Zink Fingers Figure 1 Schematic illustration of the













- Slides: 29





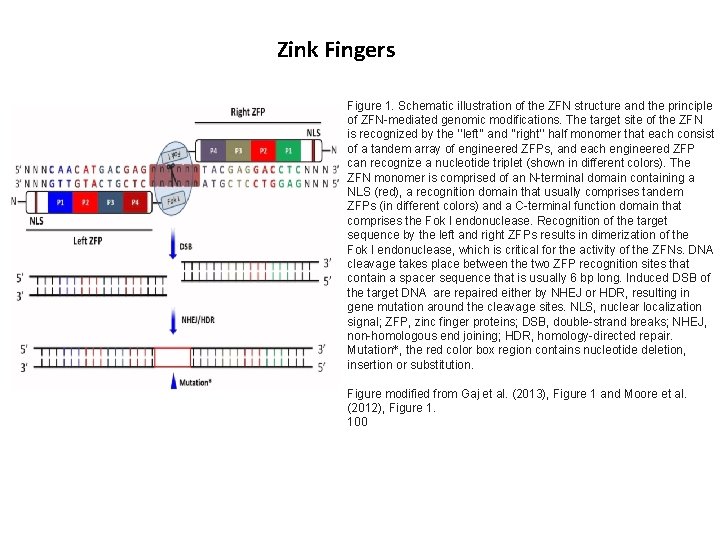
Zink Fingers Figure 1. Schematic illustration of the ZFN structure and the principle of ZFN-mediated genomic modifications. The target site of the ZFN is recognized by the ‘‘left’’ and ‘‘right’’ half monomer that each consist of a tandem array of engineered ZFPs, and each engineered ZFP can recognize a nucleotide triplet (shown in different colors). The ZFN monomer is comprised of an N-terminal domain containing a NLS (red), a recognition domain that usually comprises tandem ZFPs (in different colors) and a C-terminal function domain that comprises the Fok I endonuclease. Recognition of the target sequence by the left and right ZFPs results in dimerization of the Fok I endonuclease, which is critical for the activity of the ZFNs. DNA cleavage takes place between the two ZFP recognition sites that contain a spacer sequence that is usually 6 bp long. Induced DSB of the target DNA are repaired either by NHEJ or HDR, resulting in gene mutation around the cleavage sites. NLS, nuclear localization signal; ZFP, zinc finger proteins; DSB, double-strand breaks; NHEJ, non-homologous end joining; HDR, homology-directed repair. Mutation*, the red color box region contains nucleotide deletion, insertion or substitution. Figure modified from Gaj et al. (2013), Figure 1 and Moore et al. (2012), Figure 1. 100

TALEN Figure 2. The structure of TALEN and the principle of TALENmediated genomic modifications. The target site of TALEN is recognized by the ‘‘left’’ and ‘‘right’’ half monomer that each consist of a tandem repeat of TALE repeats. Each TALE repeat comprises a 34 amino acid (aa) unit that differs at two hypervariable aa located at the 12 th and 13 th position, known as RVD, which determine the recognition specificity of each repeat. The TALEN monomer consists of an N-terminal domain containing a nuclear localization signal (NLS, red), a recognition domain typically composed of tandem TALE repeats (in different colors), and a C-terminal function domain that comprises the Fok I endonuclease. Simultaneous bindings of the left and right TALE enable dimerization of the Fok I cleavage domain, resulting in DSBs of the target DNA. Induced DSBs of the target DNA are repaired either by NHEJ or HDR resulting in gene mutations that include nucleotide insertion, deletion, or substitution around the cleavage site. TALE, transcription activator-like effector; NLS, nuclear localization signal; RVD, repeat-variable di-residues; DSB, double-strand breaks; NHEJ, non-homologous end joining; HDR, homologydirected repair. Mutation*, red color box regions contain nucleotide deletion, insertion or substitution. Figure modified from Gaj et al. (2013), Figure 1 and Moore et al. (2012), Figure 1. 100

