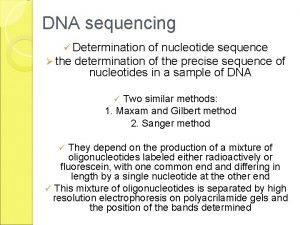
Sanger Sequencing DNA is fragmented Cloned to a





- Slides: 27


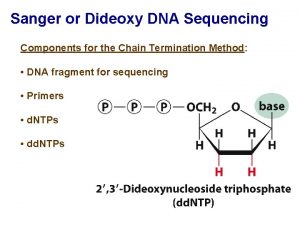
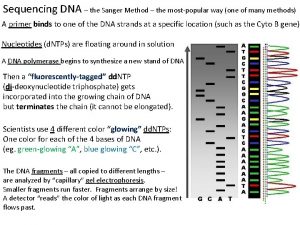
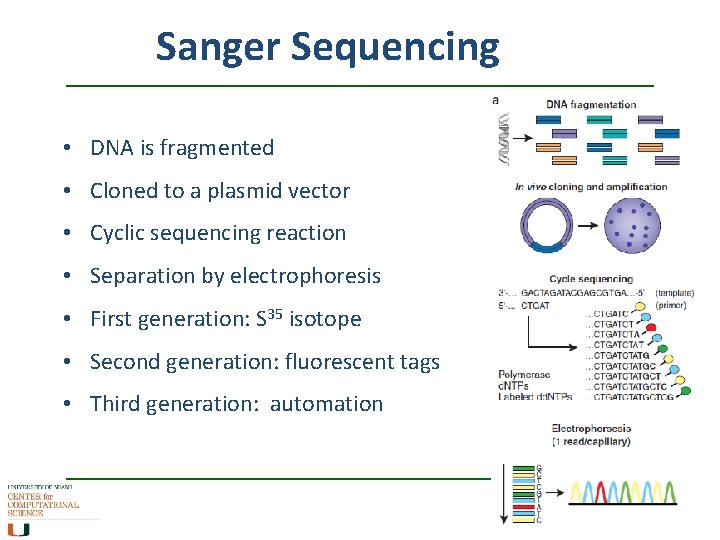
Sanger Sequencing • DNA is fragmented • Cloned to a plasmid vector • Cyclic sequencing reaction • Separation by electrophoresis • First generation: S 35 isotope • Second generation: fluorescent tags • Third generation: automation

“Next” generation sequencing • Not Sanger based biochemistry • DNA is fragmented and sequenced directly • Much more chemistry, physics, and computer science • Characterized by – Parallel Sequencing – High Throughput – Cost • Generates billions of sequences per experiment, highly computational

Human Genome Project ENCODE project Hap. Map project SNP consortium Individual human genomes James Watson, Craig Venter, 3 asian gentlemen

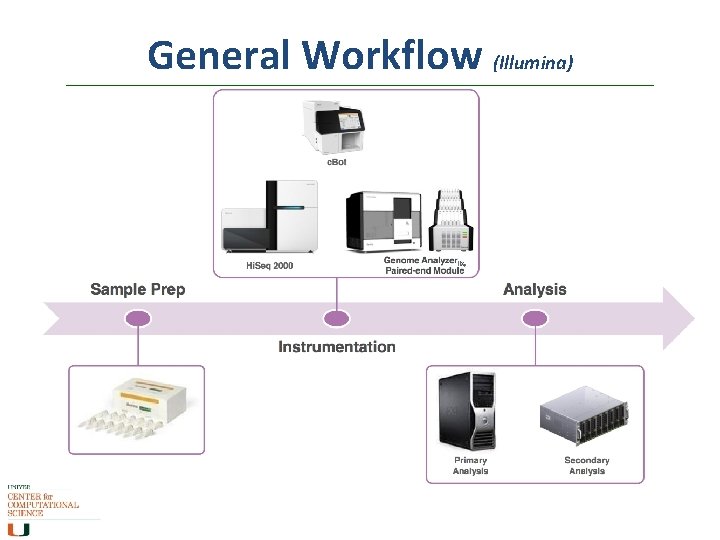
General Workflow (Illumina)

