Sanger or Dideoxy DNA Sequencing Components for the
- Slides: 18

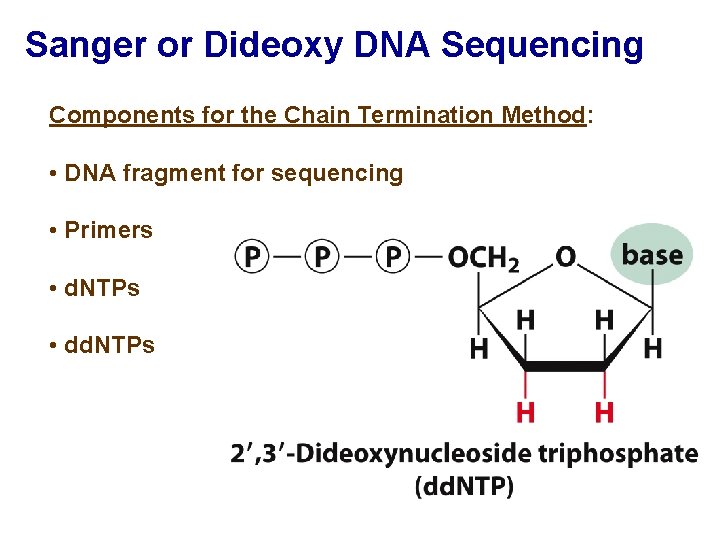
Sanger or Dideoxy DNA Sequencing Components for the Chain Termination Method: • DNA fragment for sequencing • Primers • d. NTPs • dd. NTPs

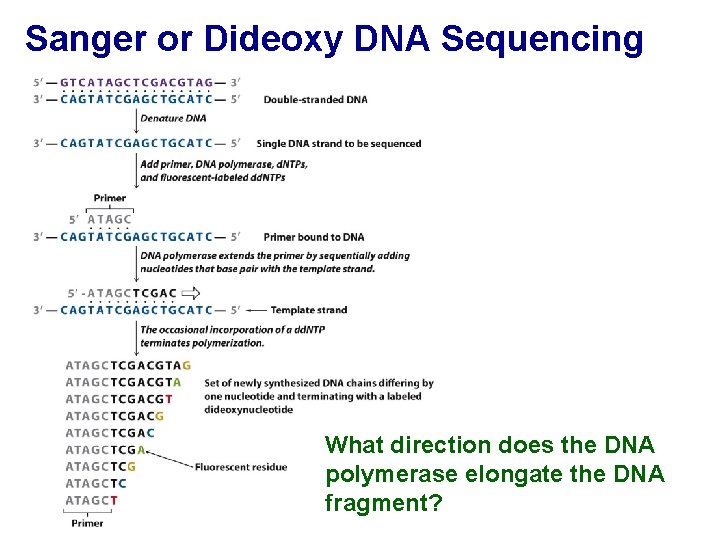
Sanger or Dideoxy DNA Sequencing What direction does the DNA polymerase elongate the DNA fragment?

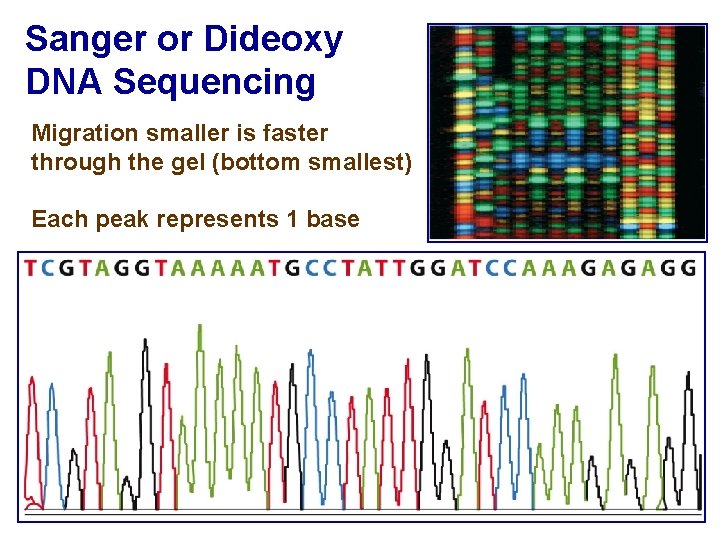
Sanger or Dideoxy DNA Sequencing Migration smaller is faster through the gel (bottom smallest) Each peak represents 1 base