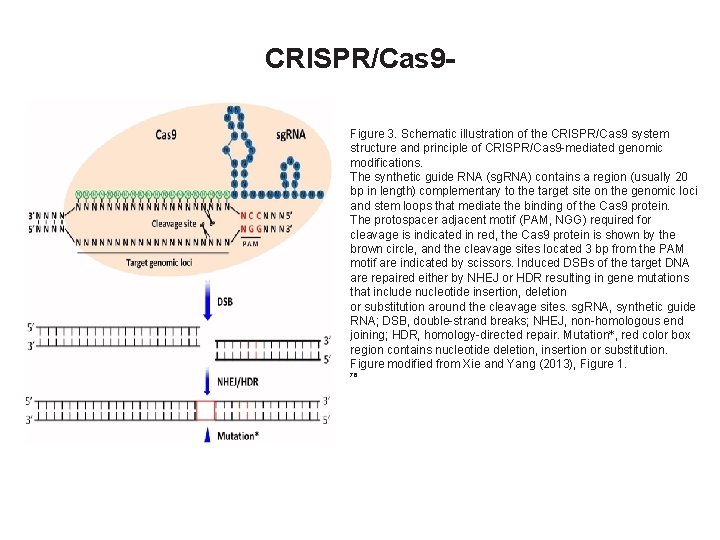
CRISPR/Cas 9 Figure 3. Schematic illustration of the CRISPR/Cas 9 system structure and principle of CRISPR/Cas 9 -mediated genomic modifications. The synthetic guide RNA (sg. RNA) contains a region (usually 20 bp in length) complementary to the target site on the genomic loci and stem loops that mediate the binding of the Cas 9 protein. The protospacer adjacent motif (PAM, NGG) required for cleavage is indicated in red, the Cas 9 protein is shown by the brown circle, and the cleavage sites located 3 bp from the PAM motif are indicated by scissors. Induced DSBs of the target DNA are repaired either by NHEJ or HDR resulting in gene mutations that include nucleotide insertion, deletion or substitution around the cleavage sites. sg. RNA, synthetic guide RNA; DSB, double-strand breaks; NHEJ, non-homologous end joining; HDR, homology-directed repair. Mutation*, red color box region contains nucleotide deletion, insertion or substitution. Figure modified from Xie and Yang (2013), Figure 1. 76

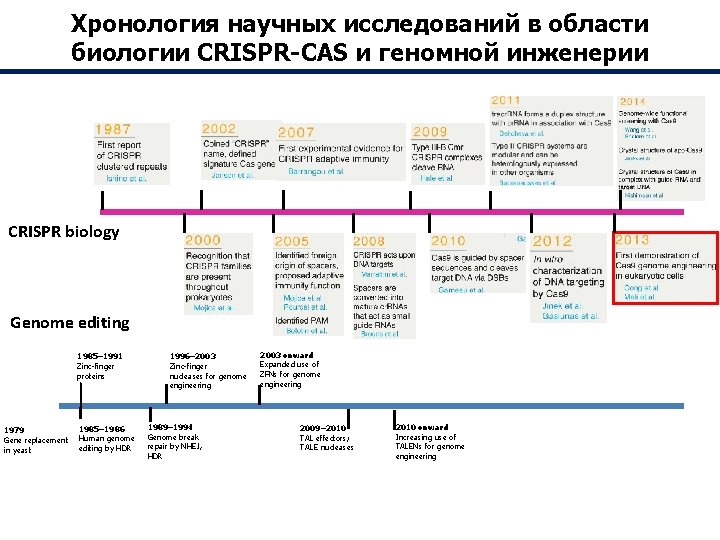
Хронология научных исследований в области биологии CRISPR-CAS и геномной инженерии CRISPR biology Genome editing 1985– 1991 Zinc-finger proteins 1979 Gene replacement in yeast 1985– 1986 Human genome editing by HDR 1996– 2003 Zinc-finger nucleases for genome engineering 1989– 1994 Genome break repair by NHEJ, HDR 2003 onward Expanded use of ZFNs for genome engineering 2009– 2010 TAL effectors; TALE nucleases 2010 onward Increasing use of TALENs for genome engineering


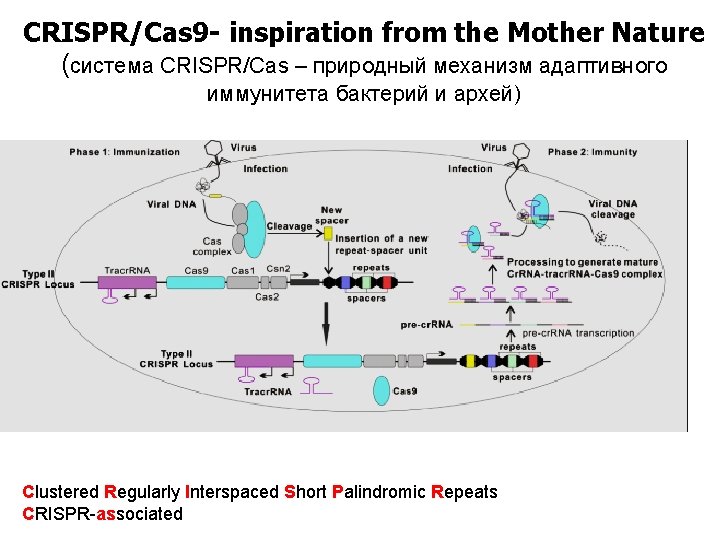
CRISPR/Cas 9 - inspiration from the Mother Nature (система CRISPR/Cas – природный механизм адаптивного иммунитета бактерий и архей) Clustered Regularly Interspaced Short Palindromic Repeats CRISPR-associated