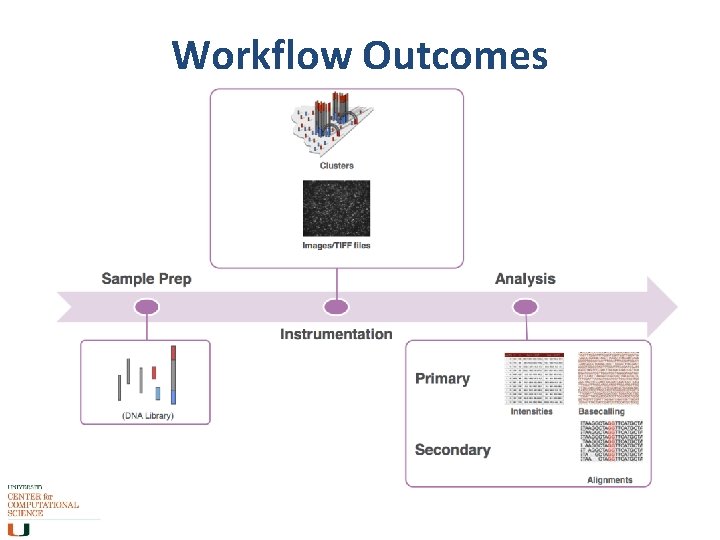
Workflow Outcomes

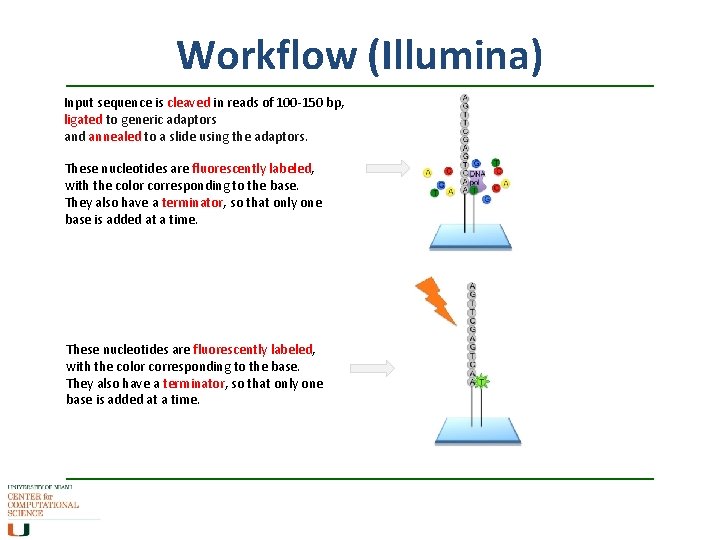
Workflow (Illumina) Input sequence is cleaved in reads of 100 -150 bp, ligated to generic adaptors and annealed to a slide using the adaptors. These nucleotides are fluorescently labeled, with the color corresponding to the base. They also have a terminator, so that only one base is added at a time.

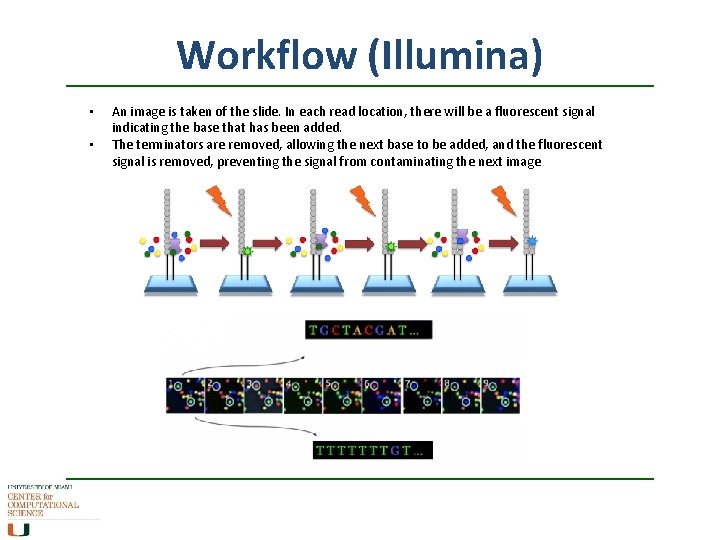
Workflow (Illumina) • • An image is taken of the slide. In each read location, there will be a fluorescent signal indicating the base that has been added. The terminators are removed, allowing the next base to be added, and the fluorescent signal is removed, preventing the signal from contaminating the next image

Data output • • Different technologies, though essentially similar Differences in number, length and quality of the sequences and in format of output Making data handling across platforms and analysis a challenge