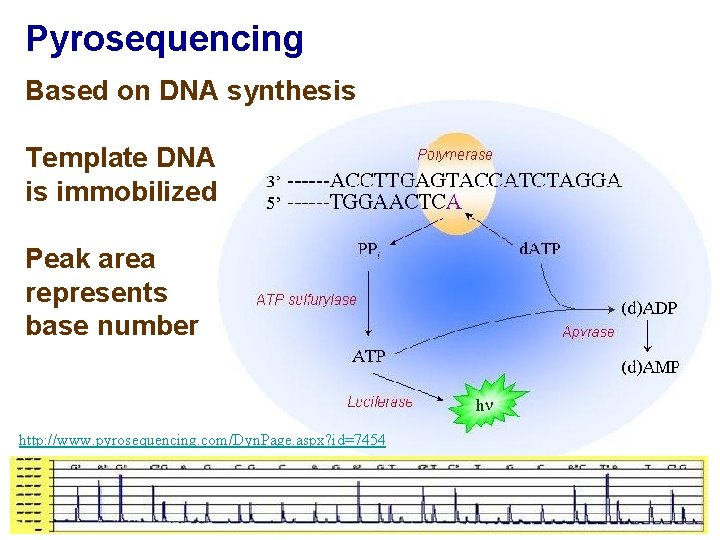
Pyrosequencing Based on DNA synthesis Template DNA is immobilized Peak area represents base number http: //www. pyrosequencing. com/Dyn. Page. aspx? id=7454

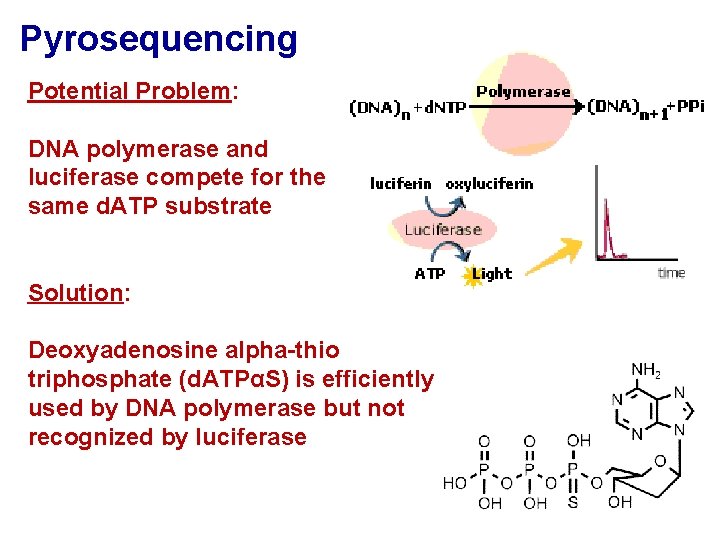
Pyrosequencing Potential Problem: DNA polymerase and luciferase compete for the same d. ATP substrate Solution: Deoxyadenosine alpha-thio triphosphate (d. ATPαS) is efficiently used by DNA polymerase but not recognized by luciferase

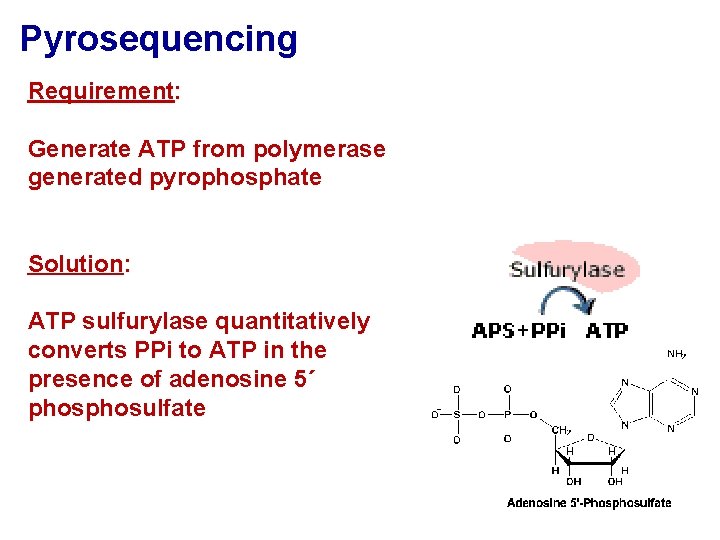
Pyrosequencing Requirement: Generate ATP from polymerase generated pyrophosphate Solution: ATP sulfurylase quantitatively converts PPi to ATP in the presence of adenosine 5´ phosulfate