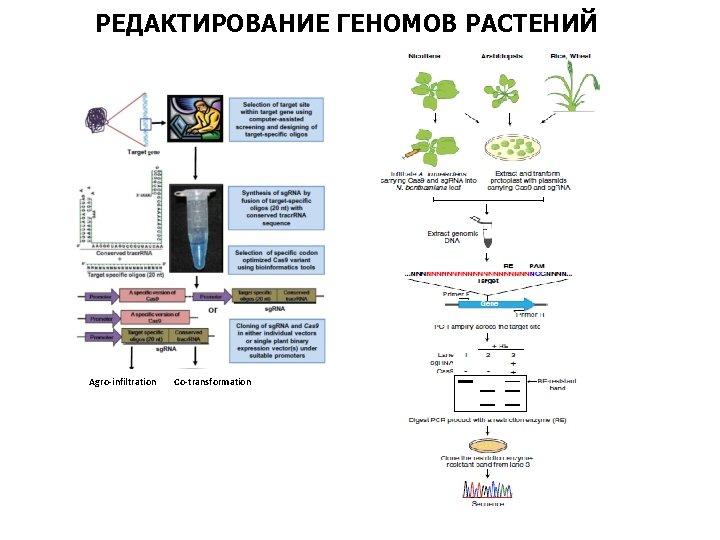
РЕДАКТИРОВАНИЕ ГЕНОМОВ РАСТЕНИЙ Agro-infiltration Co-transformation

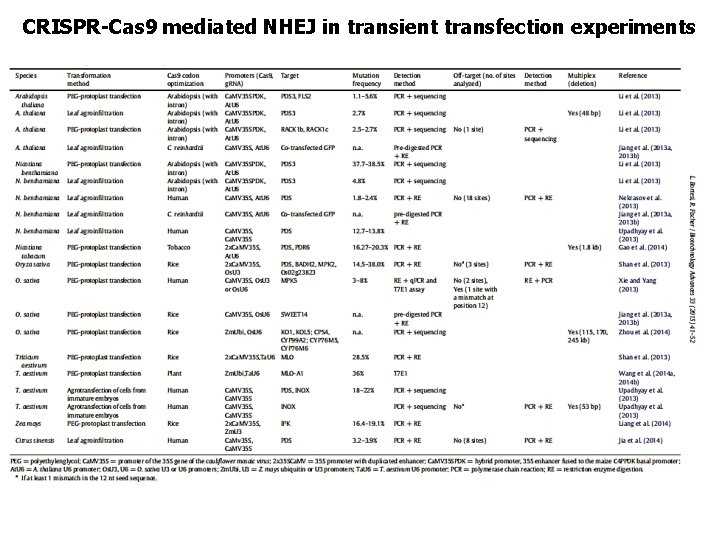
CRISPR-Cas 9 mediated NHEJ in transient transfection experiments

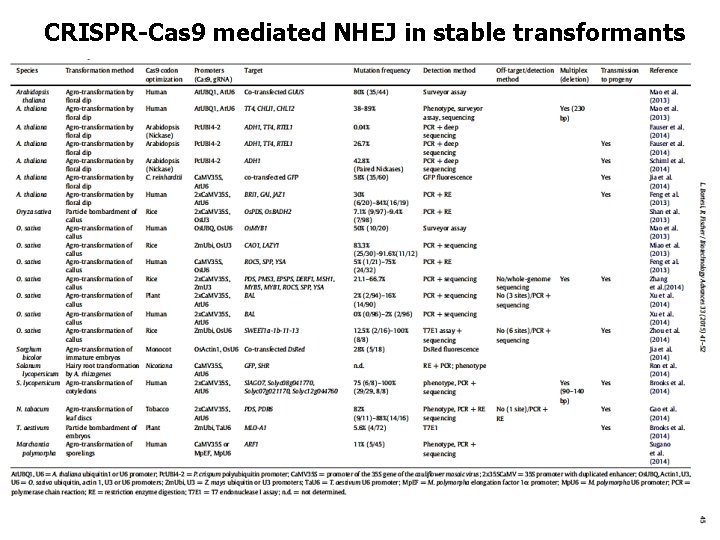
CRISPR-Cas 9 mediated NHEJ in stable transformants

