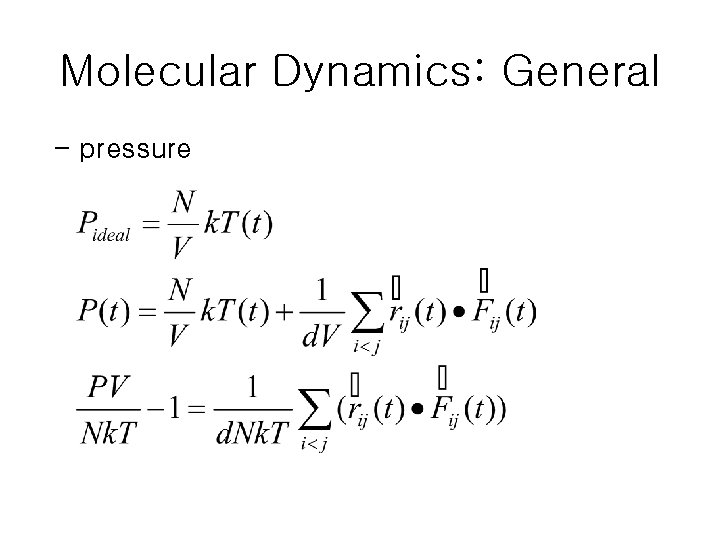




- Slides: 21

Classical Molecular Dynamics CEC, Inha University Chi-Ok Hwang

Perspectives • Empirical methods - classical molecular dynamics - tight-binding methods • First-principles methods - tight-binding methods - density-functional theory - exact methods; quantum MC

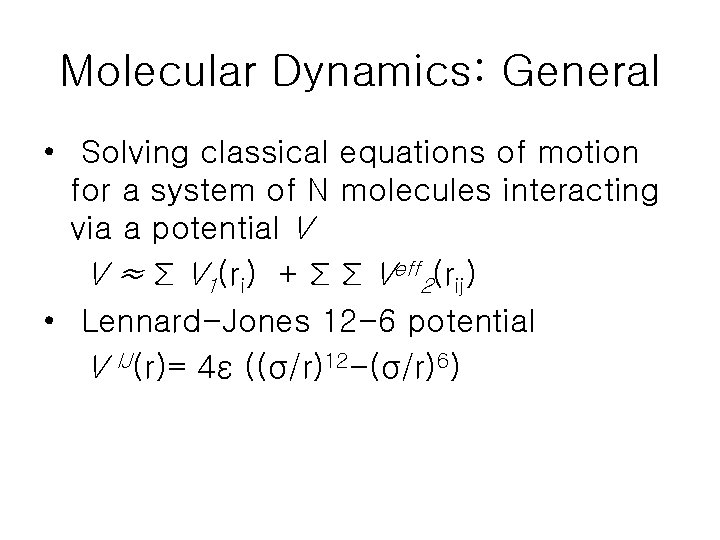
Molecular Dynamics: General • Solving classical equations of motion for a system of N molecules interacting via a potential V V ≈ Σ V 1(ri) + Σ Σ Veff 2(rij) • Lennard-Jones 12 -6 potential V IJ(r)= 4ε ((σ/r)12 -(σ/r)6)

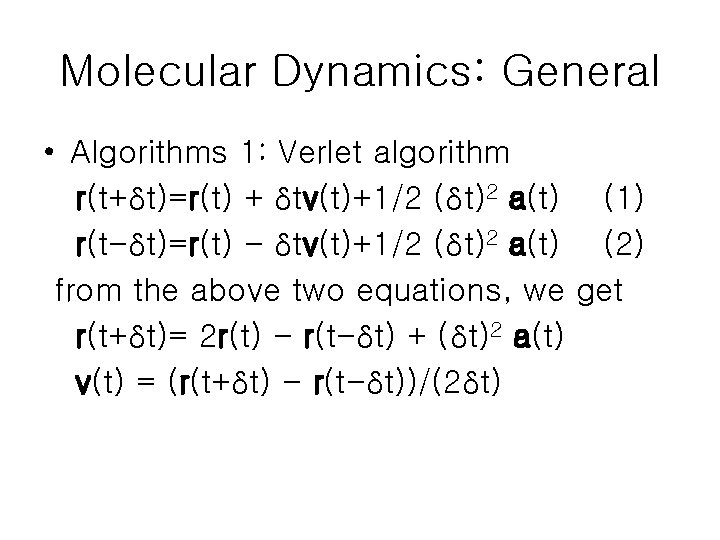
Molecular Dynamics: General • Algorithms 1: Verlet algorithm r(t+δt)=r(t) + δtv(t)+1/2 (δt)2 a(t) (1) r(t-δt)=r(t) - δtv(t)+1/2 (δt)2 a(t) (2) from the above two equations, we get r(t+δt)= 2 r(t) - r(t-δt) + (δt)2 a(t) v(t) = (r(t+δt) - r(t-δt))/(2δt)

