3 Genome Annotation Gene Prediction II Gene Prediction










- Slides: 40

3. Genome Annotation: Gene Prediction (II)

Gene Prediction: Computational Challenge • Gene: A sequence of nucleotides coding for protein • Gene Prediction Problem: Determine the beginning and end positions of genes in a genome

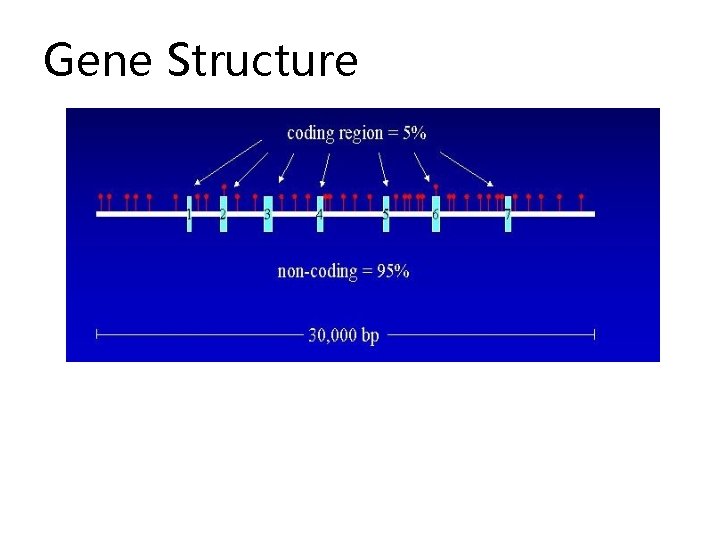
Eukaryotic gene finding • On average, vertebrate gene is about 30 KB long • Coding region takes about 1 KB • Exon sizes vary from double digit numbers to kilobases • An average 5’ UTR is about 750 bp • An average 3’UTR is about 450 bp but both can be much longer.

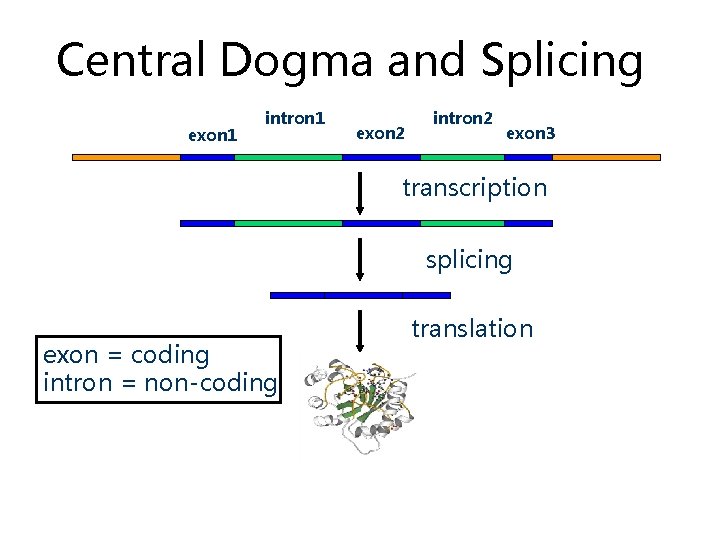
Exons and Introns • In eukaryotes, the gene is a combination of coding segments (exons) that are interrupted by non-coding segments (introns) • This makes computational gene prediction in eukaryotes even more difficult • Prokaryotes don’t have introns - Genes in prokaryotes are continuous

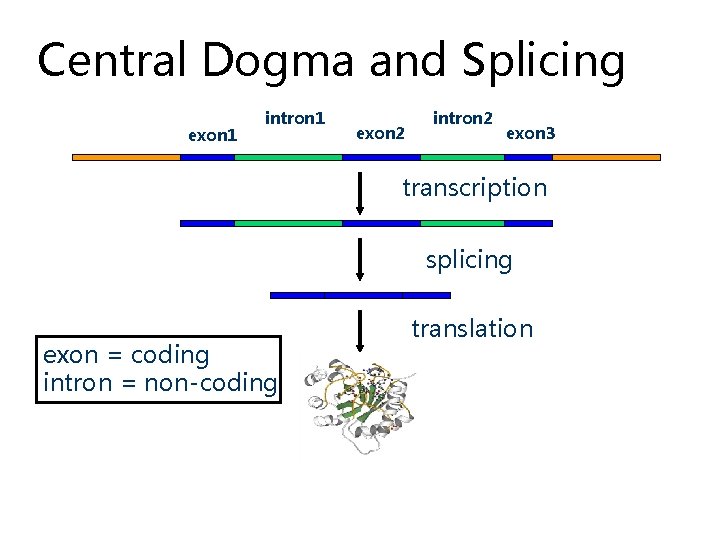
Central Dogma and Splicing exon 1 intron 1 exon 2 intron 2 exon 3 transcription splicing exon = coding intron = non-coding translation