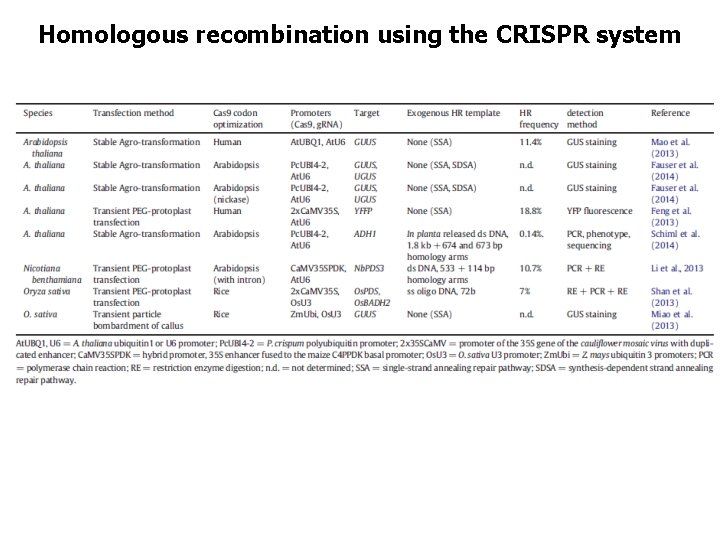
Homologous recombination using the CRISPR system

http: //www. addgene. org/search/advanced/? q=&depositor=&article=&gene=&vector=&tags=&advanced_query=&results_per_page=20&page=1&selected_f acets=vector_types_exact%3 Aplant-expression&selected_facets=vector_types_exact%3 Acrispr&sort_type=relevance


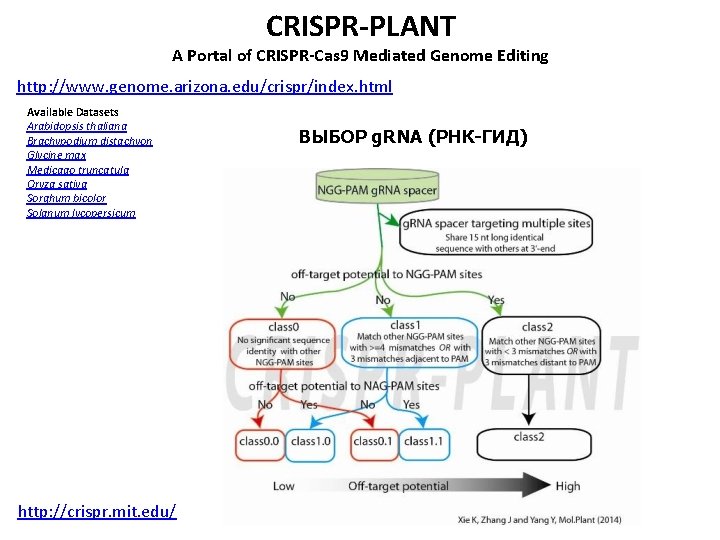
CRISPR-PLANT A Portal of CRISPR-Cas 9 Mediated Genome Editing http: //www. genome. arizona. edu/crispr/index. html Available Datasets Arabidopsis thaliana Brachypodium distachyon Glycine max Medicago truncatula Oryza sativa Sorghum bicolor Solanum lycopersicum http: //crispr. mit. edu/ ВЫБОР g. RNA (РНК-ГИД)