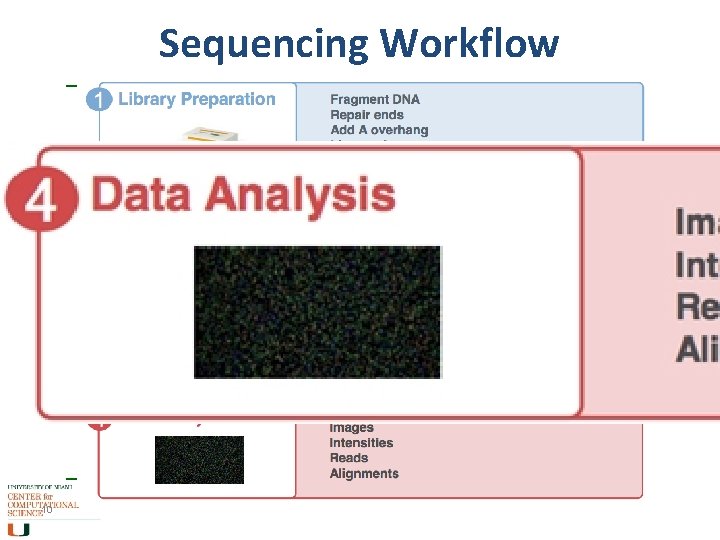
Sequencing Workflow 10

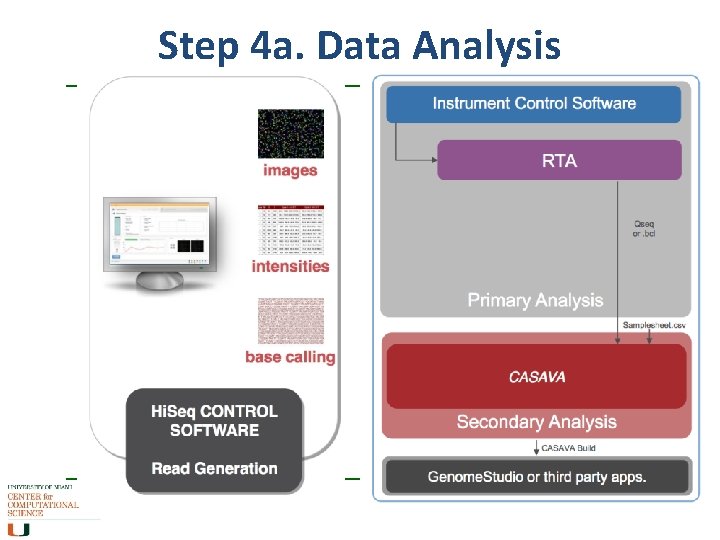
Step 4 a. Data Analysis

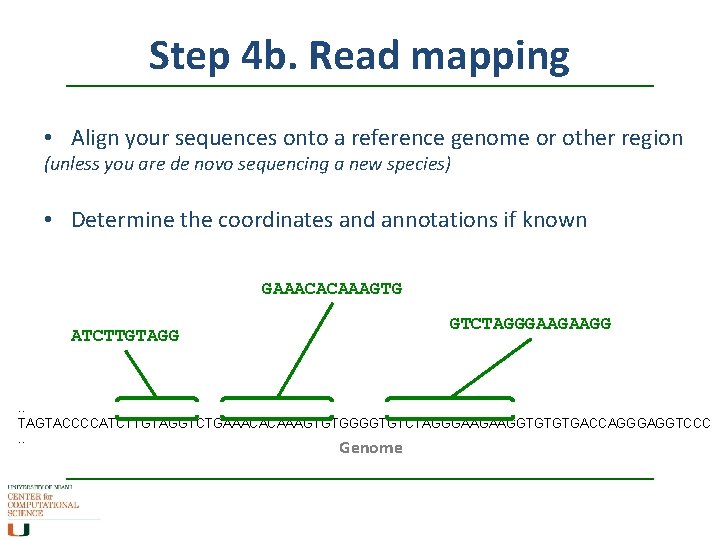
Step 4 b. Read mapping • Align your sequences onto a reference genome or other region (unless you are de novo sequencing a new species) • Determine the coordinates and annotations if known GAAACACAAAGTG GTCTAGGGAAGAAGG ATCTTGTAGG . . TAGTACCCCATCTTGTAGGTCTGAAACACAAAGTGTGGGGTGTCTAGGGAAGAAGGTGTGTGACCAGGGAGGTCCC. . Genome

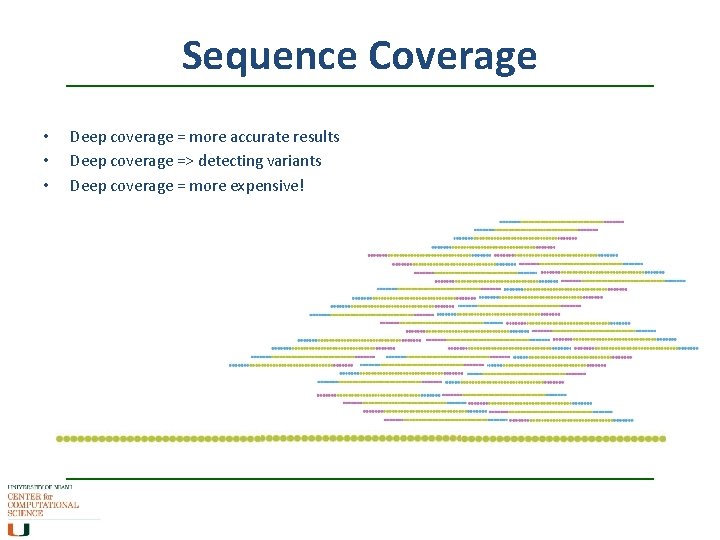
Sequence Coverage • • • Deep coverage = more accurate results Deep coverage => detecting variants Deep coverage = more expensive!