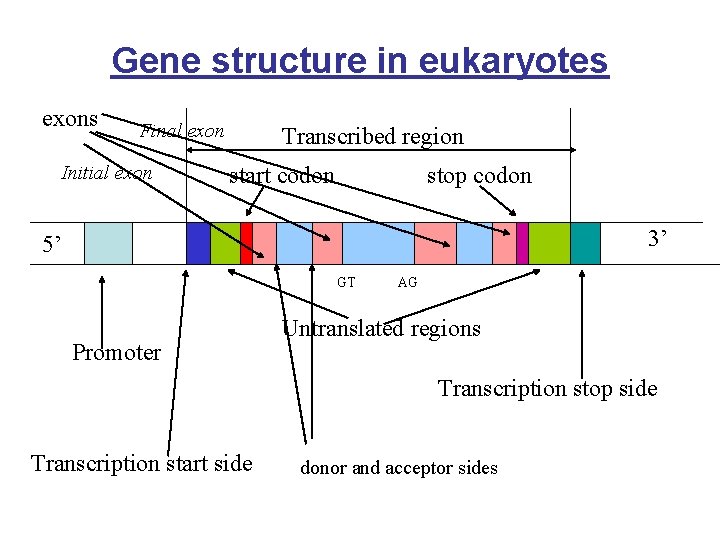
Gene Structure

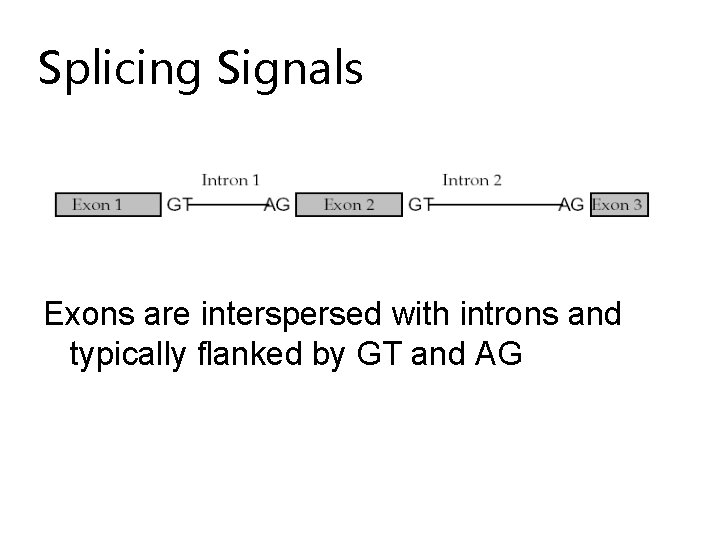
Splicing Signals Exons are interspersed with introns and typically flanked by GT and AG

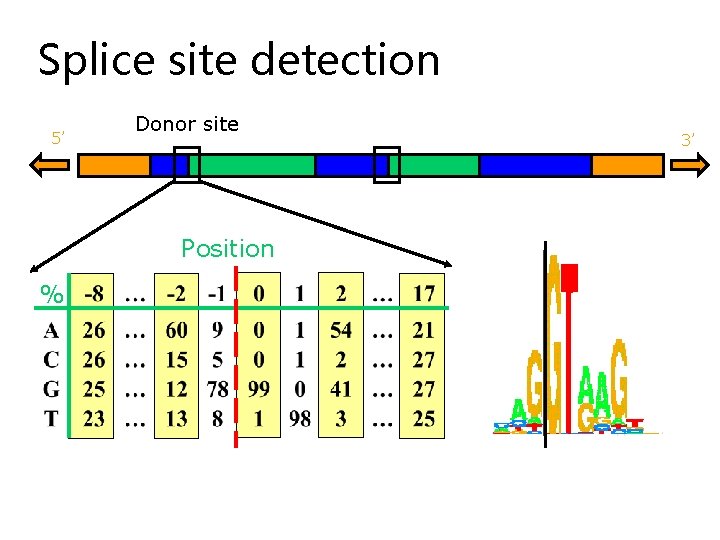
Splice site detection 5’ Donor site Position % 3’

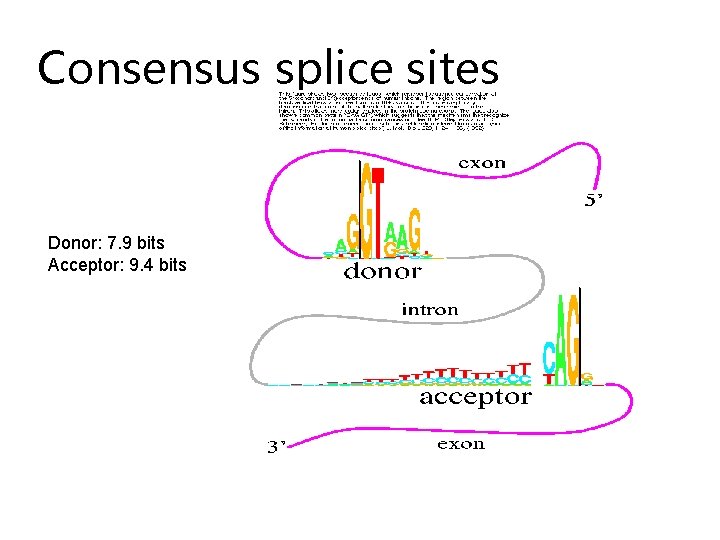
Consensus splice sites Donor: 7. 9 bits Acceptor: 9. 4 bits

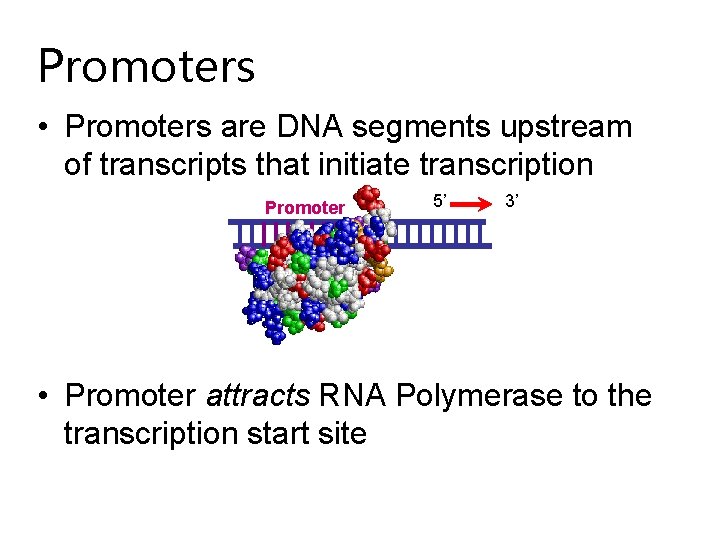
Promoters • Promoters are DNA segments upstream of transcripts that initiate transcription Promoter 5’ 3’ • Promoter attracts RNA Polymerase to the transcription start site

