Genomes Chapter 21 Genomes Sequencing of DNA Human




















- Slides: 49

Genomes Chapter 21

Genomes • • • Sequencing of DNA Human Genome Project 1990 -2003 6 countries 20 research centers

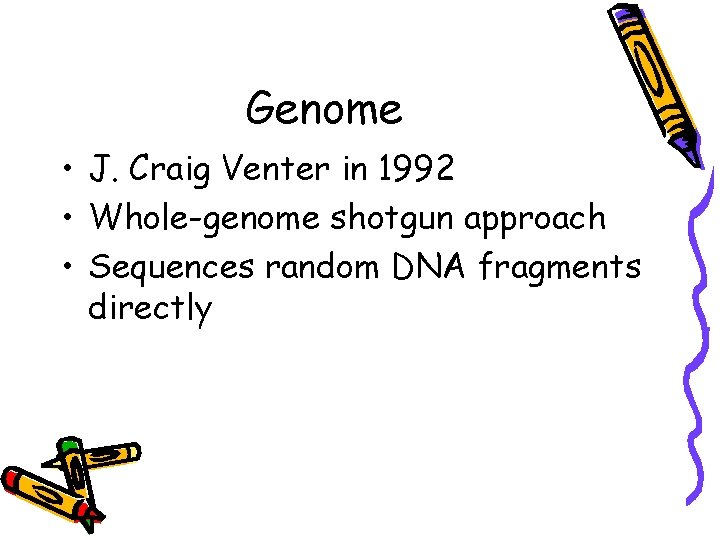
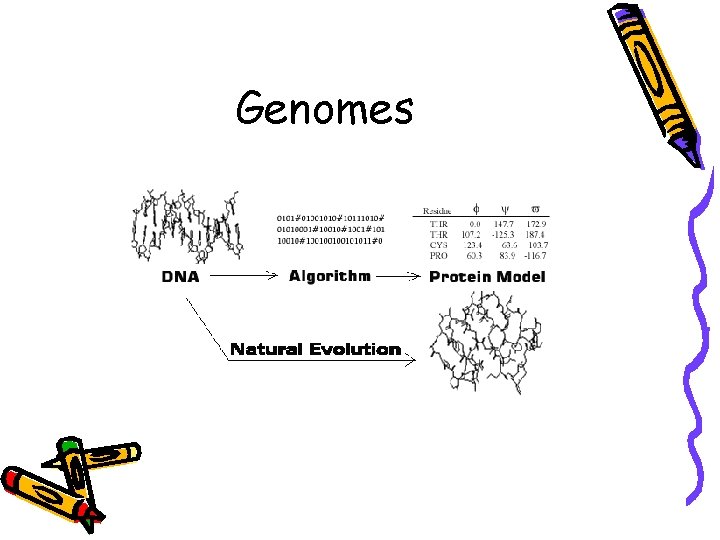
Genome • J. Craig Venter in 1992 • Whole-genome shotgun approach • Sequences random DNA fragments directly

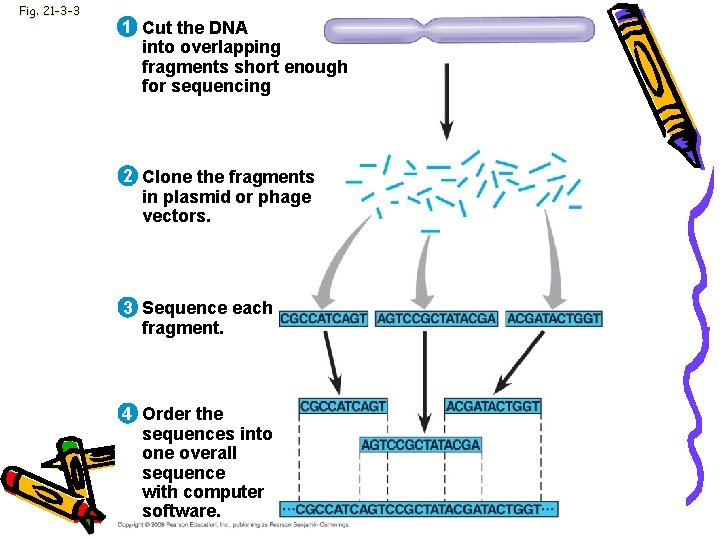
Fig. 21 -3 -3 1 Cut the DNA into overlapping fragments short enough for sequencing 2 Clone the fragments in plasmid or phage vectors. 3 Sequence each fragment. 4 Order the sequences into one overall sequence with computer software.