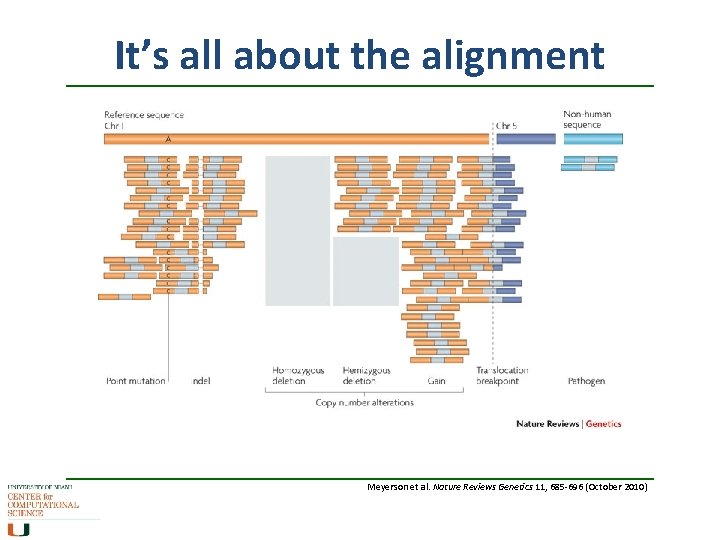
It’s all about the alignment Meyerson et al. Nature Reviews Genetics 11, 685 -696 (October 2010)

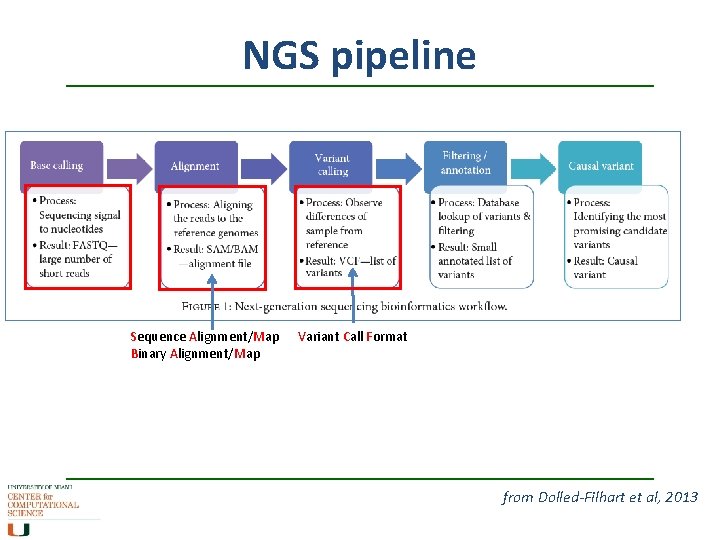
NGS pipeline Sequence Alignment/Map Binary Alignment/Map Variant Call Format from Dolled-Filhart et al, 2013

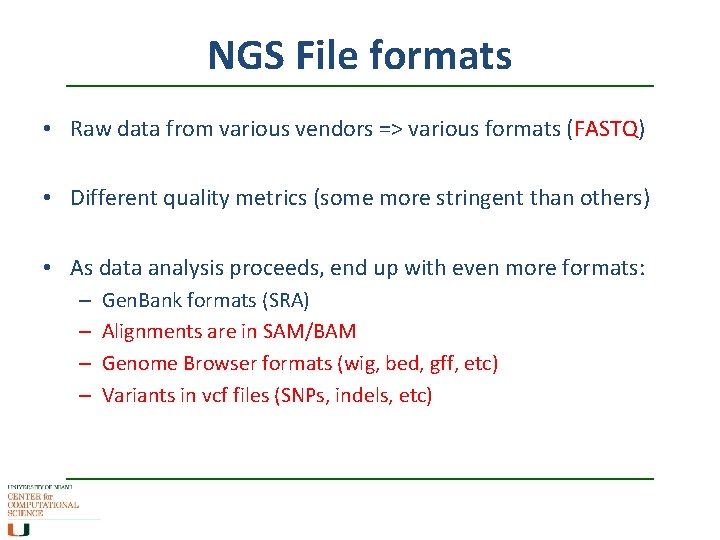
NGS File formats • Raw data from various vendors => various formats (FASTQ) • Different quality metrics (some more stringent than others) • As data analysis proceeds, end up with even more formats: – – Gen. Bank formats (SRA) Alignments are in SAM/BAM Genome Browser formats (wig, bed, gff, etc) Variants in vcf files (SNPs, indels, etc)

FASTQ format: sequence + quality sequence header identifier 1 @SRR 350953. 5 MENDEL_0047_FC 62 MN 8 AAXX: 1: 1: 1646: 938 length=152 NTCTTTTTCCTCTTTTGCCAACTTCAGCTAAATAGGAGCTACACTGATTAGGCAGAAACTTGATTAAC 2 AGGGCTTAAGGTAACCTTGTTGTAGGCCGTTTTGTAGCACTCAAAGCAATTGGTACCTCAACTGCAAAAG TCCTTGGCCC sign 3 +SRR 350953. 5 MENDEL_0047_FC 62 MN 8 AAXX: 1: 1: 1646: 938 length=152 4 raw sequence optional description +50000222 C@@@@@22: : 8888898989: : : <<<: <<<<: : : <<<<<: <: <<<IIIIIGFEEGGGG GGGII@IGDGBGGGGGGDDIIGIIEGIGG>GGGGGGDGGGGGIIHIIBIIIGIIIHIIIIGII quality string c. f. S. Brown NYU