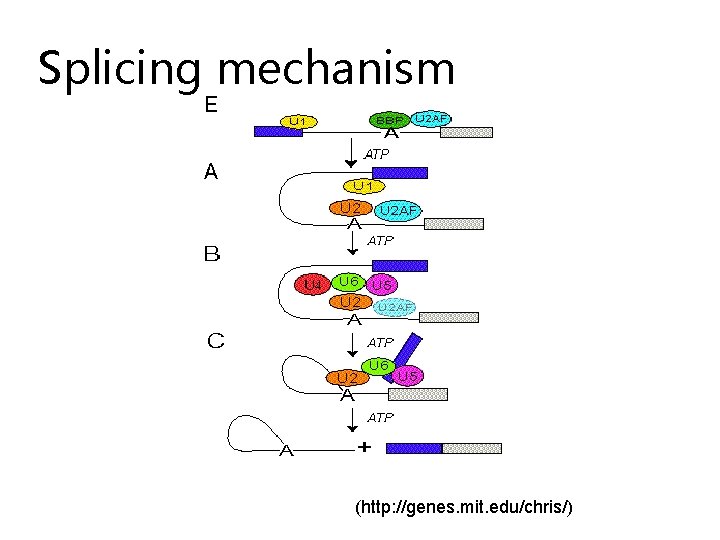
Splicing mechanism (http: //genes. mit. edu/chris/)

Splicing mechanism • Adenine recognition site marks intron • sn. RNPs bind around adenine recognition site • The spliceosome thus forms • Spliceosome excises introns in the m. RNA

Two Approaches to Eukaryotic Gene Prediction • Statistical: coding segments (exons) have typical sequences on either end and use different subwords than non-coding segments (introns). • Similarity-based: many human genes are similar to genes in mice, chicken, or even bacteria. Therefore, already known mouse, chicken, and bacterial genes may help to find human genes.

Similarity-Based Approach: Metaphor in Different Languages If you could compare the day’s news in English, side-by-side to the same news in a foreign language, some similarities may become apparent

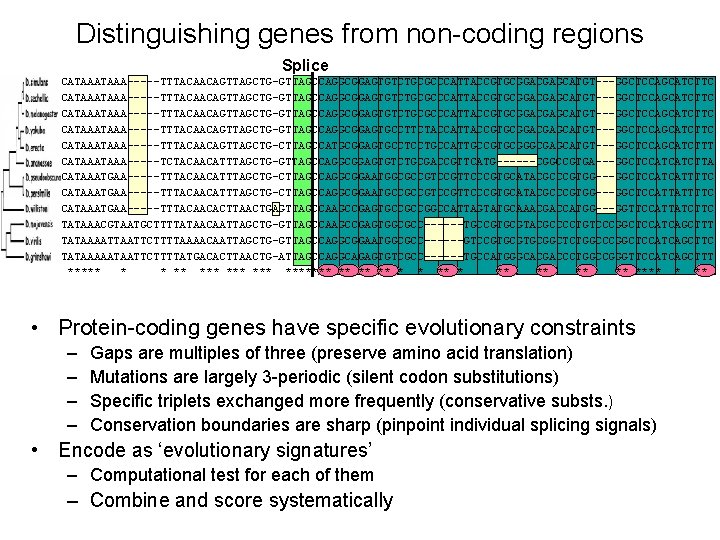
Distinguishing genes from non-coding regions Splice Dmel Dsec Dsim Dyak Dere Dana Dpse Dper Dwil Dmoj Dvir Dgri TGTTCATAAATAAA-----TTTACAACAGTTAGCTG-GTTAGCCAGGCGGAGTGTCTGCGCCCATTACCGTGCGGACGAGCATGT---GGCTCCAGCATCTTC TGTCCATAAA-----TTTACAACAGTTAGCTG-GTTAGCCAGGCGGAGTGTCTGCGCCCATTACCGTGCGGACGAGCATGT---GGCTCCAGCATCTTC TGTCCATAAA-----TTTACAACAGTTAGCTG-GTTAGCCAGGCGGAGTGCCTTCTACCATTACCGTGCGGACGAGCATGT---GGCTCCAGCATCTTC TGTCCATAAA-----TTTACAACAGTTAGCTG-CTTAGCCATGCGGAGTGCCTCCTGCCATTGCCGTGCGGGCGAGCATGT---GGCTCCAGCATCTTT TGTCCATAAA-----TCTACAACATTTAGCTG-GTTAGCCAGGCGGAGTGTCTGCGACCGTTCATG------CGGCCGTGA---GGCTCCATCATCTTA TGTCCATAAATGAA-----TTTACAACATTTAGCTG-CTTAGCCAGGCGGAATGGCGCCGTTCCCGTGCATACGCCCGTGG---GGCTCCATCATTTTC TGTCCATAAATGAA-----TTTACAACATTTAGCTG-CTTAGCCAGGCGGAATGCCGCCGTTCCCGTGCATACGCCCGTGG---GGCTCCATTATTTTC TGTTCATAAATGAA-----TTTACAACACTTAACTGAGTTAGCCAAGCCGAGTGCCGCCGGCCATTAGTATGCAAACGACCATGG---GGTTCCATTATCTTC TGATTATAAACGTAATGCTTTTATAACAATTAGCTG-GTTAGCCAAGCCGAGTGGCGCC------TGCCGTGCGTACGCCCCTGTCCCGGCTCCATCAGCTTT TGTTTATAAAATTCTTTTAAAACAATTAGCTG-GTTAGCCAGGCGGAATGGCGCC------GTCCGTGCGGCTCTGGCCCGGCTCCATCAGCTTC TGTCTATAAAAATAATTCTTTTATGACACTTAACTG-ATTAGCCAGGCAGAGTGTCGCC------TGCCATGGGCACGACCCTGGCCGGGTTCCATCAGCTTT ***** * * ** *** ******* ** ** **** * ** • Protein-coding genes have specific evolutionary constraints – – Gaps are multiples of three (preserve amino acid translation) Mutations are largely 3 -periodic (silent codon substitutions) Specific triplets exchanged more frequently (conservative substs. ) Conservation boundaries are sharp (pinpoint individual splicing signals) • Encode as ‘evolutionary signatures’ – Computational test for each of them – Combine and score systematically