Genomes • • Complete genome sequences Human, chimpanzee, E. coli, brewer’s yeast Nematode, fruit fly, house mouse,

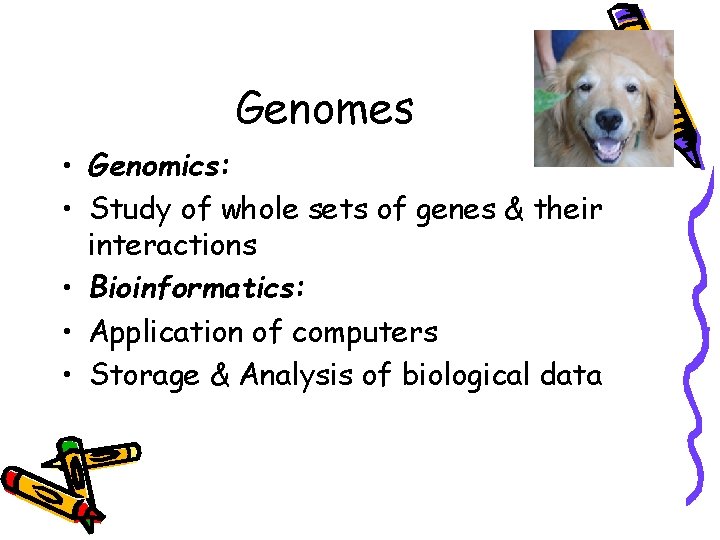
Genomes • Genomics: • Study of whole sets of genes & their interactions • Bioinformatics: • Application of computers • Storage & Analysis of biological data

Genomes • • • Metagenomics DNA Entire groups of species Environmental sample Sequenced Human “microbiome”

Figure 21. 1

Genomes • Comparison • Evolutionary history of genes • Taxonomic groups

Genome • • Phenotype to genotype Red eye fruit flies (w+w or w+w+) Computer analysis of genome Identifies sequences likely to encode proteins • Genotype to phenotype

Genomes

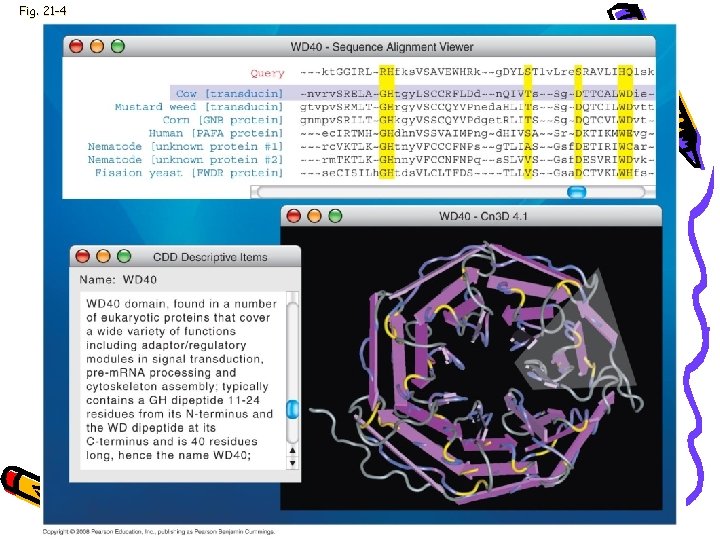
Genome • • • NCBI Genbank BLAST Compare DNA Sequences Compare predicted protein sequences Domains (known aa sequences)

Fig. 21 -4

tatggagaga ataaaagaac tgagagatct aatgtcgcag tcccgcactc gcgagatact 61 cactaagacc actgtggacc atatggccat aatcaaaaag tacacatcag gaaggcaaga 121 gaagaacccc gcactcagaa tgaagtggat gatggcaatg agatacccaa ttacagcaga 181 caagagaata atggacatga ttccagagag gaatgaacaa gggcaaaccc tctggagcaa 241 aacaaacgat gctggatcag accgagtgat ggtatcacct ctggccgtaa catggtggaa 301 taggaatggc ccaacaacaa gtacagttca ttaccctaag gtatataaaa cttatttcga 361 aaaggtcgaa aggttgaaat atggtacctt cggccctgtc cacttcagaa atcaagttaa 421 aataaggagg agagttgata caaaccctgg ccatgcagat ctcagtgcca aggaggcaca 481 ggatgtgatt atggaagttg ttttcccaaa tgaagtgggg gcaagaatac tgacatcaga 541 gtcacagctg gcaataacaa aagagaagaagagctc caggattgta aaattgctcc 601 cttgatggtg gcgtacatgc tagaaagaga attggtccgt aaaacaaggt ttctcccagt 661 agccggcgga acaggcagtg tttatattga agtgttgcac ttaacccaag ggacgtgctg 721 ggagcagatg tacactccag gaggagaagt gagaaatgat gatgttgacc aaagtttgat 781 tatcgctgct agaaacatag taagaagagc agcagtgtca gcagacccat tagcatctct 841 cttggaaatg tgccacagca cacagattgg aggagtaagg atggtggaca tccttagaca 901 gaatccaact gaggaacaag ccgtagacat atgcaaggca gcaatagggt tgaggattag 961 ctcatctttc agttttggtg ggttcacttt caaaaggaca agcggatcat cagtcaagaa

Genome • Proteomics: • Systematic study of all proteins encoded by a genome • Proteins carry out most of the cell’s activities

Application • Finding DNA sequence of organisms • Predict structure & function of new proteins & RNA sequences • Families of related proteins • Phylogenic trees evolutionary relationships

