Chapter 14 Genomes and Genomics Sequencing DNA dideoxy






- Slides: 30

Chapter 14 Genomes and Genomics

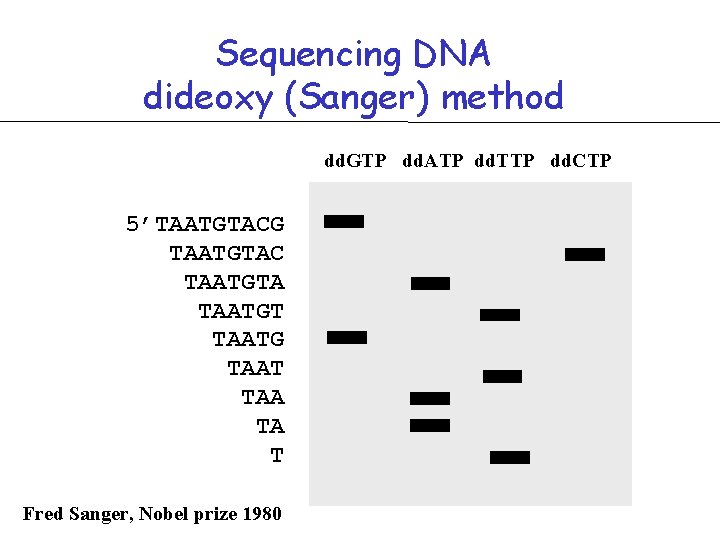
Sequencing DNA dideoxy (Sanger) method dd. GTP dd. ATP dd. TTP dd. CTP 5’TAATGTACG TAATGTAC TAATGTA TAATGT TAATG TAAT TAA TA T Fred Sanger, Nobel prize 1980

Sequencing DNA dideoxy (Sanger) method Leroy Hood, Caltech Fluorescence based sequencing Norm Dovici – Capillary electrophoresis

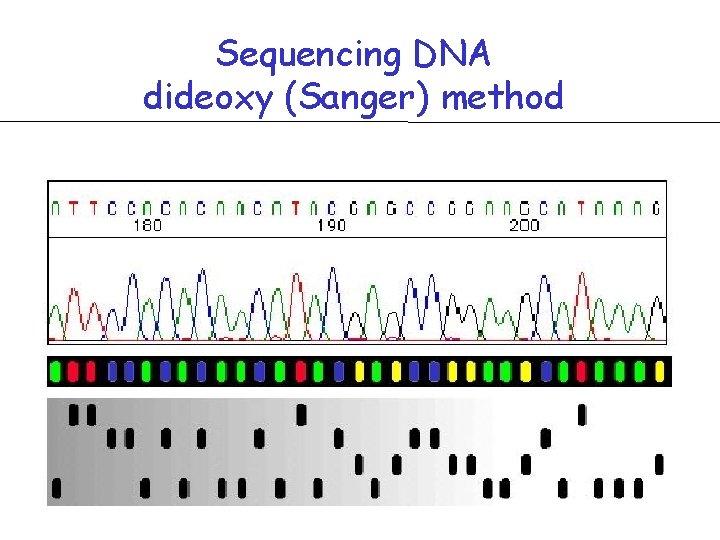
Sequencing DNA dideoxy (Sanger) method

Genomics era: High-throughput DNA sequencing The first high-throughput genomics technology was automated DNA sequencing in the early 1990. TIGR (The Institute for Genomics Research) 1995 – first whole genome sequence, H. influenza Baker’s yeast, Saccharomyces cerevisiae (15 million bp), was the first eukaryotic genome to be sequenced. In September 1999, Celera Genomics completed the sequencing of the Drosophila genome.

Genomics: Completed genomes as 2002 Currently the genome of over 600 organisms are sequenced: http: //www. genomesonline. org/ This generates large amounts of information to be handled by individual computers.