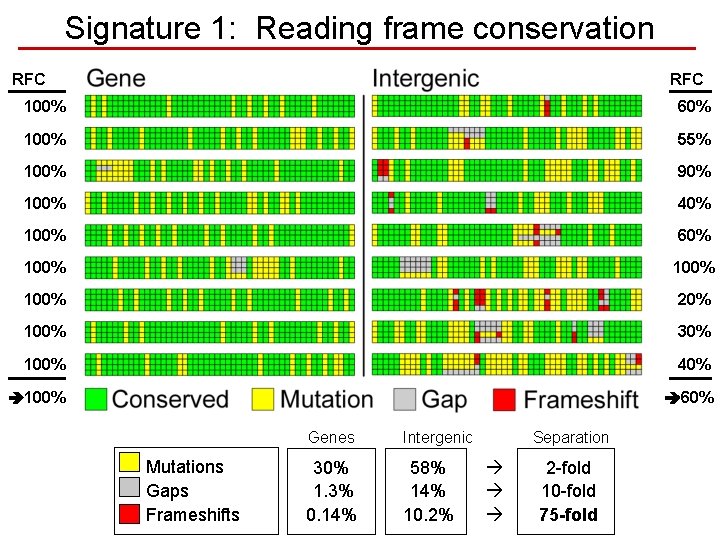
Signature 1: Reading frame conservation RFC 100% 60% 100% 55% 100% 90% 100% 40% 100% 60% 100% 20% 100% 30% 100% 40% 100% 60% Mutations Gaps Frameshifts Genes Intergenic 30% 1. 3% 0. 14% 58% 14% 10. 2% Separation 2 -fold 10 -fold 75 -fold

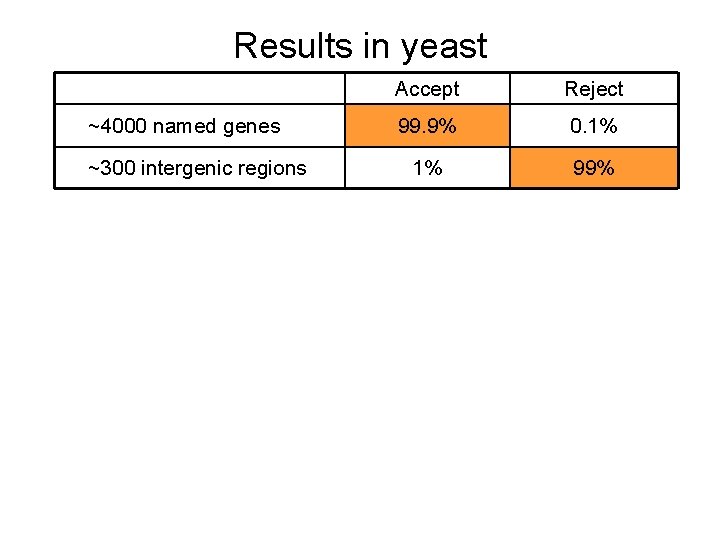
Results in yeast ~4000 named genes ~300 intergenic regions Accept Reject 99. 9% 0. 1% 1% 99%

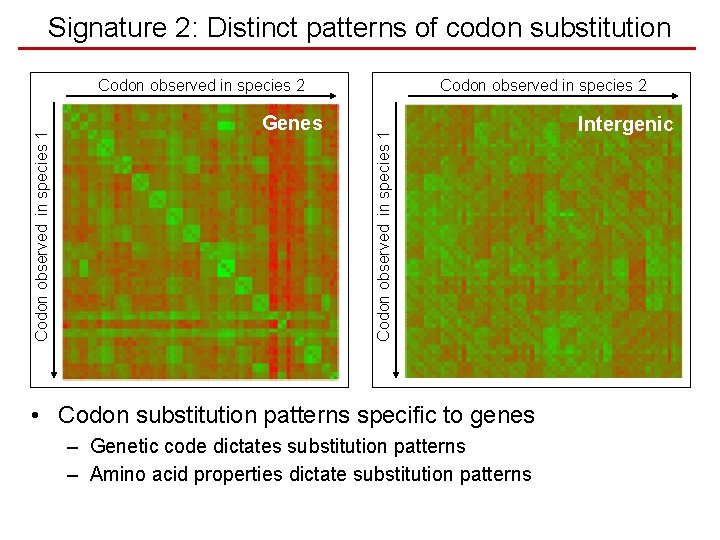
Signature 2: Distinct patterns of codon substitution Genes Codon observed in species 2 Codon observed in species 1 Codon observed in species 2 • Codon substitution patterns specific to genes – Genetic code dictates substitution patterns – Amino acid properties dictate substitution patterns Intergenic

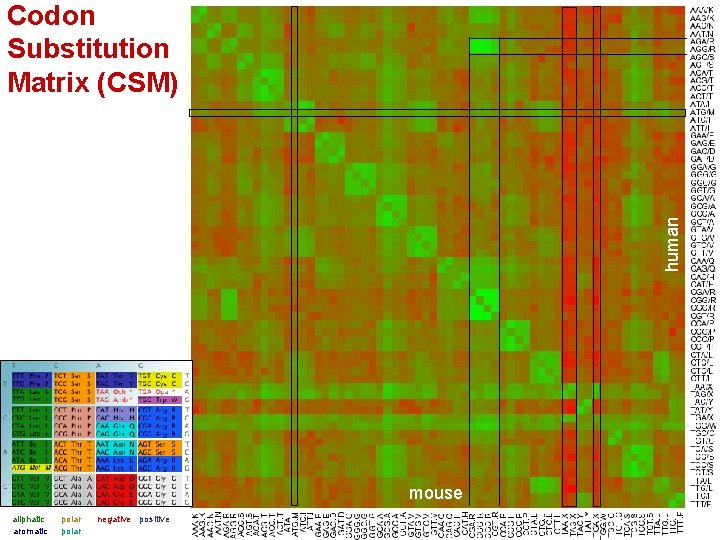
human Codon Substitution Matrix (CSM) mouse aliphatic aromatic polar negative positive