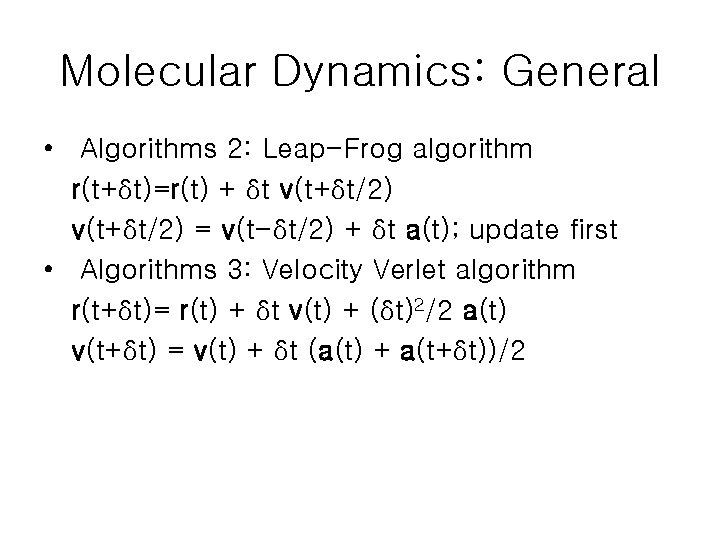
Molecular Dynamics: General • Algorithms 2: Leap-Frog algorithm r(t+δt)=r(t) + δt v(t+δt/2) = v(t-δt/2) + δt a(t); update first • Algorithms 3: Velocity Verlet algorithm r(t+δt)= r(t) + δt v(t) + (δt)2/2 a(t) v(t+δt) = v(t) + δt (a(t) + a(t+δt))/2

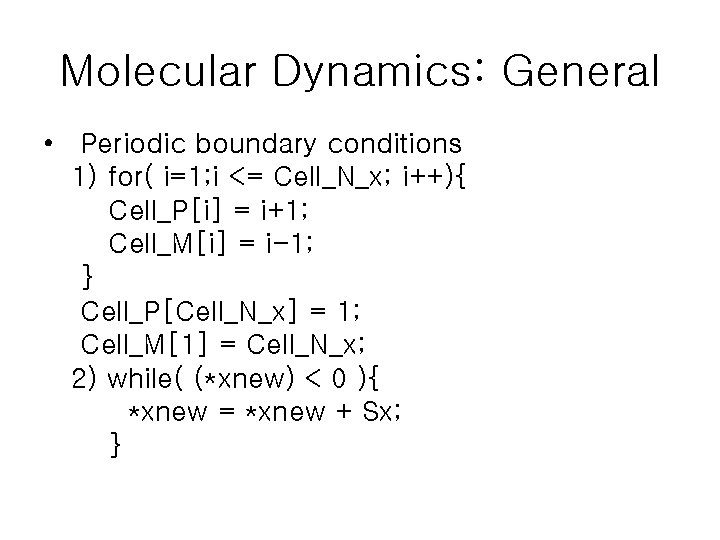
Molecular Dynamics: General • Periodic boundary conditions 1) for( i=1; i <= Cell_N_x; i++){ Cell_P[i] = i+1; Cell_M[i] = i-1; } Cell_P[Cell_N_x] = 1; Cell_M[1] = Cell_N_x; 2) while( (*xnew) < 0 ){ *xnew = *xnew + Sx; }