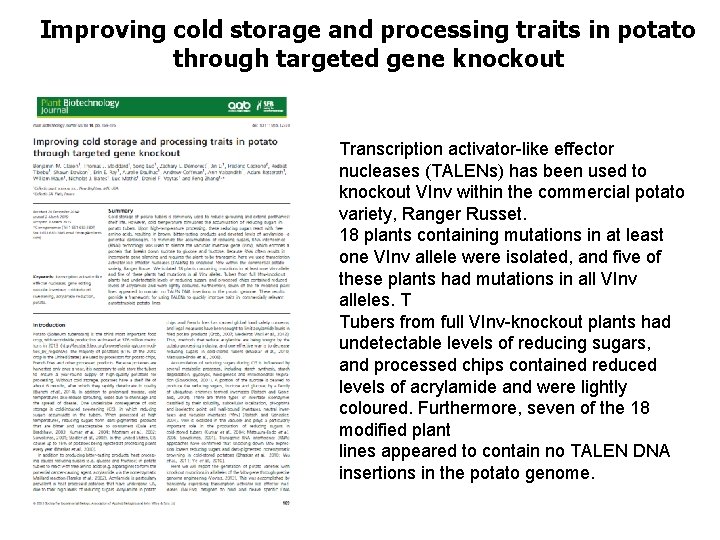
Improving cold storage and processing traits in potato through targeted gene knockout Transcription activator-like effector nucleases (TALENs) has been used to knockout VInv within the commercial potato variety, Ranger Russet. 18 plants containing mutations in at least one VInv allele were isolated, and five of these plants had mutations in all VInv alleles. T Tubers from full VInv-knockout plants had undetectable levels of reducing sugars, and processed chips contained reduced levels of acrylamide and were lightly coloured. Furthermore, seven of the 18 modified plant lines appeared to contain no TALEN DNA insertions in the potato genome.

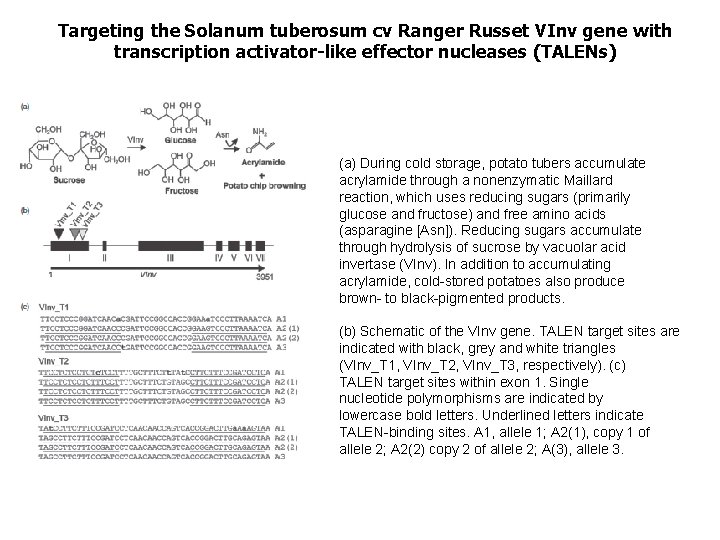
Targeting the Solanum tuberosum cv Ranger Russet VInv gene with transcription activator-like effector nucleases (TALENs) (a) During cold storage, potato tubers accumulate acrylamide through a nonenzymatic Maillard reaction, which uses reducing sugars (primarily glucose and fructose) and free amino acids (asparagine [Asn]). Reducing sugars accumulate through hydrolysis of sucrose by vacuolar acid invertase (VInv). In addition to accumulating acrylamide, cold-stored potatoes also produce brown- to black-pigmented products. (b) Schematic of the VInv gene. TALEN target sites are indicated with black, grey and white triangles (VInv_T 1, VInv_T 2, VInv_T 3, respectively). (c) TALEN target sites within exon 1. Single nucleotide polymorphisms are indicated by lowercase bold letters. Underlined letters indicate TALEN-binding sites. A 1, allele 1; A 2(1), copy 1 of allele 2; A 2(2) copy 2 of allele 2; A(3), allele 3.

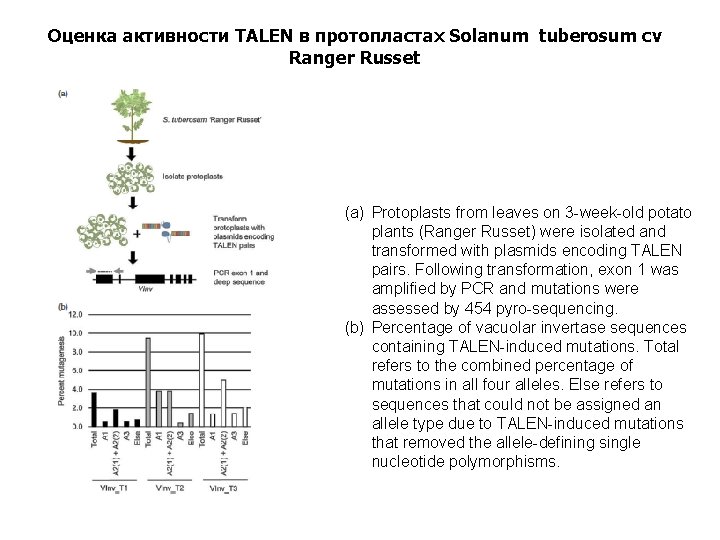
Оценка активности TALEN в протопластах Solanum tuberosum cv Ranger Russet (a) Protoplasts from leaves on 3 -week-old potato plants (Ranger Russet) were isolated and transformed with plasmids encoding TALEN pairs. Following transformation, exon 1 was amplified by PCR and mutations were assessed by 454 pyro-sequencing. (b) Percentage of vacuolar invertase sequences containing TALEN-induced mutations. Total refers to the combined percentage of mutations in all four alleles. Else refers to sequences that could not be assigned an allele type due to TALEN-induced mutations that removed the allele-defining single nucleotide polymorphisms.

