MDRANGE CEC Inha University ChiOk Hwang MDRANGE Calculating




- Slides: 8

MDRANGE CEC, Inha University Chi-Ok Hwang

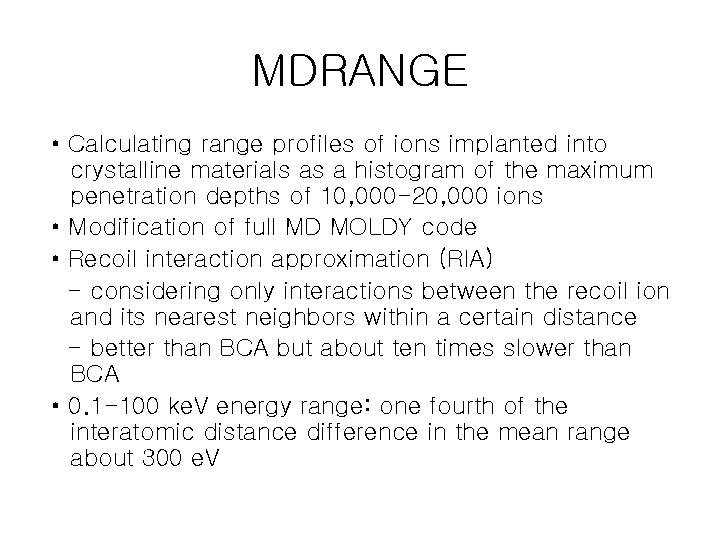
MDRANGE • Calculating range profiles of ions implanted into crystalline materials as a histogram of the maximum penetration depths of 10, 000 -20, 000 ions • Modification of full MD MOLDY code • Recoil interaction approximation (RIA) - considering only interactions between the recoil ion and its nearest neighbors within a certain distance - better than BCA but about ten times slower than BCA • 0. 1 -100 ke. V energy range: one fourth of the interatomic distance difference in the mean range about 300 e. V

MDRANGE • nuclear stopping power - ZBL-type as default - Mazzone (Morse+harmonic well) for tetrahedral semiconductors in initial state calculation - Morse-type potential for metals in initial state calculation • Electronic stopping power models - Non-local electronic stopping power read in from input file - Puska-Echenique-Nieminen-Richie (PENR) model (MDRANGE 3. 0) - Brandt-Kitakawa (BK) model (MDRANGE 3. 0)

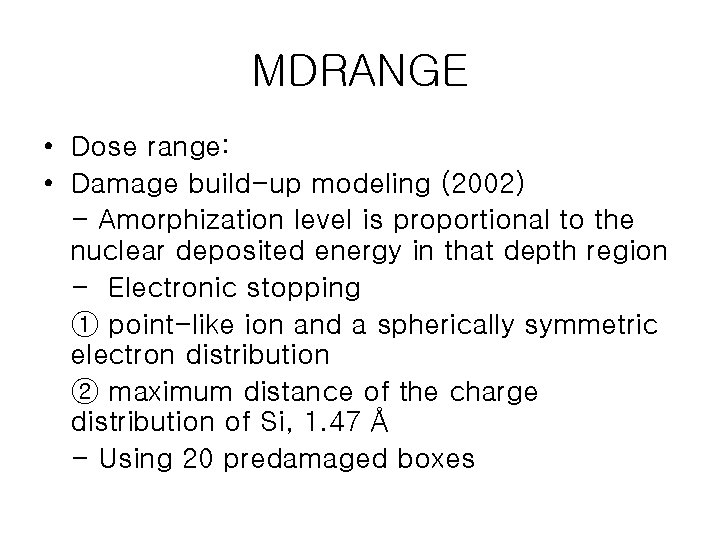
MDRANGE • Dose range: • Damage build-up modeling (2002) - Amorphization level is proportional to the nuclear deposited energy in that depth region - Electronic stopping ① point-like ion and a spherically symmetric electron distribution ② maximum distance of the charge distribution of Si, 1. 47 Å - Using 20 predamaged boxes

MDRANGE • Time step Δtmin = min( kt /v, Et /Fv, 1. 1 Δtold ) - kt, Et are proportional constants - inversely proportional to the recoil velocity - inversely proportional to the product of the total force F the recoil atom experiences and its velocity v - not to increase more than 10% from its previous value

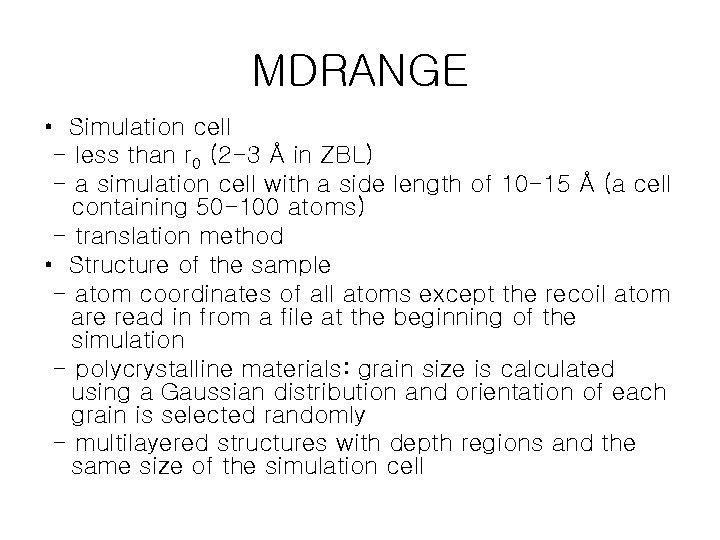
MDRANGE • Simulation cell - less than r 0 (2 -3 Å in ZBL) - a simulation cell with a side length of 10 -15 Å (a cell containing 50 -100 atoms) - translation method • Structure of the sample - atom coordinates of all atoms except the recoil atom are read in from a file at the beginning of the simulation - polycrystalline materials: grain size is calculated using a Gaussian distribution and orientation of each grain is selected randomly - multilayered structures with depth regions and the same size of the simulation cell

MDRANGE • Recoil event calculation - placing the recoil atom of desired energy and velocity a few Å outside the simulation cell with z=0 - recoil atom threshold energy, 1 e. V - electronic stopping (Se): Δv=Δt Se /m where m is the ion mass - calculating nuclear and electronic deposited energies - energy losses of the recoil atom are evaluated for each time step and stored in arrays as a function of the depth - nuclear energy loss = total energy loss - electronic energy loss

etc • Jeong-Won Kang’s local damage accumulation model Eion: deposited energy in a unit cell RD: dose rate (neglected) M 1: target material atom weight M 2: projectile atom weight RX: relaxation and recombination effects ncoil: coil events rate