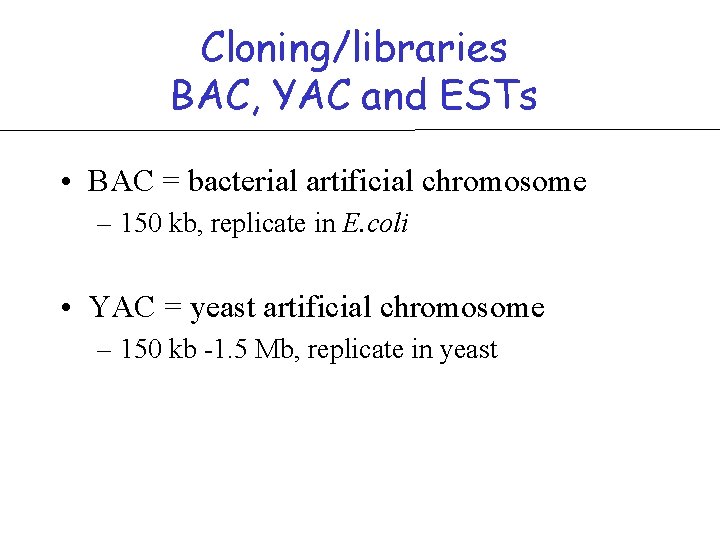
Cloning/libraries BAC, YAC and ESTs • BAC = bacterial artificial chromosome – 150 kb, replicate in E. coli • YAC = yeast artificial chromosome – 150 kb -1. 5 Mb, replicate in yeast

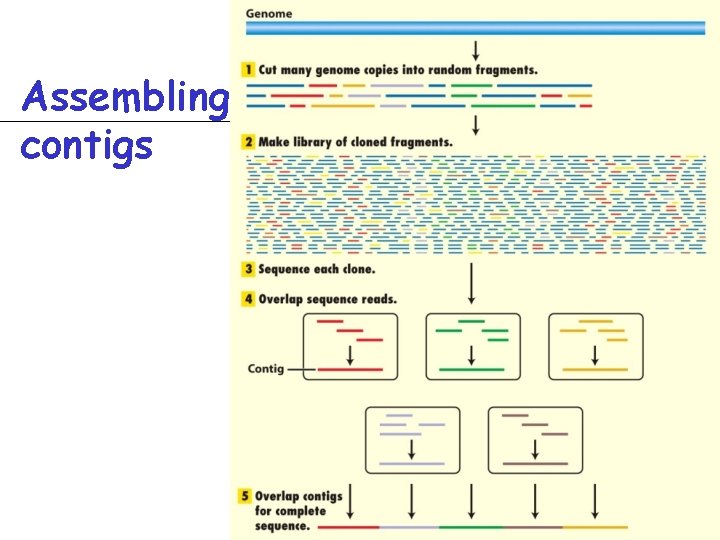
Assembling contigs

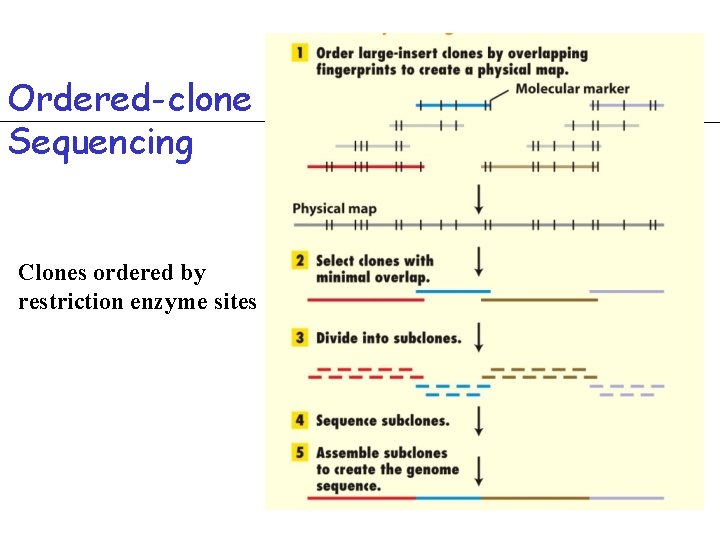
Ordered-clone Sequencing Clones ordered by restriction enzyme sites

Annotation • ORF – open reading frame • EST- Expressed sequence tag – Based on m. RNA • Comparative genomics