Gene structure in eukaryotes exons Final exon Initial exon Transcribed region start codon stop codon 3’ 5’ GT Promoter AG Untranslated regions Transcription stop side Transcription start side donor and acceptor sides

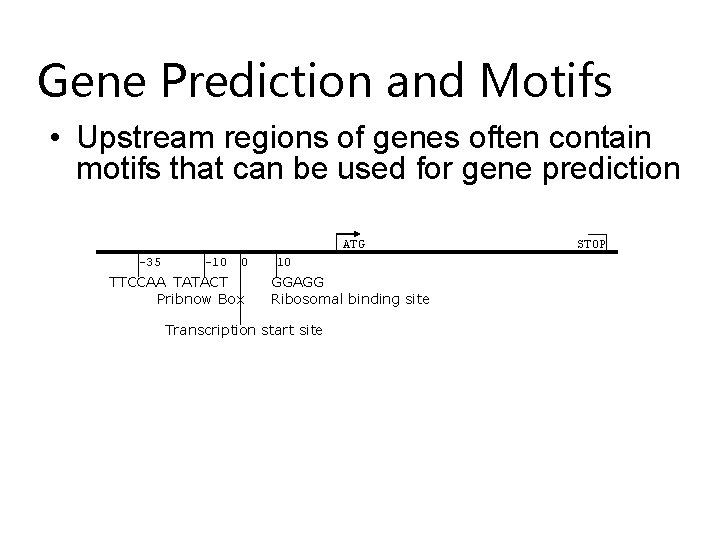
Gene Prediction and Motifs • Upstream regions of genes often contain motifs that can be used for gene prediction ATG -35 -10 0 TTCCAA TATACT Pribnow Box 10 GGAGG Ribosomal binding site Transcription start site STOP

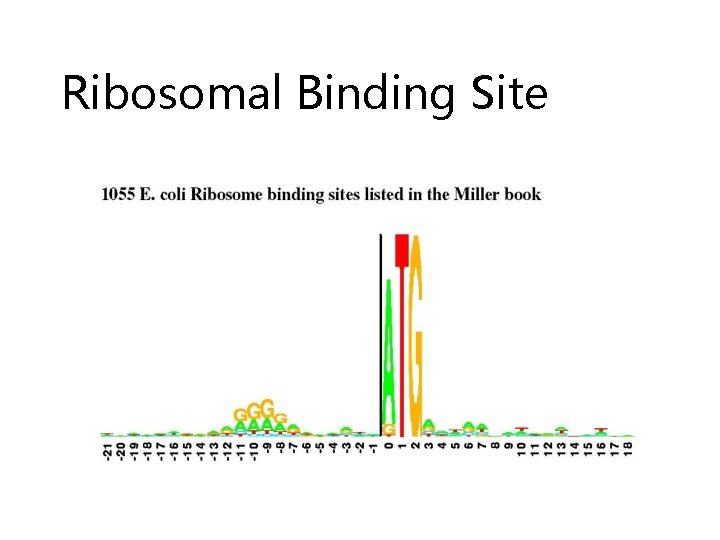
Ribosomal Binding Site

Splicing Signals • Try to recognize location of splicing signals at exon-intron junctions – This has yielded a weakly conserved donor splice site and acceptor splice site • Profiles for sites are still weak, and lends the problem to the Hidden Markov Model (HMM) approaches, which capture the statistical dependencies between sites

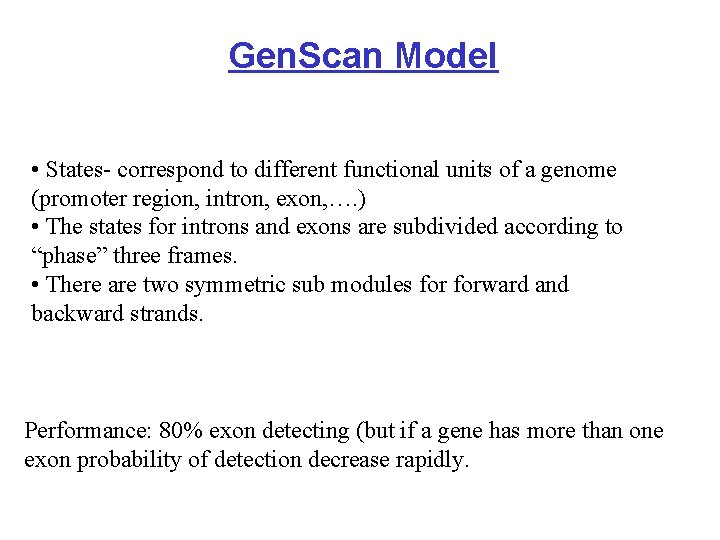
Gen. Scan Model • States- correspond to different functional units of a genome (promoter region, intron, exon, …. ) • The states for introns and exons are subdivided according to “phase” three frames. • There are two symmetric sub modules forward and backward strands. Performance: 80% exon detecting (but if a gene has more than one exon probability of detection decrease rapidly.

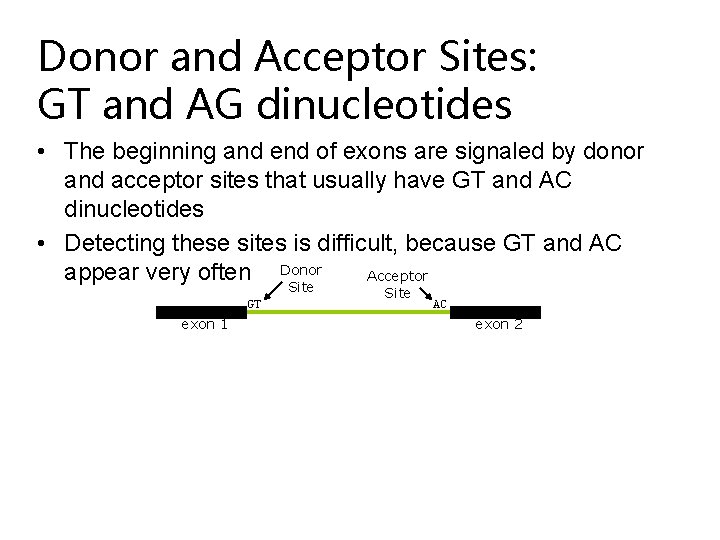
Donor and Acceptor Sites: GT and AG dinucleotides • The beginning and end of exons are signaled by donor and acceptor sites that usually have GT and AC dinucleotides • Detecting these sites is difficult, because GT and AC appear very often Donor Acceptor Site GT exon 1 Site AC exon 2

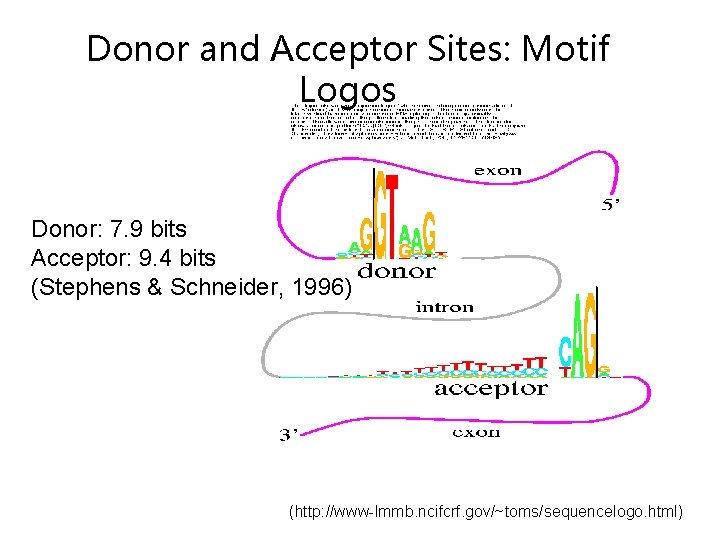
Donor and Acceptor Sites: Motif Logos Donor: 7. 9 bits Acceptor: 9. 4 bits (Stephens & Schneider, 1996) (http: //www-lmmb. ncifcrf. gov/~toms/sequencelogo. html)

Popular Gene Prediction Algorithms • GENSCAN: uses Hidden Markov Models (HMMs) • TWINSCAN – Uses both HMM and similarity (e. g. , between human and mouse genomes)

Similarity-based gene finding • Alignment of – Genomic sequence and (assembled) EST sequences – Genomic sequence and known (similar) protein sequences – Two or more similar genomic sequences

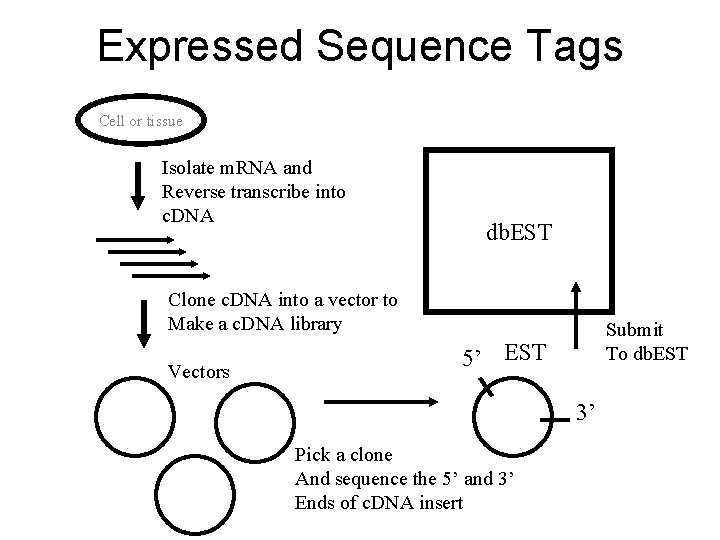
Expressed Sequence Tags Cell or tissue Isolate m. RNA and Reverse transcribe into c. DNA db. EST Clone c. DNA into a vector to Make a c. DNA library Vectors Submit To db. EST 5’ EST 3’ Pick a clone And sequence the 5’ and 3’ Ends of c. DNA insert

Central Dogma and Splicing exon 1 intron 1 exon 2 intron 2 exon 3 transcription splicing exon = coding intron = non-coding translation

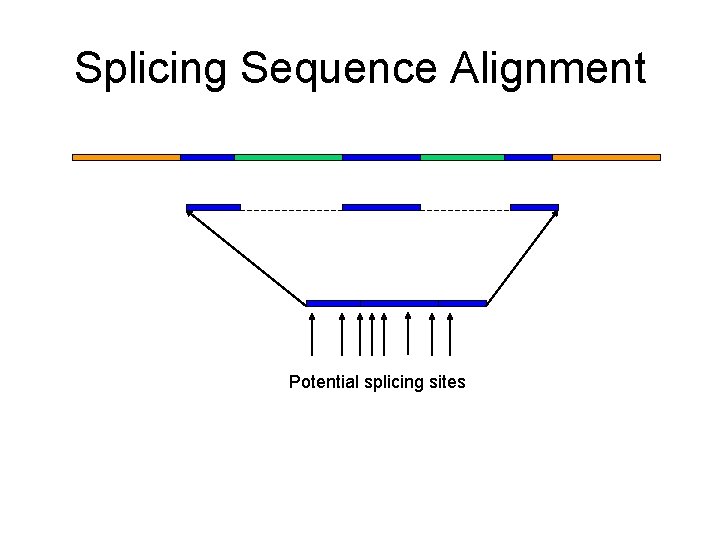
Splicing Sequence Alignment Potential splicing sites

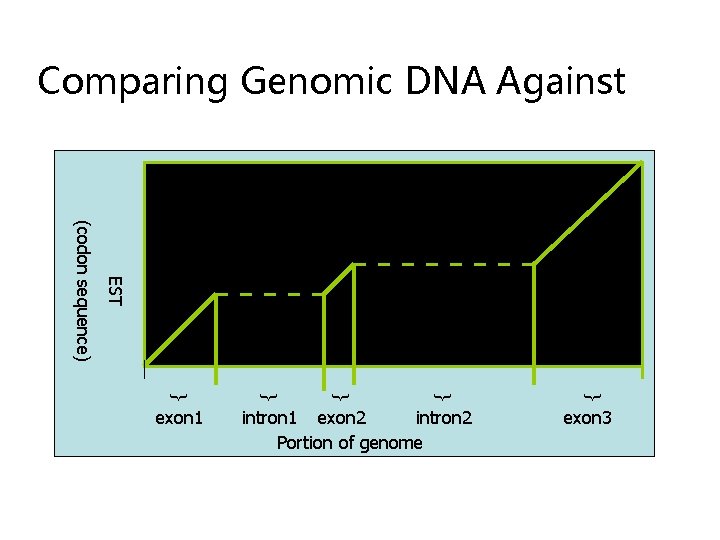
Comparing Genomic DNA Against intron 1 exon 2 intron 2 Portion of genome { { { EST (codon sequence) exon 1 exon 3

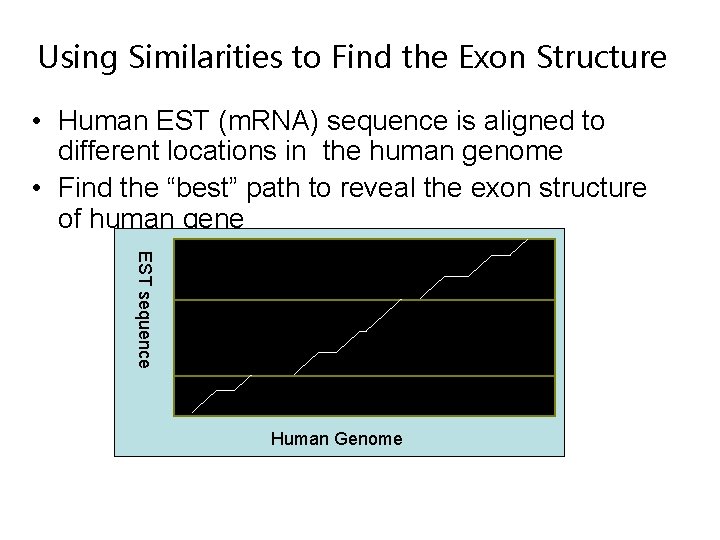
Using Similarities to Find the Exon Structure • Human EST (m. RNA) sequence is aligned to different locations in the human genome • Find the “best” path to reveal the exon structure of human gene EST sequence Human Genome

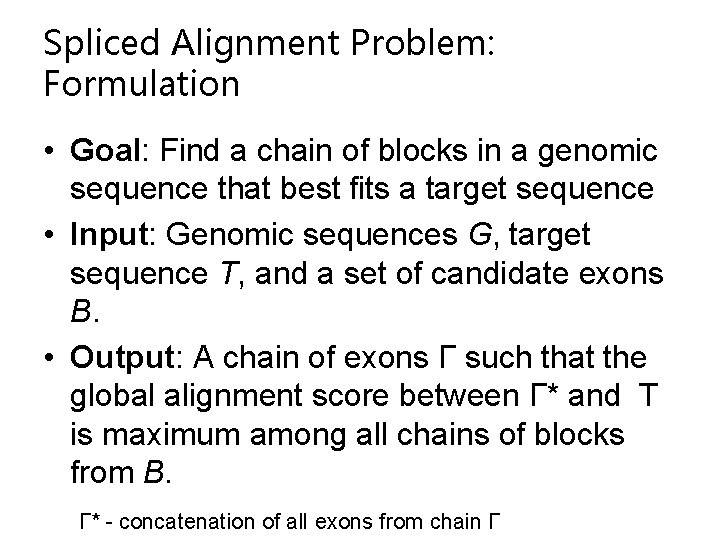
Spliced Alignment Problem: Formulation • Goal: Find a chain of blocks in a genomic sequence that best fits a target sequence • Input: Genomic sequences G, target sequence T, and a set of candidate exons B. • Output: A chain of exons Γ such that the global alignment score between Γ* and T is maximum among all chains of blocks from B. Γ* - concatenation of all exons from chain Γ

Lewis Carroll Example

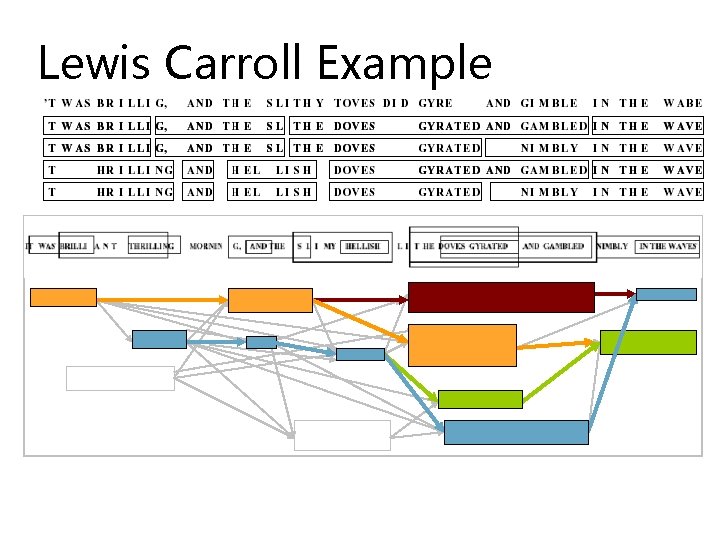
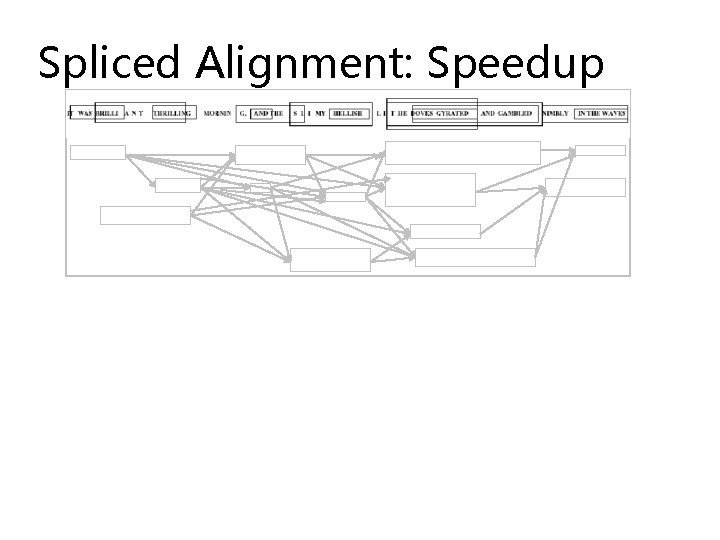
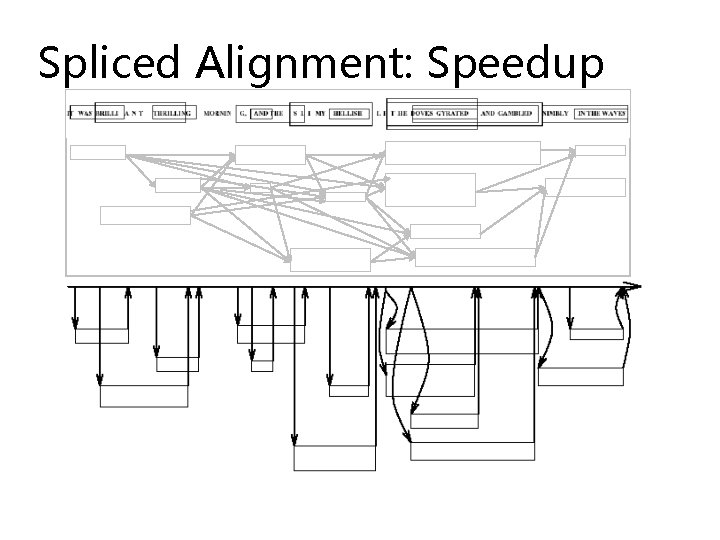
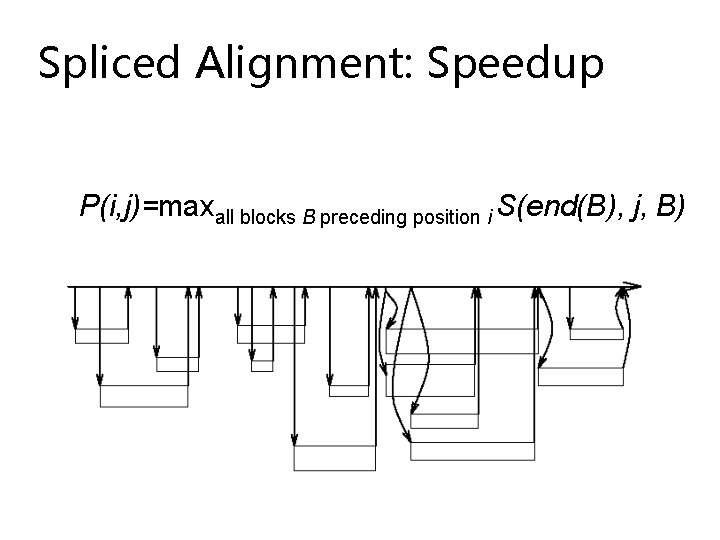
Spliced Alignment: Speedup

Spliced Alignment: Speedup

Spliced Alignment: Speedup P(i, j)=maxall blocks B preceding position i S(end(B), j, B)

EST_genome • http: //www. well. ox. ac. uk/~rmott/ESTGEN OME/est_genome. shtml

Gene finding based on multiple genomes • Twinscan • Phylo. HMM
 Genome assembly ppt
Genome assembly ppt Scale annotation pipeline
Scale annotation pipeline Tomato genome annotation
Tomato genome annotation Semi-global alignment
Semi-global alignment Gene finding
Gene finding David annotation tool
David annotation tool Gene prediction in prokaryotes and eukaryotes
Gene prediction in prokaryotes and eukaryotes Gene by gene test results
Gene by gene test results Protein power point
Protein power point Bacteriophage annotation
Bacteriophage annotation Countersink autocad
Countersink autocad Gcse art final piece evaluation template
Gcse art final piece evaluation template Annotation guide
Annotation guide Benjamin banneker letter to thomas jefferson annotation
Benjamin banneker letter to thomas jefferson annotation Nothing gold can stay connotation
Nothing gold can stay connotation Living space poem pdf
Living space poem pdf Catch annotations
Catch annotations Gcse photography annotation
Gcse photography annotation Annotation toolkit
Annotation toolkit Annotation slows down the reader to deepen understanding
Annotation slows down the reader to deepen understanding Theme of acquainted with the night
Theme of acquainted with the night David go annotation
David go annotation Dulce et decorum est annotation
Dulce et decorum est annotation The most dangerous game annotations
The most dangerous game annotations Landscape with the fall of icarus annotation
Landscape with the fall of icarus annotation What is text annotation
What is text annotation Convert labels to annotation
Convert labels to annotation Captain o captain meaning
Captain o captain meaning Valiant cousin macbeth
Valiant cousin macbeth Basking shark annotated
Basking shark annotated American law reports annotation
American law reports annotation Valediction forbidding mourning analysis
Valediction forbidding mourning analysis Jumanji
Jumanji Morphological annotation
Morphological annotation Text symbols for annotating text
Text symbols for annotating text Richard xiao
Richard xiao Shall i compare thee to a summer's day annotation
Shall i compare thee to a summer's day annotation Patricia pogson yesterday
Patricia pogson yesterday Nbis
Nbis Critical annotation
Critical annotation Acquainted with the night annotation
Acquainted with the night annotation