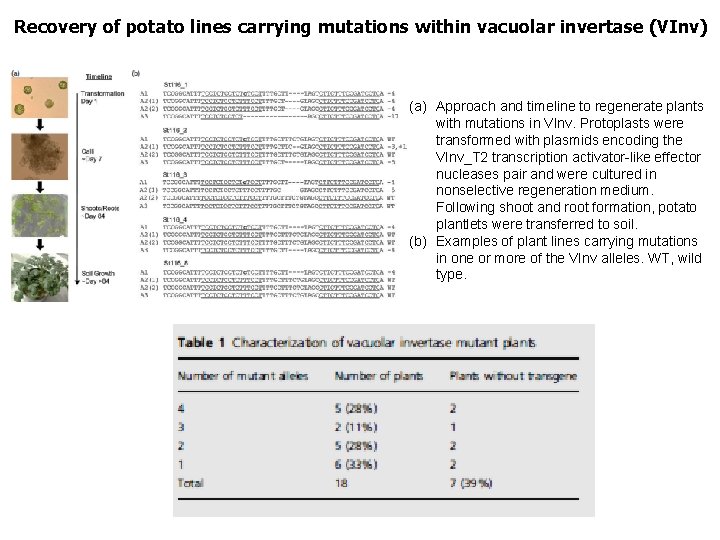
Recovery of potato lines carrying mutations within vacuolar invertase (VInv) (a) Approach and timeline to regenerate plants with mutations in VInv. Protoplasts were transformed with plasmids encoding the VInv_T 2 transcription activator-like effector nucleases pair and were cultured in nonselective regeneration medium. Following shoot and root formation, potato plantlets were transferred to soil. (b) Examples of plant lines carrying mutations in one or more of the VInv alleles. WT, wild type.

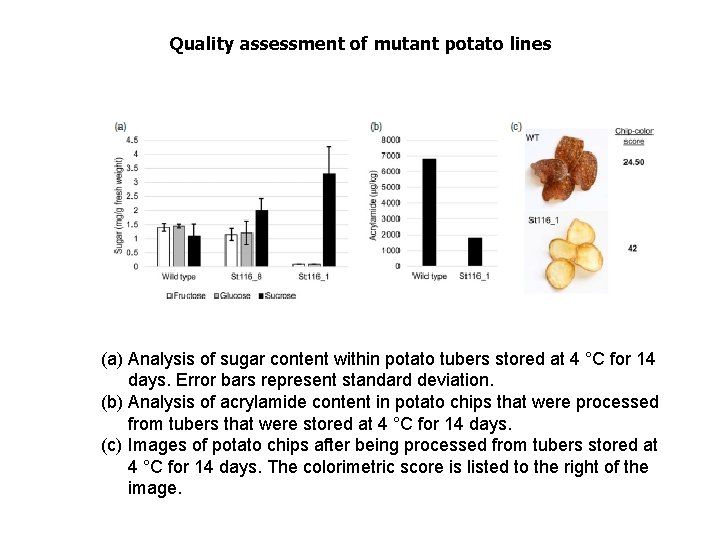
Quality assessment of mutant potato lines (a) Analysis of sugar content within potato tubers stored at 4 °C for 14 days. Error bars represent standard deviation. (b) Analysis of acrylamide content in potato chips that were processed from tubers that were stored at 4 °C for 14 days. (c) Images of potato chips after being processed from tubers stored at 4 °C for 14 days. The colorimetric score is listed to the right of the image.