Application • The Cancer Genome Atlas project • Monitors 2, 000 genes in cancer cells for changes • Mutations & rearrangements • Lung, ovarian and glioblastoma • Compare to normal cells

Application • DNA sequencing • Highlight diseases • Specialize tx

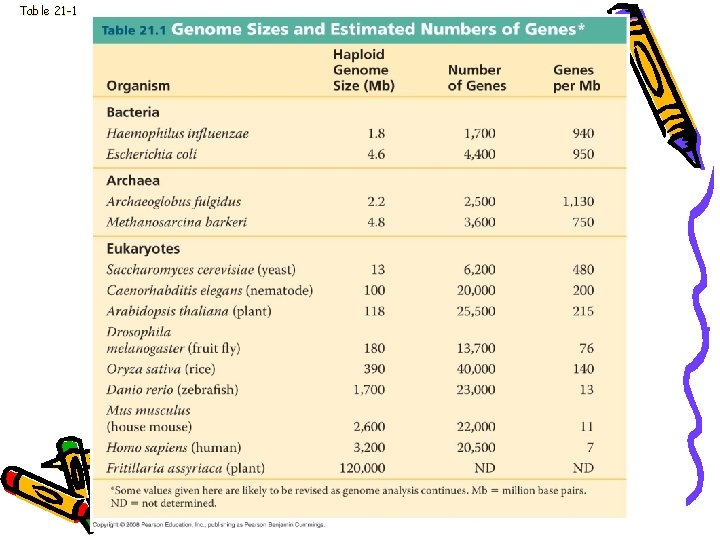
Genome size • Bacteria range from 1 to 6 million base pairs (Mb) • Eukaryotes usually larger • Humans have 3, 200 Mb

Table 21 -1

Fig. 21 -UN 1 Bacteria Archaea Genome size Most are 1– 6 Mb Number of genes 1, 500– 7, 500 Gene density Introns Other noncoding DNA Higher than in eukaryotes None in Present in protein-coding some genes Very little Eukarya Most are 10– 4, 000 Mb, but a few are much larger 5, 000– 40, 000 Lower than in prokaryotes (Within eukaryotes, lower density is correlated with larger genomes. ) Unicellular eukaryotes: present, but prevalent only in some species Multicellular eukaryotes: present in most genes Can be large amounts; generally more repetitive noncoding DNA in multicellular eukaryotes

Genome • • • Gene density: Number of genes in a given length of DNA Humans & other mammals-lowest Multicellular eukaryotes have many introns “Junk DNA”

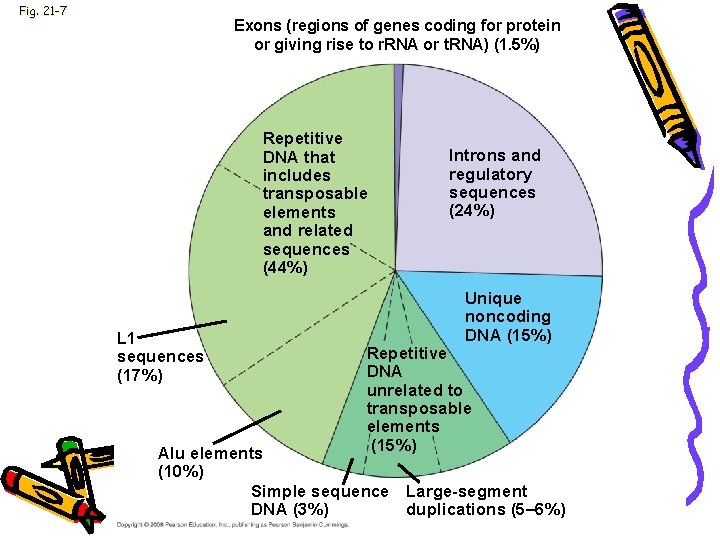
Genome • Genomes of humans, rats, & mice • 500 noncoding regions-are the same • 98. 5% of the genome does not code for proteins, r. RNAs, or t. RNAs • 24% regulatory sequences & introns

Fig. 21 -7 Exons (regions of genes coding for protein or giving rise to r. RNA or t. RNA) (1. 5%) Repetitive DNA that includes transposable elements and related sequences (44%) L 1 sequences (17%) Introns and regulatory sequences (24%) Unique noncoding DNA (15%) Repetitive DNA unrelated to transposable elements (15%) Alu elements (10%) Simple sequence Large-segment DNA (3%) duplications (5– 6%)

Genome • • Pseudogene: Former genes, mutated Repetitive genes: Sequences in multiple copies