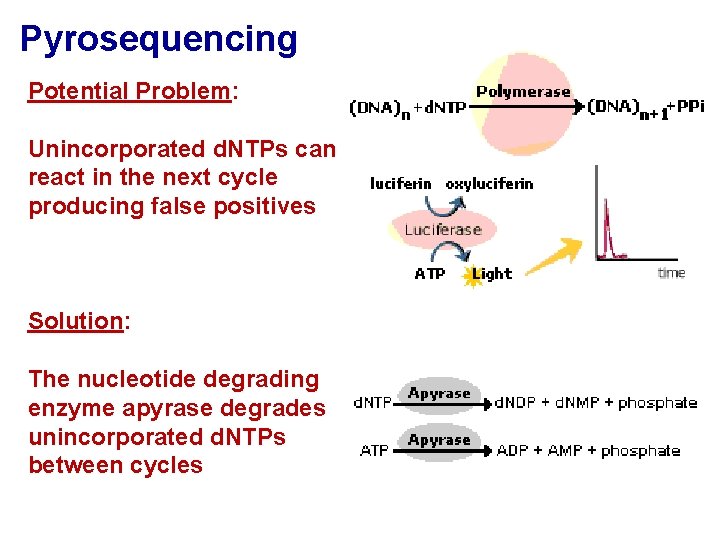
Pyrosequencing Potential Problem: Unincorporated d. NTPs can react in the next cycle producing false positives Solution: The nucleotide degrading enzyme apyrase degrades unincorporated d. NTPs between cycles

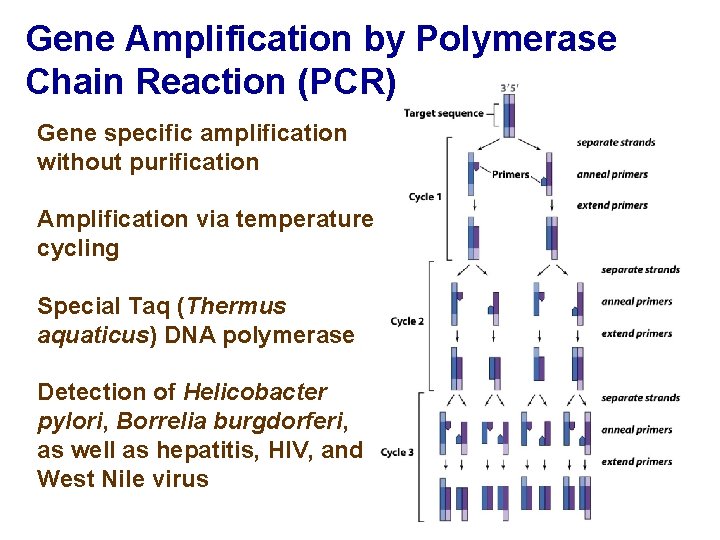
Gene Amplification by Polymerase Chain Reaction (PCR) Gene specific amplification without purification Amplification via temperature cycling Special Taq (Thermus aquaticus) DNA polymerase Detection of Helicobacter pylori, Borrelia burgdorferi, as well as hepatitis, HIV, and West Nile virus

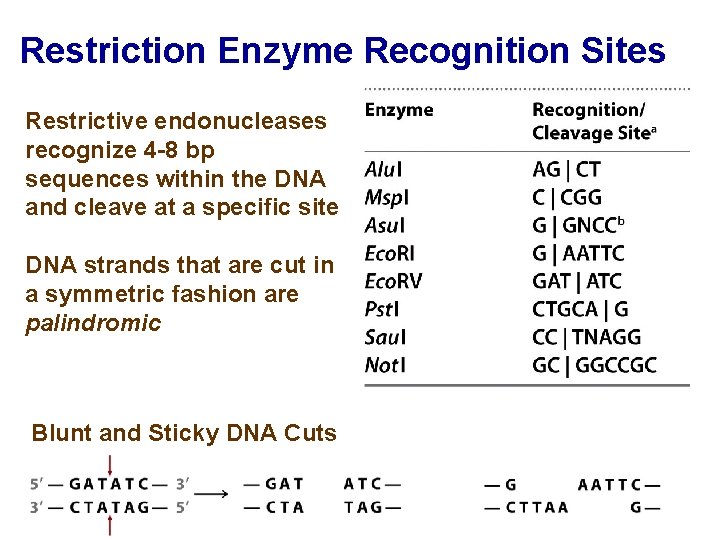
Restriction Enzyme Recognition Sites Restrictive endonucleases recognize 4 -8 bp sequences within the DNA and cleave at a specific site DNA strands that are cut in a symmetric fashion are palindromic Blunt and Sticky DNA Cuts