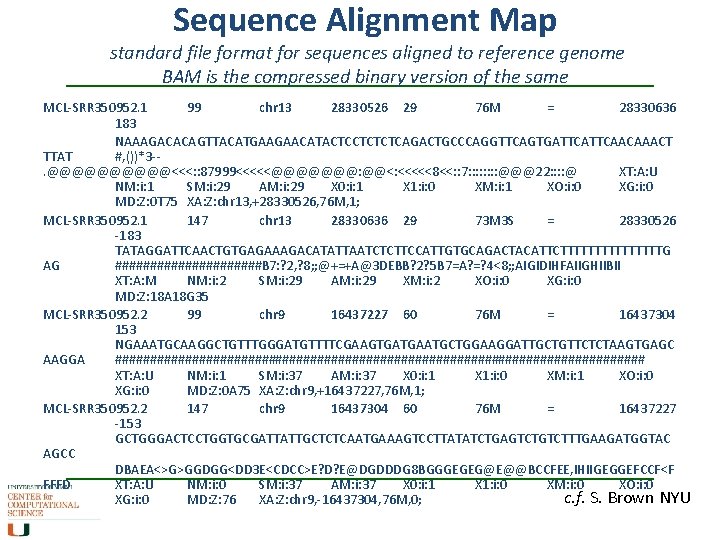
Sequence Alignment Map standard file format for sequences aligned to reference genome BAM is the compressed binary version of the same MCL-SRR 350952. 1 99 chr 13 28330526 29 76 M = 28330636 183 NAAAGACACAGTTACATGAAGAACATACTCCTCTCTCAGACTGCCCAGGTTCAGTGATTCAACAAACT TTAT #, ())*3 -. @@@@@<<<: : 87999<<<<<@@@@@@@: @@<: <<<<<8<<: : 7: : : : @@@22: : @ XT: A: U NM: i: 1 SM: i: 29 AM: i: 29 X 0: i: 1 X 1: i: 0 XM: i: 1 XO: i: 0 XG: i: 0 MD: Z: 0 T 75 XA: Z: chr 13, +28330526, 76 M, 1; MCL-SRR 350952. 1 147 chr 13 28330636 29 73 M 3 S = 28330526 -183 TATAGGATTCAACTGTGAGAAAGACATATTAATCTCTTCCATTGTGCAGACTACATTCTTTTTTTTG AG ###########B 7: ? 2, ? 8; ; @+=+A@3 DEBB? 2? 5 B 7=A? =? 4<8; ; AIGIDIHFAIIGHIIBII XT: A: M NM: i: 2 SM: i: 29 AM: i: 29 XM: i: 2 XO: i: 0 XG: i: 0 MD: Z: 18 A 18 G 35 MCL-SRR 350952. 2 99 chr 9 16437227 60 76 M = 16437304 153 NGAAATGCAAGGCTGTTTGGGATGTTTTCGAAGTGATGAATGCTGGAAGGATTGCTGTTCTCTAAGTGAGC AAGGA ###################################### XT: A: U NM: i: 1 SM: i: 37 AM: i: 37 X 0: i: 1 X 1: i: 0 XM: i: 1 XO: i: 0 XG: i: 0 MD: Z: 0 A 75 XA: Z: chr 9, +16437227, 76 M, 1; MCL-SRR 350952. 2 147 chr 9 16437304 60 76 M = 16437227 -153 GCTGGGACTCCTGGTGCGATTATTGCTCTCAATGAAAGTCCTTATATCTGAGTCTTTGAAGATGGTAC AGCC DBAEA<>G>GGDGG<DD 3 E<CDCC>E? D? E@DGDDDG 8 BGGGEGEG@E@@BCCFEE, IHIIGEGGEFCCF<F FFFD XT: A: U NM: i: 0 SM: i: 37 AM: i: 37 X 0: i: 1 X 1: i: 0 XM: i: 0 XO: i: 0 c. f. S. Brown NYU XG: i: 0 MD: Z: 76 XA: Z: chr 9, -16437304, 76 M, 0;