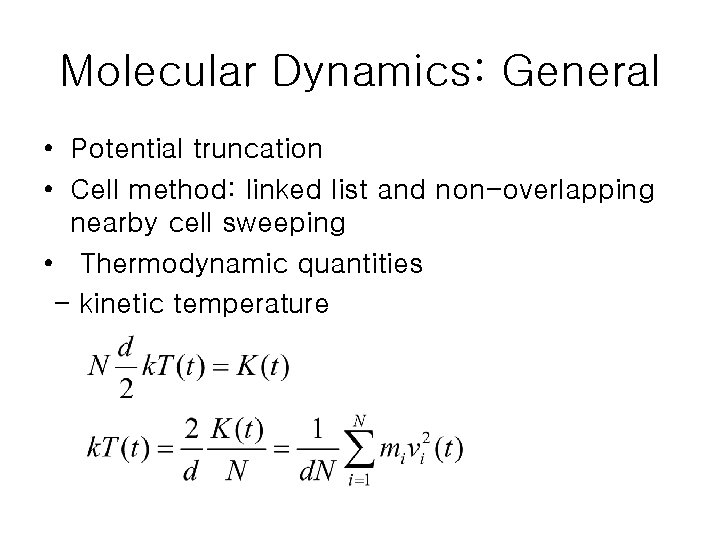
Molecular Dynamics: General • Potential truncation • Cell method: linked list and non-overlapping nearby cell sweeping • Thermodynamic quantities - kinetic temperature
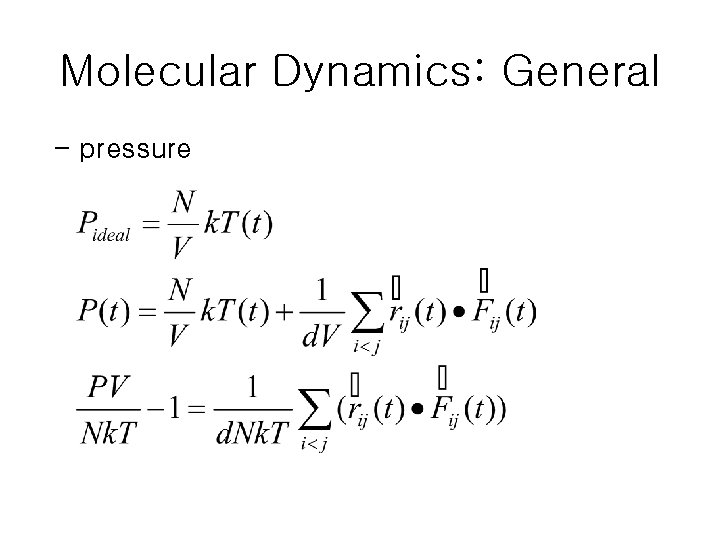
Molecular Dynamics: General - pressure

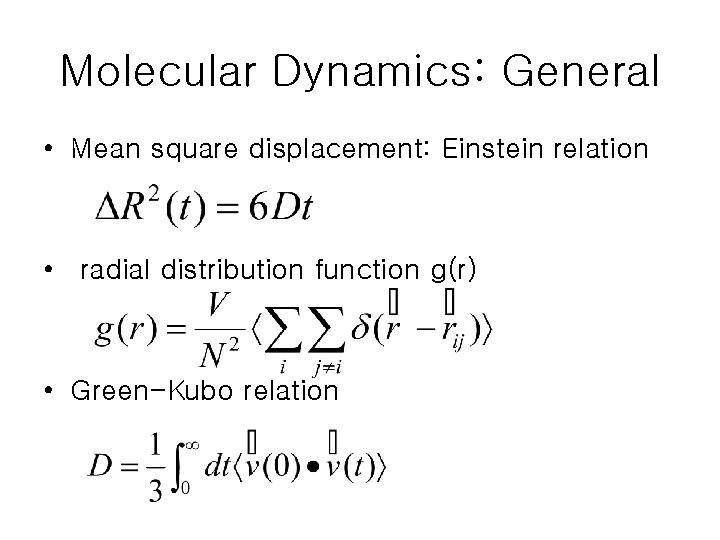
Molecular Dynamics: General • Mean square displacement: Einstein relation • radial distribution function g(r) • Green-Kubo relation