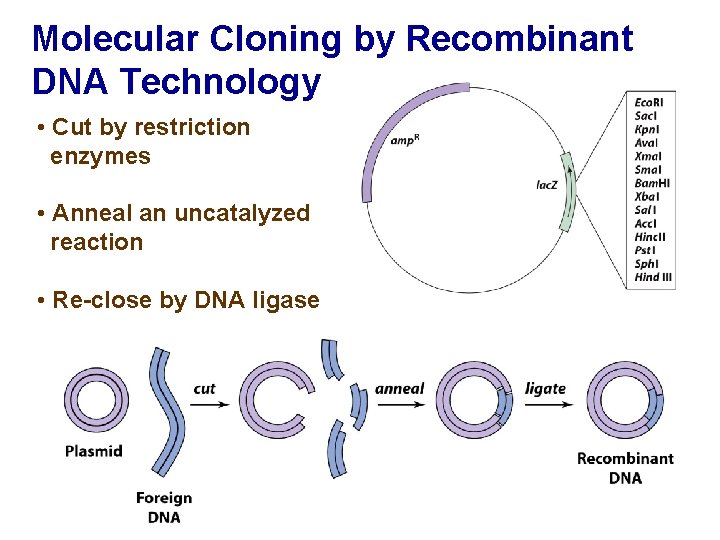
Molecular Cloning by Recombinant DNA Technology • Cut by restriction enzymes • Anneal an uncatalyzed reaction • Re-close by DNA ligase

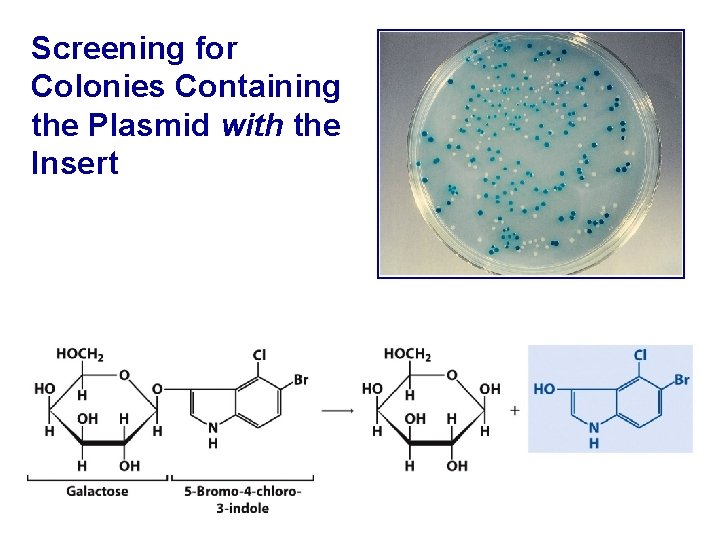
Screening for Colonies Containing the Plasmid with the Insert