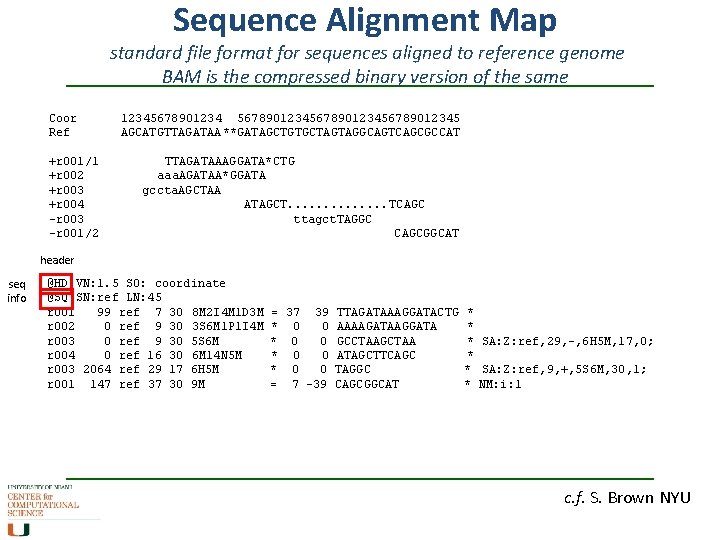
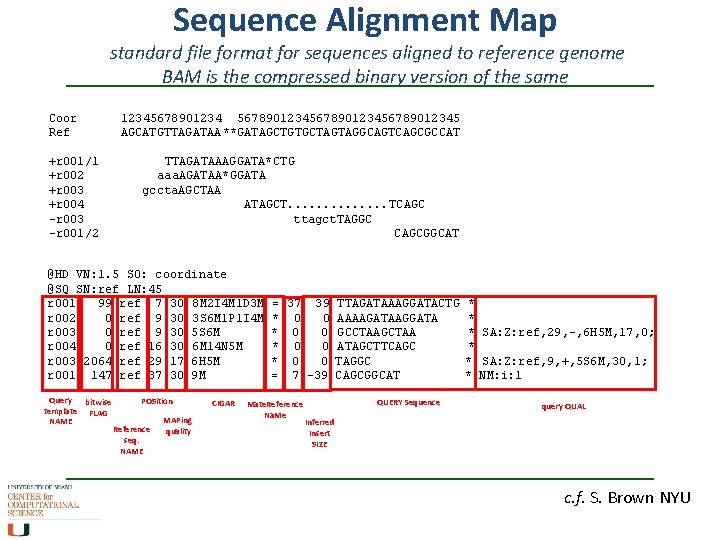
Sequence Alignment Map standard file format for sequences aligned to reference genome BAM is the compressed binary version of the same Coor Ref +r 001/1 +r 002 +r 003 +r 004 -r 003 -r 001/2 1234567890123456789012345 AGCATGTTAGATAA **GATAGCTGTGCTAGTAGGCAGTCAGCGCCAT TTAGATAAAGGATA*CTG aaa. AGATAA*GGATA gccta. AGCTAA ATAGCT. . . TCAGC ttagct. TAGGC CAGCGGCAT header seq info @HD VN: 1. 5 SO: coordinate @SQ SN: ref LN: 45 r 001 99 ref 7 30 8 M 2 I 4 M 1 D 3 M r 002 0 ref 9 30 3 S 6 M 1 P 1 I 4 M r 003 0 ref 9 30 5 S 6 M r 004 0 ref 16 30 6 M 14 N 5 M r 003 2064 ref 29 17 6 H 5 M r 001 147 ref 37 30 9 M = 37 39 * 0 0 = 7 -39 TTAGATAAAGGATACTG AAAAGATAAGGATA GCCTAAGCTAA ATAGCTTCAGC TAGGC CAGCGGCAT * * * SA: Z: ref, 29, -, 6 H 5 M, 17, 0; * * SA: Z: ref, 9, +, 5 S 6 M, 30, 1; * NM: i: 1 c. f. S. Brown NYU

Sequence Alignment Map standard file format for sequences aligned to reference genome BAM is the compressed binary version of the same Coor Ref +r 001/1 +r 002 +r 003 +r 004 -r 003 -r 001/2 1234567890123456789012345 AGCATGTTAGATAA **GATAGCTGTGCTAGTAGGCAGTCAGCGCCAT TTAGATAAAGGATA*CTG aaa. AGATAA*GGATA gccta. AGCTAA ATAGCT. . . TCAGC ttagct. TAGGC CAGCGGCAT @HD VN: 1. 5 SO: coordinate @SQ SN: ref LN: 45 r 001 99 ref 7 30 8 M 2 I 4 M 1 D 3 M r 002 0 ref 9 30 3 S 6 M 1 P 1 I 4 M r 003 0 ref 9 30 5 S 6 M r 004 0 ref 16 30 6 M 14 N 5 M r 003 2064 ref 29 17 6 H 5 M r 001 147 ref 37 30 9 M Query bitwise template FLAG NAME POSition Reference seq. NAME MAPing quality CIGAR = 37 39 * 0 0 = 7 -39 Mate. Reference Na. Me TTAGATAAAGGATACTG AAAAGATAAGGATA GCCTAAGCTAA ATAGCTTCAGC TAGGC CAGCGGCAT QUERY Sequence * * * SA: Z: ref, 29, -, 6 H 5 M, 17, 0; * * SA: Z: ref, 9, +, 5 S 6 M, 30, 1; * NM: i: 1 query QUAL Inferred Insert SIZE c. f. S. Brown NYU