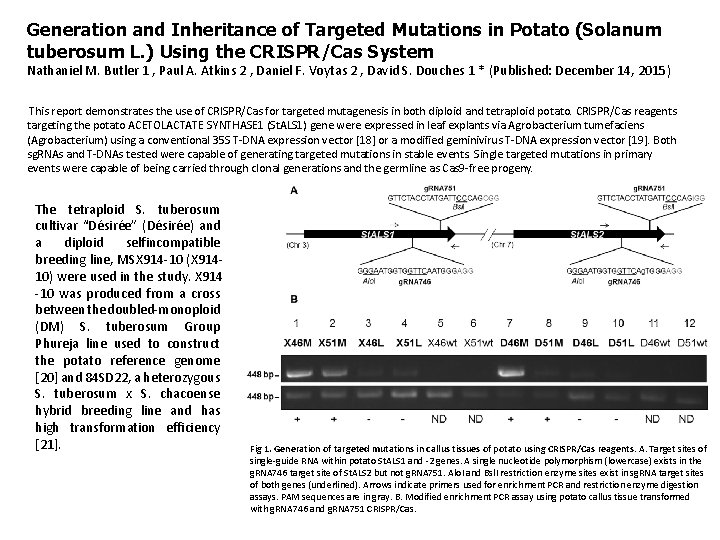
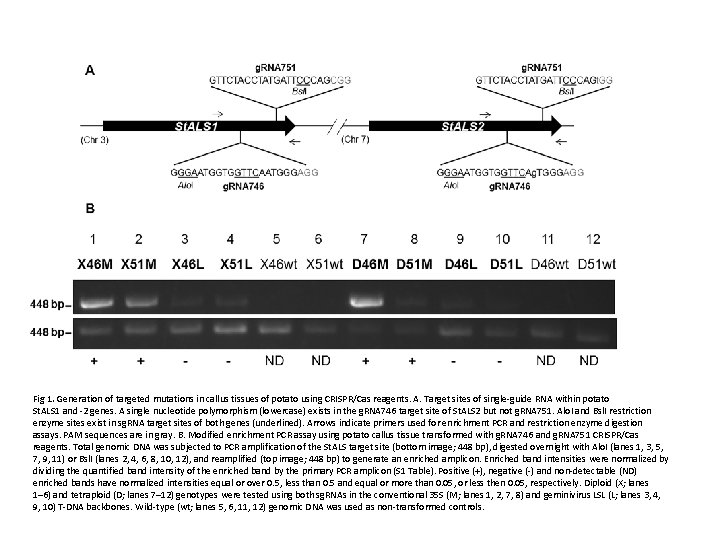
Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L. ) Using the CRISPR/Cas System Nathaniel M. Butler 1 , Paul A. Atkins 2 , Daniel F. Voytas 2 , David S. Douches 1 * (Published: December 14, 2015) This report demonstrates the use of CRISPR/Cas for targeted mutagenesis in both diploid and tetraploid potato. CRISPR/Cas reagents targeting the potato ACETOLACTATE SYNTHASE 1 (St. ALS 1) gene were expressed in leaf explants via Agrobacterium tumefaciens (Agrobacterium) using a conventional 35 S T-DNA expression vector [18] or a modified geminivirus T-DNA expression vector [19]. Both sg. RNAs and T-DNAs tested were capable of generating targeted mutations in stable events. Single targeted mutations in primary events were capable of being carried through clonal generations and the germline as Cas 9 -free progeny. The tetraploid S. tuberosum cultivar “Désirée” (Désirée) and a diploid selfincompatible breeding line, MSX 914 -10 (X 91410) were used in the study. X 914 -10 was produced from a cross between the doubled-monoploid (DM) S. tuberosum Group Phureja line used to construct the potato reference genome [20] and 84 SD 22, a heterozygous S. tuberosum x S. chacoense hybrid breeding line and has high transformation efficiency [21]. Fig 1. Generation of targeted mutations in callus tissues of potato using CRISPR/Cas reagents. A. Target sites of single-guide RNA within potato St. ALS 1 and -2 genes. A single nucleotide polymorphism (lowercase) exists in the g. RNA 746 target site of St. ALS 2 but not g. RNA 751. Alo. I and Bsl. I restriction enzyme sites exist in sg. RNA target sites of both genes (underlined). Arrows indicate primers used for enrichment PCR and restriction enzyme digestion assays. PAM sequences are in gray. B. Modified enrichment PCR assay using potato callus tissue transformed with g. RNA 746 and g. RNA 751 CRISPR/Cas.