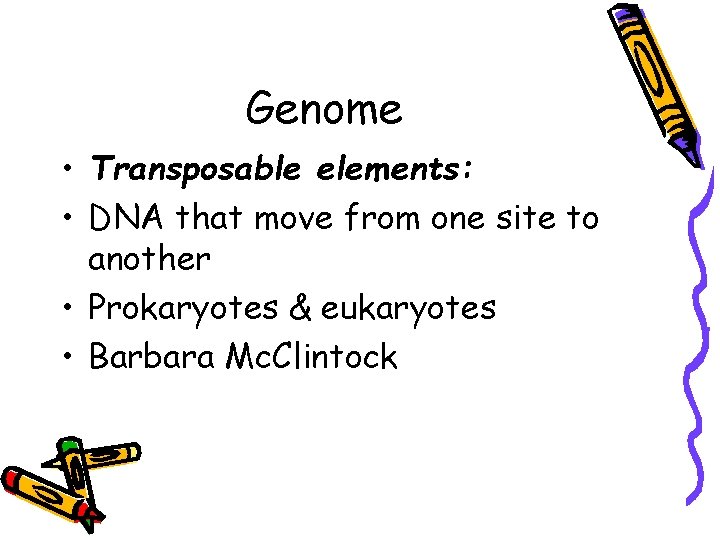
Genome • Transposable elements: • DNA that move from one site to another • Prokaryotes & eukaryotes • Barbara Mc. Clintock

Fig. 21 -8

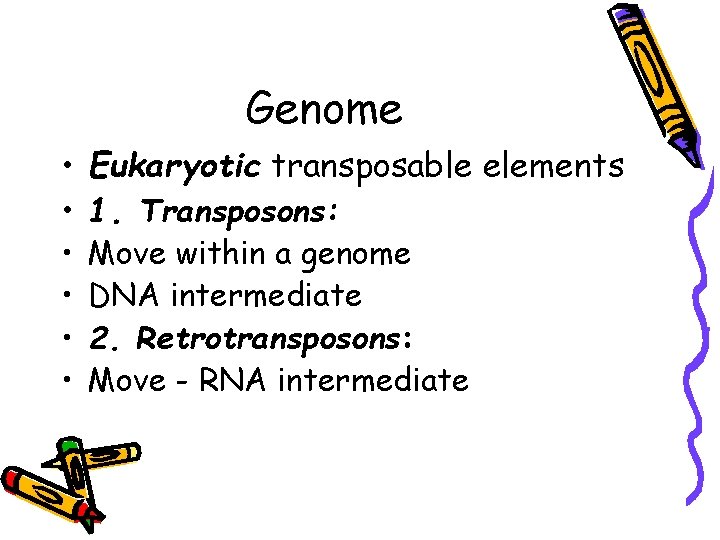
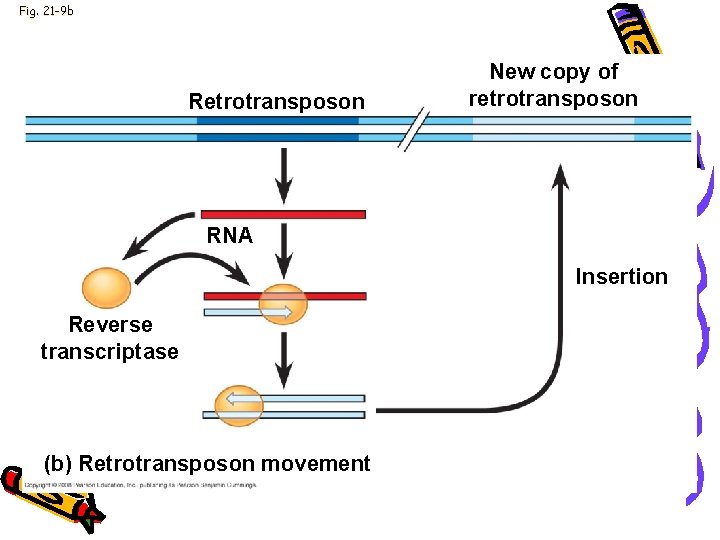
Genome • Eukaryotic transposable elements • 1. Transposons: • • Move within a genome DNA intermediate 2. Retrotransposons: Move - RNA intermediate

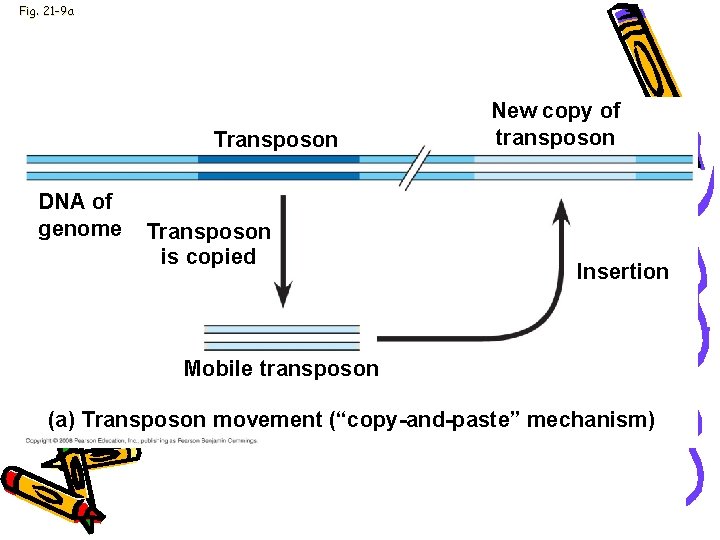
Fig. 21 -9 a Transposon DNA of genome Transposon is copied New copy of transposon Insertion Mobile transposon (a) Transposon movement (“copy-and-paste” mechanism)