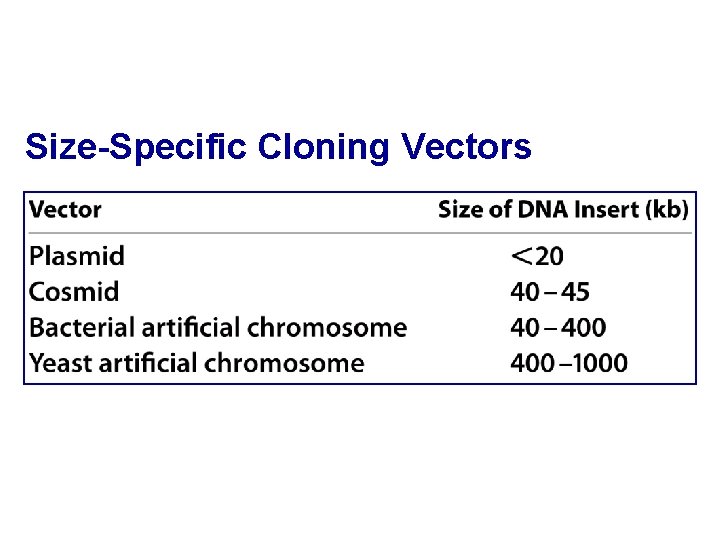
Size-Specific Cloning Vectors

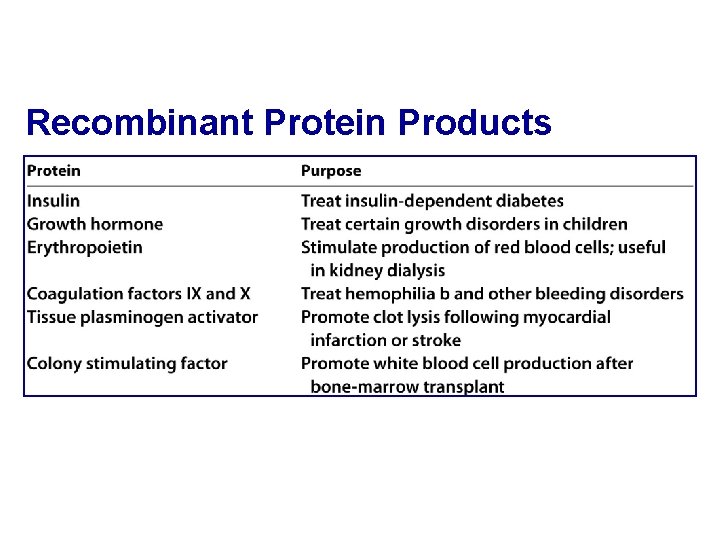
Recombinant Protein Products

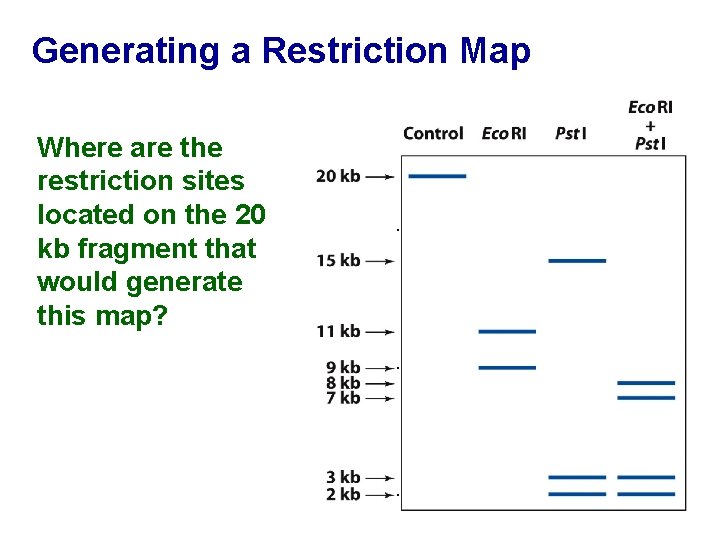
Generating a Restriction Map Where are the restriction sites located on the 20 kb fragment that would generate this map? . . .

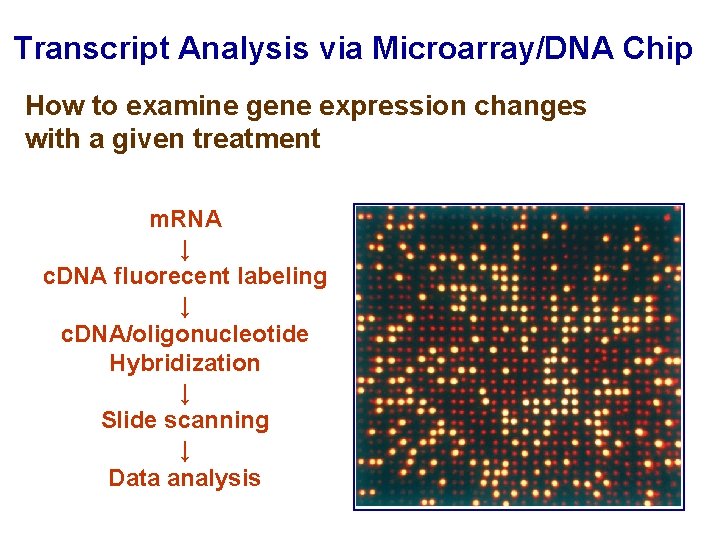
Transcript Analysis via Microarray/DNA Chip How to examine gene expression changes with a given treatment m. RNA ↓ c. DNA fluorecent labeling ↓ c. DNA/oligonucleotide Hybridization ↓ Slide scanning ↓ Data analysis