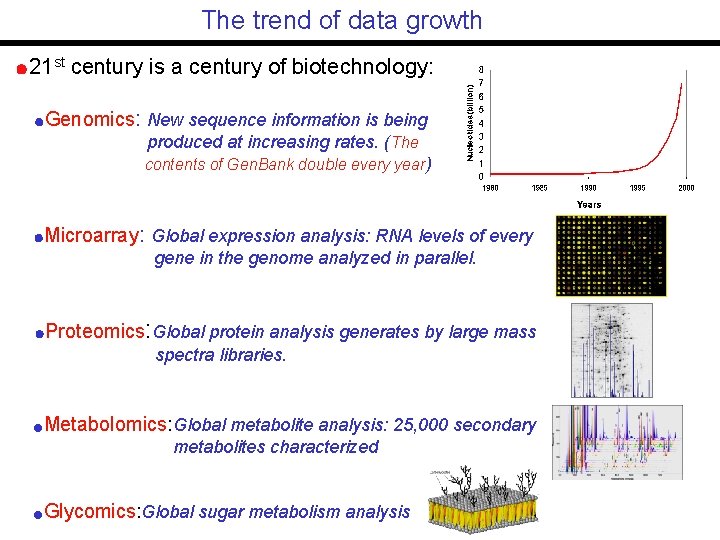
The trend of data growth 21 st century is a century of biotechnology: Genomics: New sequence information is being produced at increasing rates. (The contents of Gen. Bank double every year) Microarray: Global expression analysis: RNA levels of every gene in the genome analyzed in parallel. Proteomics: Global protein analysis generates by large mass spectra libraries. Metabolomics: Global metabolite analysis: 25, 000 secondary metabolites characterized Glycomics: Global sugar metabolism analysis

How to handle the large amount of information? Drew Sheneman, New Jersey--The Newark Star Ledger Answer: bioinformatics and Internet

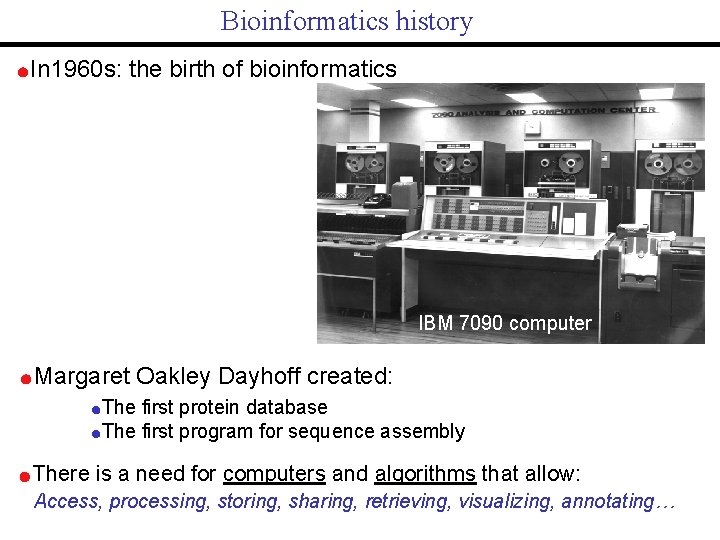
Bioinformatics history In 1960 s: the birth of bioinformatics IBM 7090 computer Margaret Oakley Dayhoff created: The first protein database The first program for sequence assembly There is a need for computers and algorithms that allow: Access, processing, storing, sharing, retrieving, visualizing, annotating…

DNA (nucleotide sequences) databases They are big databases and searching either one should produce similar results because they exchange information routinely. -Gen. Bank (NCBI): www. ncbi. nlm. nih. gov -Arabidopsis: (TAIR) www. arabidopsis. org Specialized databases: Tissues, species… -ESTs (Expressed Sequence Tags) ~at NCBI ~at TIGR -. . . many more!

Comparative genomics BLAST – basic local alignment and search tool (http: //www. ncbi. nlm. nih. gov/) Homologs orthologs paralogs

Question You are a researcher who has tentatively identified a human homolog of a yeast gene. You determine the DNA sequence of c. DNAs of both your yeast gene and the human gene and decide to compare the gene sequences, as well as the predicted protein sequence of each, using alignment software. You would expect the greatest sequence identity from comparisons of the: a. b. c. d. e. c. DNA sequences Protein sequences Genomic DNA sequences Both (a) and (b) will give you equivalent sequence similarity All will give equivalent sequence similarity

What is a microarray?