Fig. 21 -9 b Retrotransposon New copy of retrotransposon RNA Insertion Reverse transcriptase (b) Retrotransposon movement

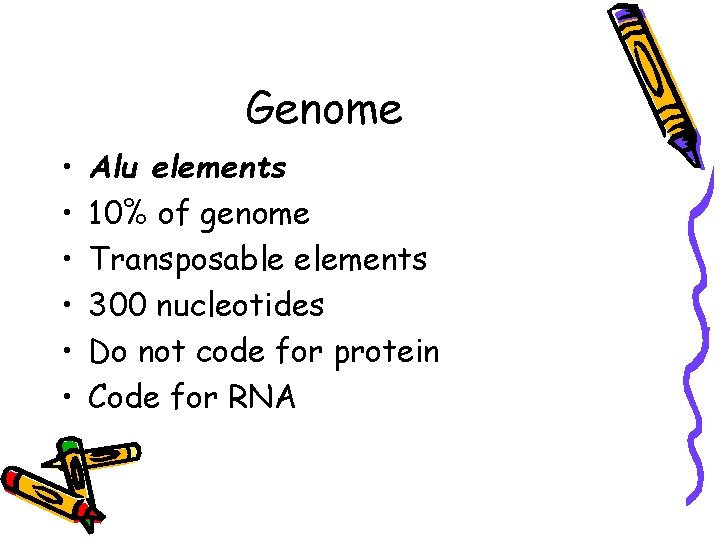
Genome • • • Alu elements 10% of genome Transposable elements 300 nucleotides Do not code for protein Code for RNA

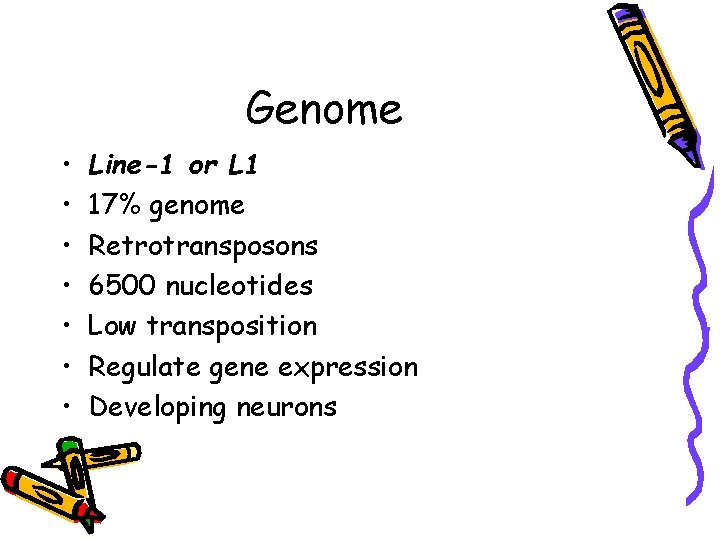
Genome • • Line-1 or L 1 17% genome Retrotransposons 6500 nucleotides Low transposition Regulate gene expression Developing neurons

Genome • • • Repetitive DNA not transposons 15% 1. Long sequences of DNA 2. Simple sequence DNA Many copies of repeated short sequences • GTTACGTTACGTTAC

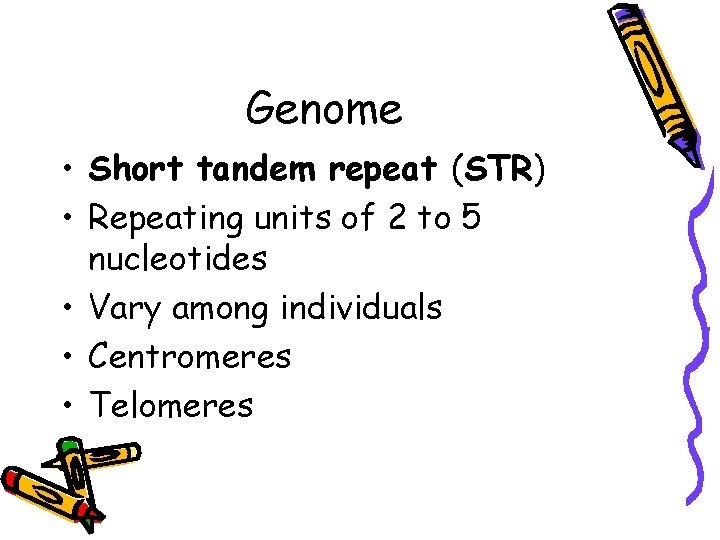
Genome • Short tandem repeat (STR) • Repeating units of 2 to 5 nucleotides • Vary among individuals • Centromeres • Telomeres