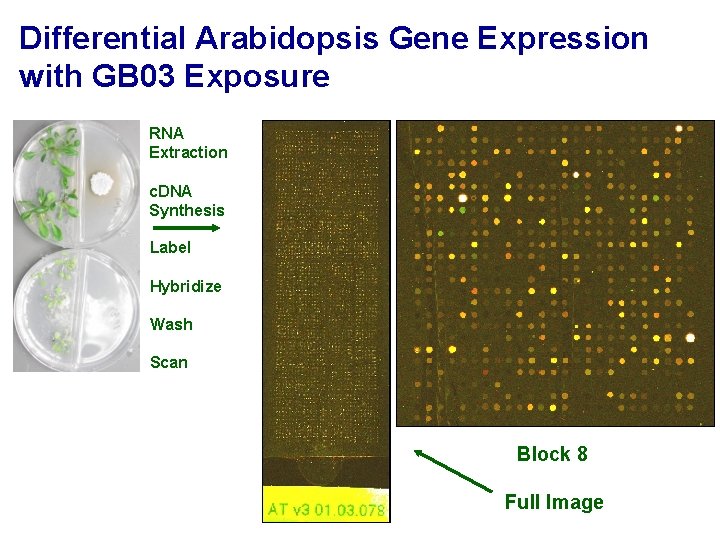
Differential Arabidopsis Gene Expression with GB 03 Exposure RNA Extraction c. DNA Synthesis Label Hybridize Wash Scan Block 8 Full Image

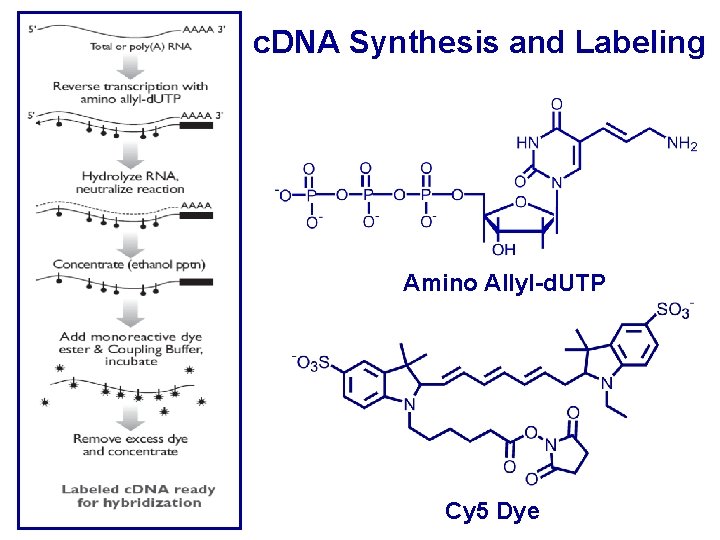
c. DNA Synthesis and Labeling Amino Allyl-d. UTP Cy 5 Dye

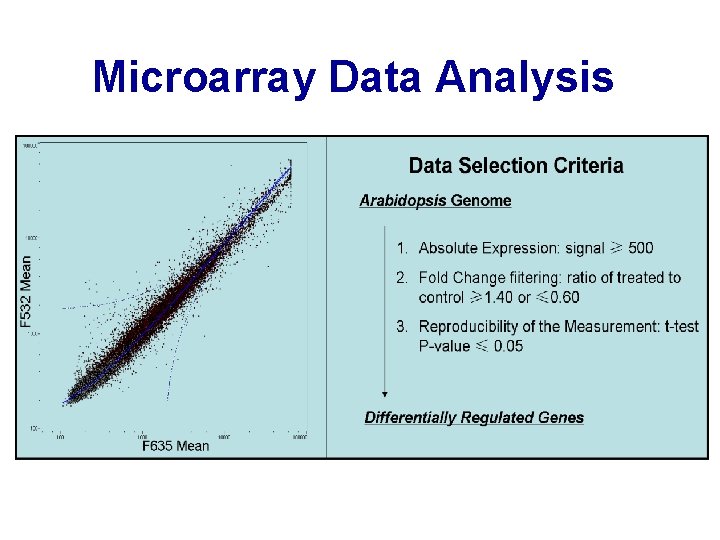
Microarray Data Analysis