Types of Arrays • Expression Arrays – c. DNA – Genome • Affymetrix (Gene. Chip®) • Agilent • Tiling arrays

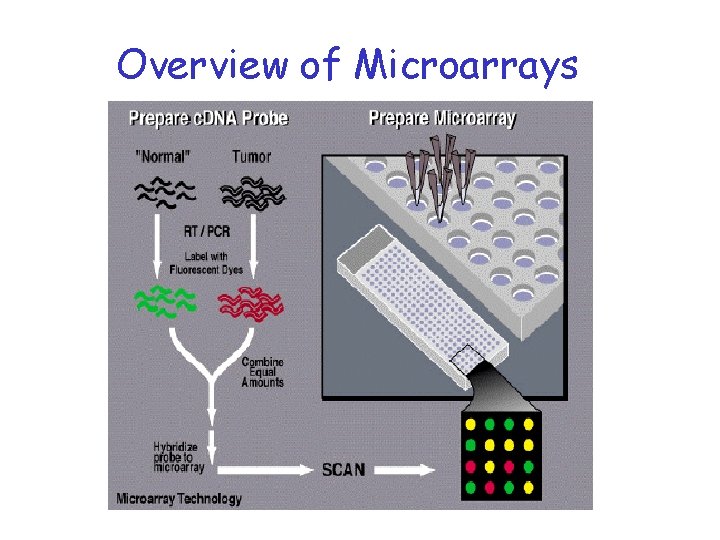
Overview of Microarrays

Transcription Profiling of a mutant WT

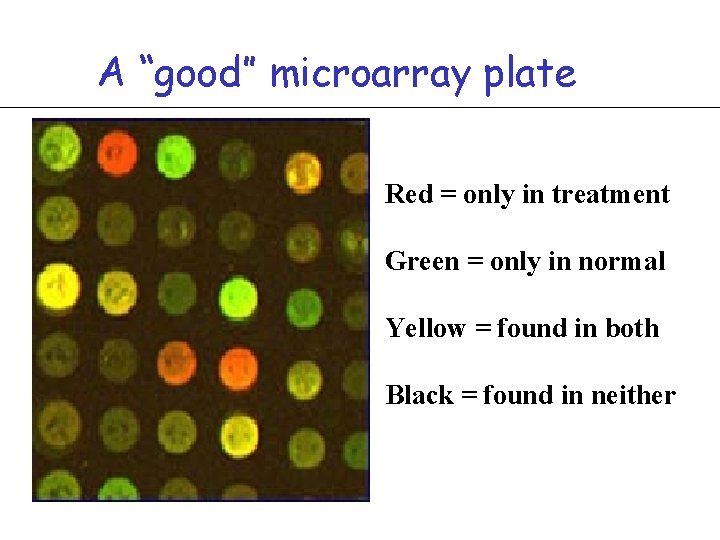
A “good” microarray plate Red = only in treatment Green = only in normal Yellow = found in both Black = found in neither

Results 100’s of genes identified, those turned on, those turned off

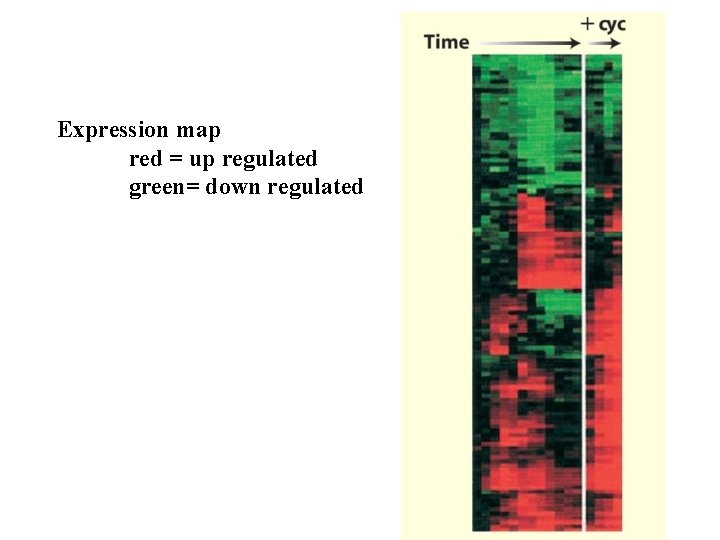
Expression map red = up regulated green= down regulated

Question Microarray technology directly involves: a. b. c. d. e. PCR DNA sequencing Hybridization RFLP detection None of the above