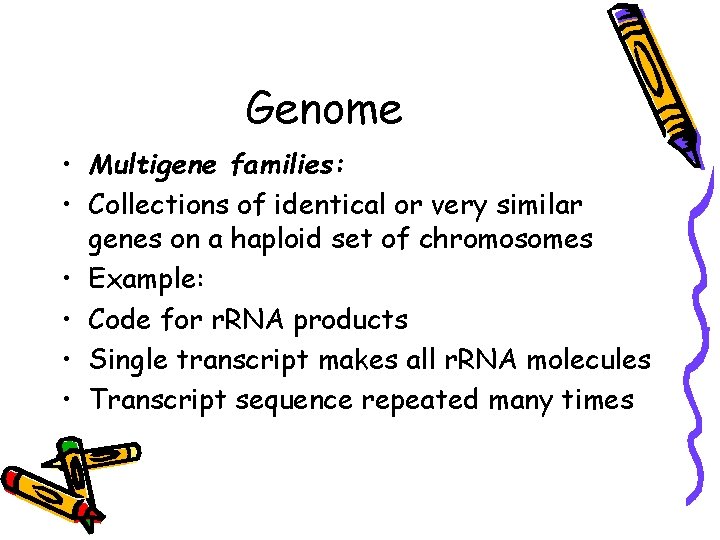
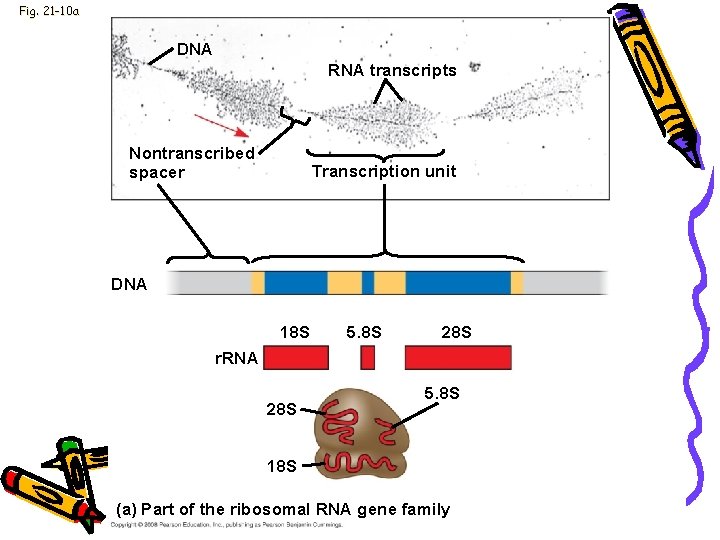
Genome • Multigene families: • Collections of identical or very similar genes on a haploid set of chromosomes • Example: • Code for r. RNA products • Single transcript makes all r. RNA molecules • Transcript sequence repeated many times

Fig. 21 -10 a DNA RNA transcripts Nontranscribed spacer Transcription unit DNA 18 S 5. 8 S 28 S r. RNA 28 S 5. 8 S 18 S (a) Part of the ribosomal RNA gene family

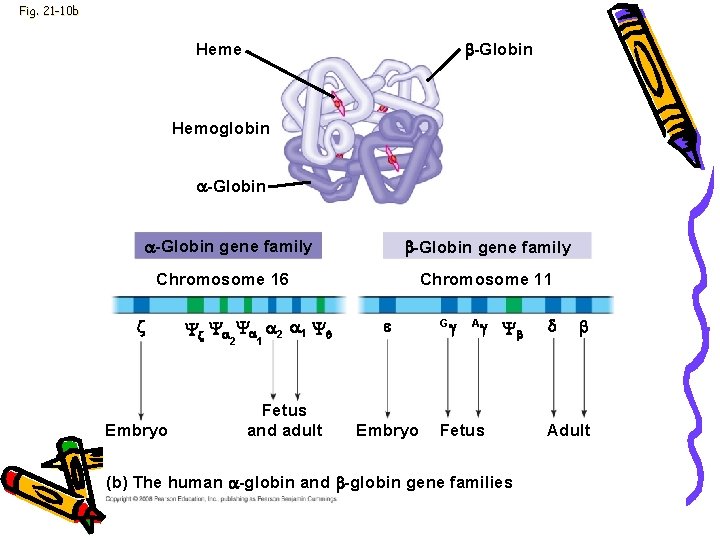
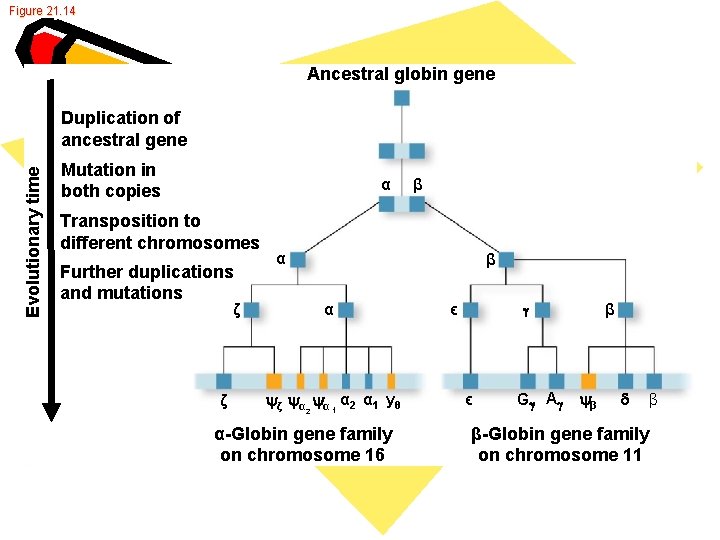
Genome • • • Nonidentical genes Hemoglobin Chromosome 16 -α globulin Chromosome 11 -ß globulin Code separately Animal development