Ion Implantation • Simulation size: cascade size (10 -25 cm 3 (M. -J. Caturla etc, PRB 54, 16683, 1996) ) - 1000 atoms (J. B. Gibson etc, PR 120(4), 1229, 1960) - a few hundreds of thousands of atoms (J. Frantz etc, PRB 64, 125313 , 2001) • Time scales - thermal vibration periods of atoms in solids: 0. 1 ps (10 -13 sec) or longer - cascade lifetime: 10 ps (M. -J. Caturla etc, PRB 54, 16683, 1996) • Si density: 5 x 1022 /cm 3 (5. 43Å unit cell, 8/unit cell) • ion dose: 1017 ions/cm 2

Ion Implantation • ion implantation Potential: BCA - nuclear stopping power; elastic collision Vij(r) = Zi Zje 2 /r Φ(r); screening of the nuclei due to the electron cloud ① Thomas-Fermi ② ZBL; universal screening potential - electronic stopping power; frictional force ③ Stillinger-Weber potential

Ion Implantation • Simulations of ion implantation - Full MD - Recoil Interaction Approximation (RIA) (1 -100 ke. V) - BCA: valid for low-mass ions at incident energies from 1 -15 ke. V (M. -J. Caturla, etc, PRB 54, 16683, 1996)

Ion Implantation Three phases of collision cascade - collisional phase (0. 1 -1 ps) - thermal spike (1 ns) - relaxation phase (a few thousands of fs) • Measurements of depth profiling - Rutherford Backscattering Spectroscopy (RBS) - Secondary Ion Mass Spectroscopy (SIMS) - (Energy-Filtered) Transmission Electron Microscopy ((EF)TEM) •

MDRANGE • Calculating range profiles of ions implanted into crystalline materials as a histogram of the maximum penetration depths of 10, 000 -20, 000 ions • Modification of full MD MOLDY code • Recoil interaction approximation (RIA) - considering only interactions between the recoil ion and its nearest neighbors within a certain distance - better than BCA but about ten times slower than BCA • 0. 1 -100 ke. V energy range: one fourth of the interatomic distance difference in the mean range about 300 e. V

MDRANGE • nuclear stopping power - ZBL-type as default - Mazzone (Morse+harmonic well) for tetrahedral semiconductors in initial state calculation - Morse-type potential for metals in initial state calculation • Electronic stopping power models - Non-local electronic stopping power read in from input file - Puska-Echenique-Nieminen-Richie (PENR) model (MDRANGE 3. 0) - Brandt-Kitakawa (BK) model (MDRANGE 3. 0)

MDRANGE • Dose range: • Damage build-up modeling (2002) - Amorphization level is proportional to the nuclear deposited energy in that depth region - Electronic stopping ① point-like ion and a spherically symmetric electron distribution ② maximum distance of the charge distribution of Si, 1. 47 Å - Using 20 predamaged boxes

MDRANGE • Time step Δtmin = min( kt /v, Et /Fv, 1. 1 Δtold ) - kt, Et are proportional constants - inversely proportional to the recoil velocity - inversely proportional to the product of the total force F the recoil atom experiences and its velocity v - not to increase more than 10% from its previous value

MDRANGE • Simulation cell - less than r 0 (2 -3 Å in ZBL) - a simulation cell with a side length of 10 -15 Å (a cell containing 50 -100 atoms) - translation method • Structure of the sample - atom coordinates of all atoms except the recoil atom are read in from a file at the beginning of the simulation - polycrystalline materials: grain size is calculated using a Gaussian distribution and orientation of each grain is selected randomly - multilayered structures with depth regions and the same size of the simulation cell

MDRANGE • Recoil event calculation - placing the recoil atom of desired energy and velocity a few Å outside the simulation cell with z=0 - recoil atom threshold energy, 1 e. V - electronic stopping (Se): Δv=Δt Se /m where m is the ion mass - calculating nuclear and electronic deposited energies - energy losses of the recoil atom are evaluated for each time step and stored in arrays as a function of the depth - nuclear energy loss = total energy loss - electronic energy loss

etc • Jeong-Won Kang’s local damage accumulation model Eion: deposited energy in a unit cell RD: dose rate (neglected) M 1: target material atom weight M 2: projectile atom weight RX: relaxation and recombination effects ncoil: coil events rate

Future Studies • Area and ion dose criteria where local accumulated damages affect implanted ion range • Damage accumulation model • Different stopping powers • Full MD criteria for ultra-low energy implantation • Ion-beam amorphization modeling (of silicon) • Multi-ion recoil approximation