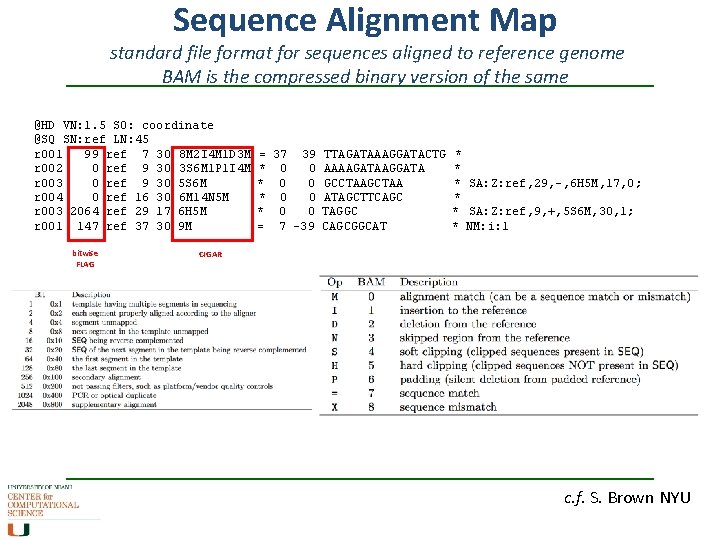
Sequence Alignment Map standard file format for sequences aligned to reference genome BAM is the compressed binary version of the same @HD VN: 1. 5 SO: coordinate @SQ SN: ref LN: 45 r 001 99 ref 7 30 8 M 2 I 4 M 1 D 3 M r 002 0 ref 9 30 3 S 6 M 1 P 1 I 4 M r 003 0 ref 9 30 5 S 6 M r 004 0 ref 16 30 6 M 14 N 5 M r 003 2064 ref 29 17 6 H 5 M r 001 147 ref 37 30 9 M bitwise FLAG = 37 39 * 0 0 = 7 -39 TTAGATAAAGGATACTG AAAAGATAAGGATA GCCTAAGCTAA ATAGCTTCAGC TAGGC CAGCGGCAT * * * SA: Z: ref, 29, -, 6 H 5 M, 17, 0; * * SA: Z: ref, 9, +, 5 S 6 M, 30, 1; * NM: i: 1 CIGAR c. f. S. Brown NYU

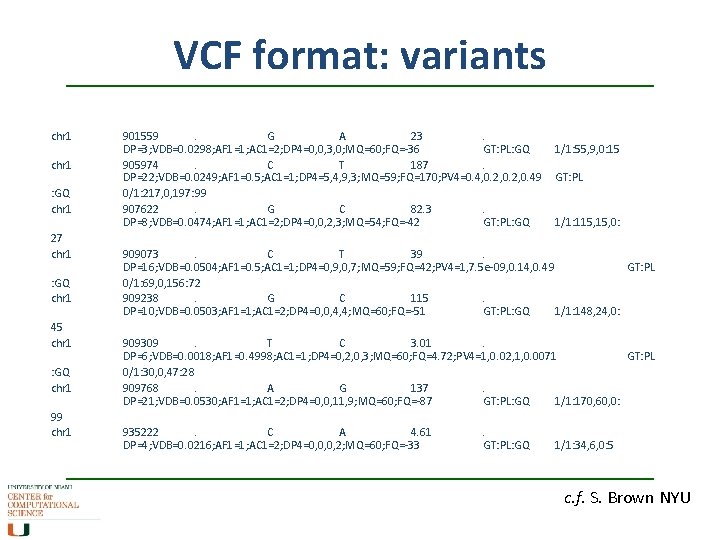
VCF format: variants chr 1 : GQ chr 1 27 chr 1 : GQ chr 1 45 chr 1 : GQ chr 1 99 chr 1 901559. G A 23. DP=3; VDB=0. 0298; AF 1=1; AC 1=2; DP 4=0, 0, 3, 0; MQ=60; FQ=-36 GT: PL: GQ 905974. C T 187. DP=22; VDB=0. 0249; AF 1=0. 5; AC 1=1; DP 4=5, 4, 9, 3; MQ=59; FQ=170; PV 4=0. 4, 0. 2, 0. 49 0/1: 217, 0, 197: 99 907622. G C 82. 3. DP=8; VDB=0. 0474; AF 1=1; AC 1=2; DP 4=0, 0, 2, 3; MQ=54; FQ=-42 GT: PL: GQ 1/1: 55, 9, 0: 15 GT: PL 1/1: 115, 0: 909073. C T 39. DP=16; VDB=0. 0504; AF 1=0. 5; AC 1=1; DP 4=0, 9, 0, 7; MQ=59; FQ=42; PV 4=1, 7. 5 e-09, 0. 14, 0. 49 GT: PL 0/1: 69, 0, 156: 72 909238. G C 115. DP=10; VDB=0. 0503; AF 1=1; AC 1=2; DP 4=0, 0, 4, 4; MQ=60; FQ=-51 GT: PL: GQ 1/1: 148, 24, 0: 909309. T C 3. 01. DP=6; VDB=0. 0018; AF 1=0. 4998; AC 1=1; DP 4=0, 2, 0, 3; MQ=60; FQ=4. 72; PV 4=1, 0. 02, 1, 0. 0071 GT: PL 0/1: 30, 0, 47: 28 909768. A G 137. DP=21; VDB=0. 0530; AF 1=1; AC 1=2; DP 4=0, 0, 11, 9; MQ=60; FQ=-87 GT: PL: GQ 1/1: 170, 60, 0: 935222. C A 4. 61 DP=4; VDB=0. 0216; AF 1=1; AC 1=2; DP 4=0, 0, 0, 2; MQ=60; FQ=-33 . GT: PL: GQ 1/1: 34, 6, 0: 5 c. f. S. Brown NYU