Fig. 21 -10 b -Globin Heme Hemoglobin -Globin gene family Chromosome 16 Embryo 2 1 Fetus and adult Chromosome 11 Embryo G A Fetus (b) The human -globin and -globin gene families Adult

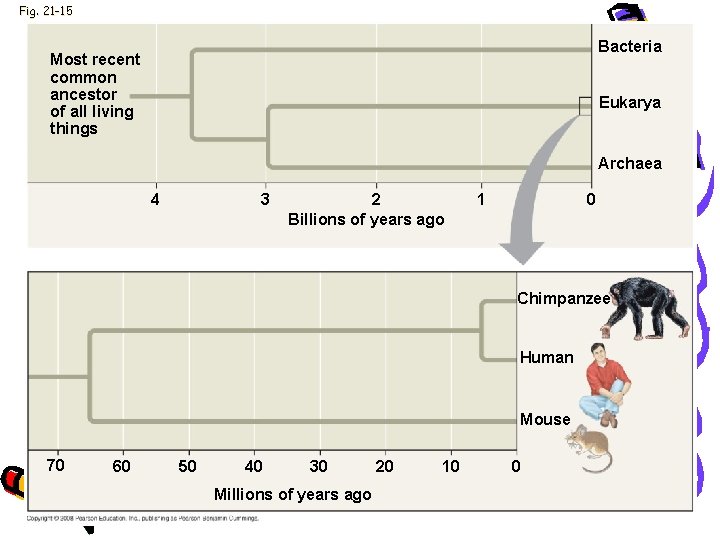
Evolution • • Human & chimpanzee genomes differ by 1. 2% More Alu elements in humans Several genes are evolving faster in humans Genes involved in defense against malaria and tuberculosis • Gene that regulations brain size • Genes that code for transcription factors

Fig. 21 -15 Bacteria Most recent common ancestor of all living things Eukarya Archaea 4 3 2 Billions of years ago 1 0 Chimpanzee Human Mouse 70 60 50 40 30 Millions of years ago 20 10 0

Evolution • FOXP 2 gene • Vocalization • Mutation causes speech impairment • 2 aa difference chimps and humans

Evolution • Humans 23 pairs of chromosomes • Chimpanzees 24 pairs • Humans & chimpanzees diverged from a common ancestor • 2 ancestral chromosomes fused in humans • Duplications & inversions result from mistakes during meiotic recombination

Figure 21. 11 Human chromosome Chimpanzee chromosomes Telomere sequences Centromere sequences Telomere-like sequences 12 Centromere-like sequences 2 13

Figure 21. 12 Human chromosome 16 Mouse chromosomes 7 8 16 17

Figure 21. 14 Ancestral globin gene Evolutionary time Duplication of ancestral gene Mutation in both copies α Transposition to different chromosomes Further duplications and mutations α β α ζ ζ β ζ α α 1 α 2 α 1 y θ 2 α-Globin gene family on chromosome 16 ϵ β ϵ G A β β-Globin gene family on chromosome 11 β

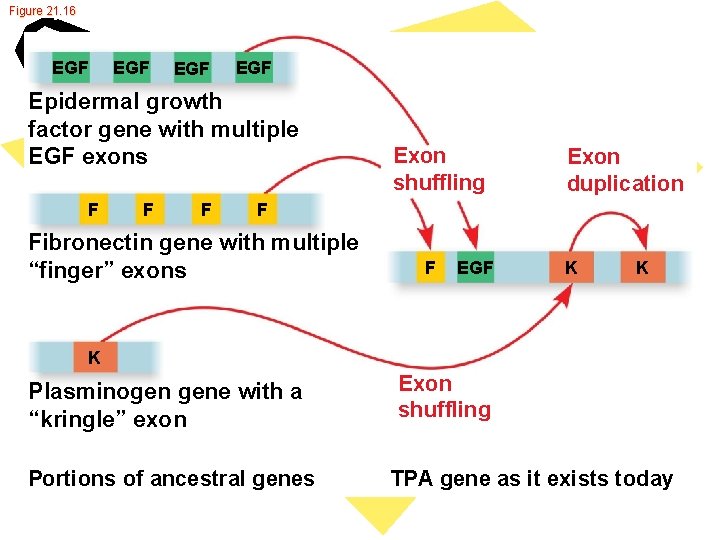
Figure 21. 16 EGF EGF Epidermal growth factor gene with multiple EGF exons F F F Exon shuffling Exon duplication F Fibronectin gene with multiple “finger” exons F EGF K K K Plasminogen gene with a “kringle” exon Portions of ancestral genes Exon shuffling TPA gene as it exists today

Evolution • Evo-devo • Evolutionary developmental biology • Developmental processes in multicellular organisms • Genomic information shows minor differences in gene sequence or regulation • Results in major differences in form

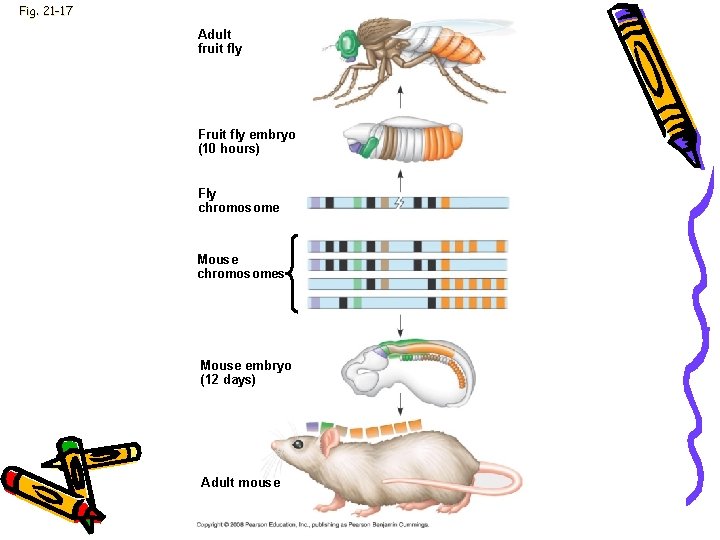
Evolution • • • Homeotic genes Body segments (fruit fly) 180 -nucleotide sequence Homeobox Related homeobox sequences have been found in regulatory genes of yeasts, plants, and even eukaryotes

Fig. 21 -17 Adult fruit fly Fruit fly embryo (10 hours) Fly chromosome Mouse chromosomes Mouse embryo (12 days) Adult mouse
 Chapter 18 genomes and their evolution
Chapter 18 genomes and their evolution Computational biology: genomes, networks, evolution
Computational biology: genomes, networks, evolution Pyrogram
Pyrogram 3rd generation dna sequencing
3rd generation dna sequencing Dna sequencing methods
Dna sequencing methods Dna sequencing
Dna sequencing Dna sequencing
Dna sequencing Dna sequencing applications
Dna sequencing applications Human genome project
Human genome project Dna polymerase function in dna replication
Dna polymerase function in dna replication Bioflix activity dna replication dna replication diagram
Bioflix activity dna replication dna replication diagram Coding dna and non coding dna
Coding dna and non coding dna What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Human needs and human development chapter 8
Human needs and human development chapter 8 Chapter 8 human needs and human development
Chapter 8 human needs and human development Human dna sequence example
Human dna sequence example Higher biology dna replication
Higher biology dna replication Sequencing ap csp
Sequencing ap csp Exome sequencing project
Exome sequencing project Sequence or process writing
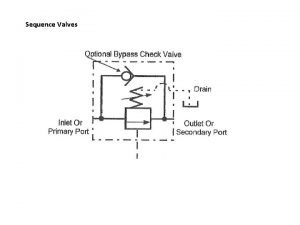
Sequence or process writing Pressure sequence valve
Pressure sequence valve Sequence words
Sequence words Johnson's rule of sequencing
Johnson's rule of sequencing Sam file
Sam file Sequence, selection, and iteration
Sequence, selection, and iteration What is microprogram sequencer
What is microprogram sequencer Johnsons rule
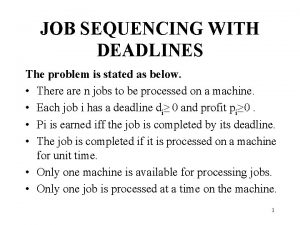
Johnsons rule Job sequencing with deadlines
Job sequencing with deadlines Reversible terminator
Reversible terminator Cask of amontillado quiz
Cask of amontillado quiz What does norman ask melinda?
What does norman ask melinda? Signing naturally homework 5:8
Signing naturally homework 5:8 Pyrosequencing animation
Pyrosequencing animation Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Rooster's off to see the world sequencing
Rooster's off to see the world sequencing History of sequencing
History of sequencing Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Genome sequencing
Genome sequencing History of human genome project
History of human genome project Scheduling loading sequencing and monitoring
Scheduling loading sequencing and monitoring Are you my mother sequencing cards
Are you my mother sequencing cards Calculation priority sequence
Calculation priority sequence Wilkes control unit
Wilkes control unit Sequencing suki
Sequencing suki Greedy algorithm for job sequencing with deadlines
Greedy algorithm for job sequencing with deadlines Sequencing batch reactor
Sequencing batch reactor Panfacial fractures sequencing
Panfacial fractures sequencing Basil khuder
Basil khuder Address sequencing in computer architecture
Address sequencing in computer architecture