Fig 1. Generation of targeted mutations in callus tissues of potato using CRISPR/Cas reagents. A. Target sites of single-guide RNA within potato St. ALS 1 and -2 genes. A single nucleotide polymorphism (lowercase) exists in the g. RNA 746 target site of St. ALS 2 but not g. RNA 751. Alo. I and Bsl. I restriction enzyme sites exist in sg. RNA target sites of both genes (underlined). Arrows indicate primers used for enrichment PCR and restriction enzyme digestion assays. PAM sequences are in gray. B. Modified enrichment PCR assay using potato callus tissue transformed with g. RNA 746 and g. RNA 751 CRISPR/Cas reagents. Total genomic DNA was subjected to PCR amplification of the St. ALS target site (bottom image; 448 bp), digested overnight with Alo. I (lanes 1, 3, 5, 7, 9, 11) or Bsl. I (lanes 2, 4, 6, 8, 10, 12), and reamplified (top image; 448 bp) to generate an enriched amplicon. Enriched band intensities were normalized by dividing the quantified band intensity of the enriched band by the primary PCR amplicon (S 1 Table). Positive (+), negative (-) and non-detectable (ND) enriched bands have normalized intensities equal or over 0. 5, less than 0. 5 and equal or more than 0. 05, or less then 0. 05, respectively. Diploid (X; lanes 1– 6) and tetraploid (D; lanes 7– 12) genotypes were tested using both sg. RNAs in the conventional 35 S (M; lanes 1, 2, 7, 8) and geminivirus LSL (L; lanes 3, 4, 9, 10) T-DNA backbones. Wild-type (wt; lanes 5, 6, 11, 12) genomic DNA was used as non-transformed controls.

Fig 2. Generation and cloning of targeted mutations in primary events of potato using CRISPR/Cas reagents. A. Restriction enzyme digestion assay of diploid (X; lanes 2– 4) and tetraploid (D; lanes 5– 8) primary events. Total genomic DNA from primary events was subjected to PCR amplification of the St. ALS target site and digested overnight with Alo. I yielding a 448 bp resistant band 326 bp and 122 bp digested bands. Wild-type X 914 -10 (WT; lane 1) and Désirée (Fig 3 A) genomic DNA were used as a negative controls. B. Cloned targeted mutations in primary events of potato. Diploid (X) and tetraploid (D) events constitutively expressing g. RNA 746 (46) and g. RNA 751 (51) CRISPR/Cas reagents were used for cloning. Resistant bands from restriction enzyme digestion assays were excised from 2. 0% agarose gels, purified, and subcloned for Sanger sequencing. Sanger reads from each event were aligned to St. ALS 1 and -2 wild-type sequence (WT) from each sg. RNA target site (g. RNA 746; top alignments, g. RNA 751; bottom alignments). The lengths of deletions (-) or insertions (+) are in parenthesis to the left of each cloned mutation and the number of reads generated in the primary event (T 0 ) or first clonal generation (CG 1 ) are in brackets on the right. All targeted mutations were cloned from St. ALS 1 unless indicated on the right. PAM sequences are in gray.



 Aluminium kemisk beteckning
Aluminium kemisk beteckning Sam byron
Sam byron Kimberly zink
Kimberly zink Steckbrief zink
Steckbrief zink Saltframställning
Saltframställning Intrapulmonary pressure
Intrapulmonary pressure Fingers poet x
Fingers poet x Niccolo paganini fingers
Niccolo paganini fingers Rods airway
Rods airway Vulcan fingers genetics
Vulcan fingers genetics Heberden's nodes
Heberden's nodes Best phone for fat fingers
Best phone for fat fingers Chapter 15 section 3: shaping evolutionary theory
Chapter 15 section 3: shaping evolutionary theory Absorptive atelectasis
Absorptive atelectasis Superficial pulp spaces of fingers
Superficial pulp spaces of fingers Chapter 24 the forearm, wrist, hand, and, fingers
Chapter 24 the forearm, wrist, hand, and, fingers Sole horn nail
Sole horn nail Fingers fing
Fingers fing Infection of fascial spaces of palm
Infection of fascial spaces of palm Radial pulse 3 fingers
Radial pulse 3 fingers Victory waits on your fingers poster meaning
Victory waits on your fingers poster meaning Truman show fingers crossed meaning
Truman show fingers crossed meaning Medium fidelity prototype
Medium fidelity prototype Hoot roy
Hoot roy Elbows off the table fingers off the food song
Elbows off the table fingers off the food song Dawson fingers ms
Dawson fingers ms ,,
,, 5 fingers of evolution
5 fingers of evolution Supinator origin and insertion
Supinator origin and insertion Fred fingers phonics
Fred fingers phonics