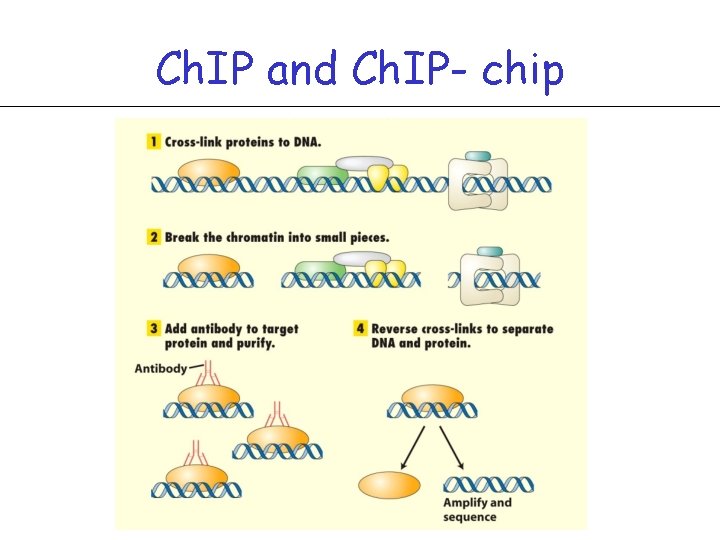
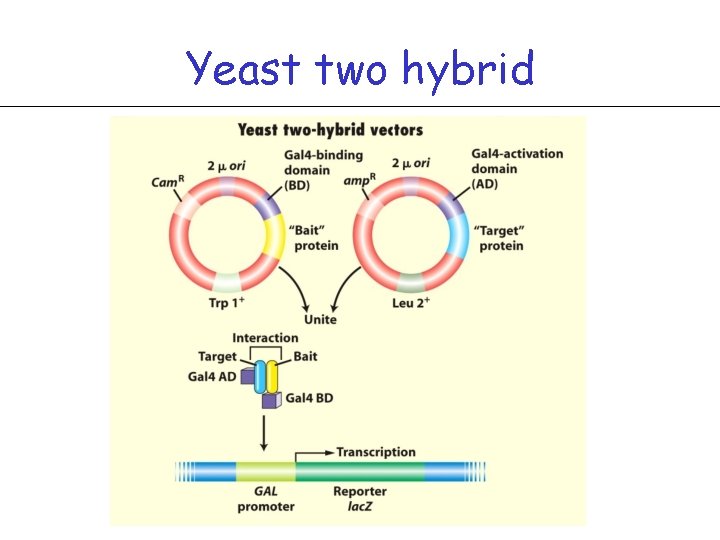
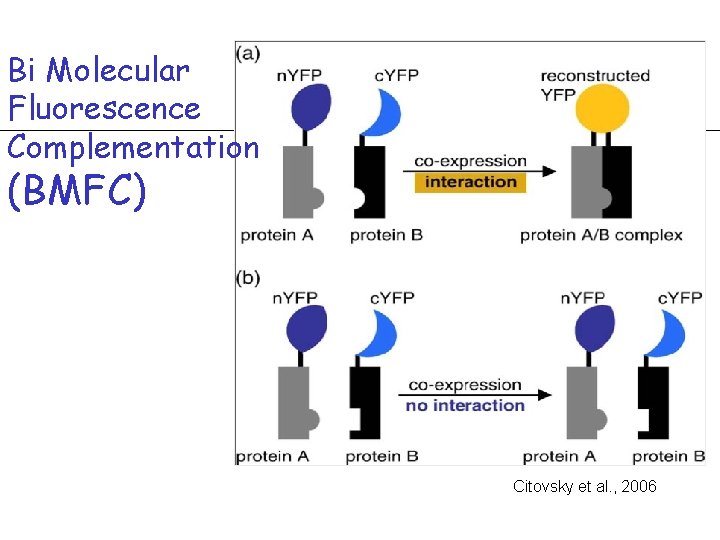
Protein – protein interactions • Ch. IP (chomatin immunoprecipitation) • Yeast two hybrid • Bi Molecular Fluorescence Complementation (BMFC)

Ch. IP and Ch. IP- chip

Yeast two hybrid

Bi Molecular Fluorescence Complementation (BMFC) Citovsky et al. , 2006

Reverse genetics • • Gene knockouts RNAi Overexpression Altered expression

Summary • DNA Sequencing and the rise of genomics • Annotation of genome sequence – Comparative genomics – Functional genomics • Protein-protein interactions • ESTs • Reverse genetics