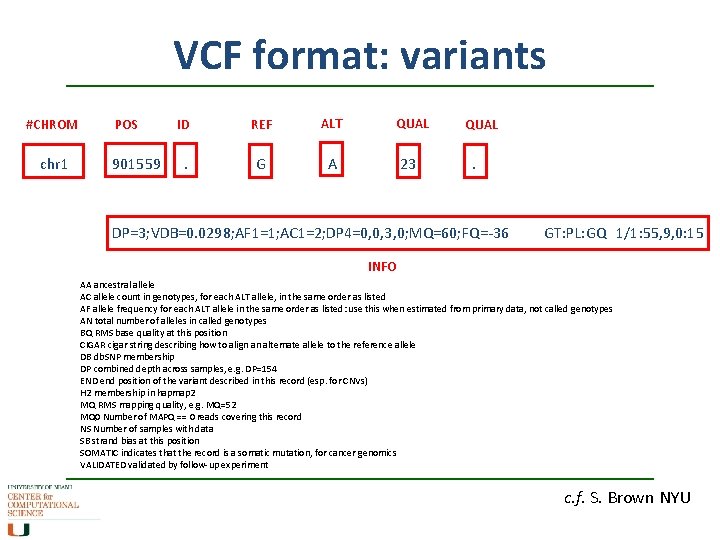
VCF format: variants #CHROM chr 1 POS 901559 ID REF ALT QUAL . G A 23 QUAL . DP=3; VDB=0. 0298; AF 1=1; AC 1=2; DP 4=0, 0, 3, 0; MQ=60; FQ=-36 GT: PL: GQ 1/1: 55, 9, 0: 15 INFO AA ancestral allele AC allele count in genotypes, for each ALT allele, in the same order as listed AF allele frequency for each ALT allele in the same order as listed: use this when estimated from primary data, not called genotypes AN total number of alleles in called genotypes BQ RMS base quality at this position CIGAR cigar string describing how to align an alternate allele to the reference allele DB db. SNP membership DP combined depth across samples, e. g. DP=154 END end position of the variant described in this record (esp. for CNVs) H 2 membership in hapmap 2 MQ RMS mapping quality, e. g. MQ=52 MQ 0 Number of MAPQ == 0 reads covering this record NS Number of samples with data SB strand bias at this position SOMATIC indicates that the record is a somatic mutation, for cancer genomics VALIDATED validated by follow-up experiment c. f. S. Brown NYU

Alignment or NGS What are the challenges? c. f. S. Brown NYU

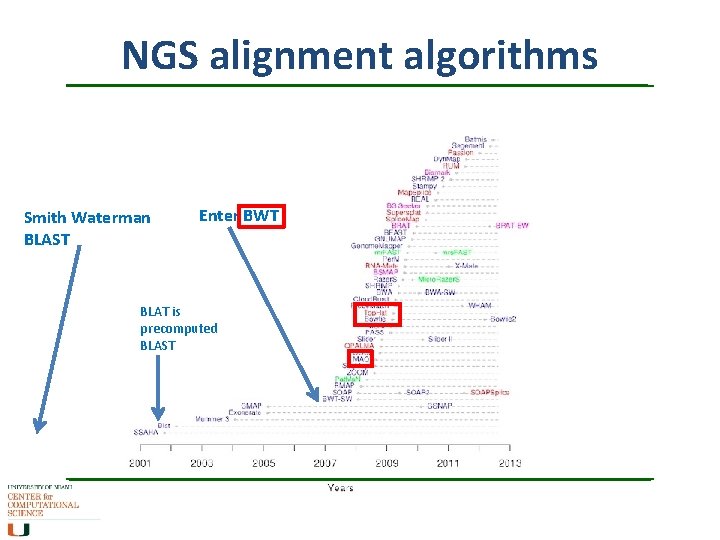
NGS alignment algorithms Smith Waterman BLAST Enter BWT BLAT is precomputed BLAST
 Dna sequencing importance
Dna sequencing importance Sanger vs maxam gilbert sequencing
Sanger vs maxam gilbert sequencing Sanger vs ngs
Sanger vs ngs Sanger sequencing
Sanger sequencing Helioscope sequencing
Helioscope sequencing Contigs
Contigs Dna sequencing
Dna sequencing 3rd generation dna sequencing
3rd generation dna sequencing Dna sequencing applications
Dna sequencing applications Planned parenthood is done margaret sanger
Planned parenthood is done margaret sanger Hvorfor feirer vi 17 mai
Hvorfor feirer vi 17 mai Hristrose
Hristrose Sanger and edman method
Sanger and edman method Sanger
Sanger Fragmented care definition
Fragmented care definition An embryonic industry is one that
An embryonic industry is one that Noun cluster adalah
Noun cluster adalah Fragmented logistics structure
Fragmented logistics structure Elizabeth picks up the clothes from her bedroom floor
Elizabeth picks up the clothes from her bedroom floor What year did darcydi nucci coined it as fragmented future?
What year did darcydi nucci coined it as fragmented future? Mind mapping kesehatan
Mind mapping kesehatan Disadvantages of perforated states
Disadvantages of perforated states Fragmented logistics structure
Fragmented logistics structure Medulla patterns
Medulla patterns Mature industry
Mature industry Faulty parallel structure
Faulty parallel structure Coding dna and non coding dna
Coding dna and non coding dna What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna?