Use of Open Earth Tools to make net


- Slides: 45

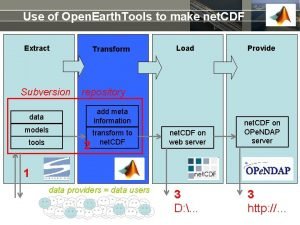
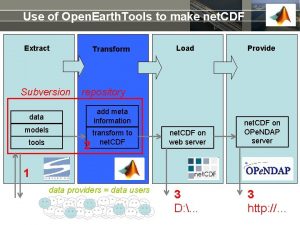
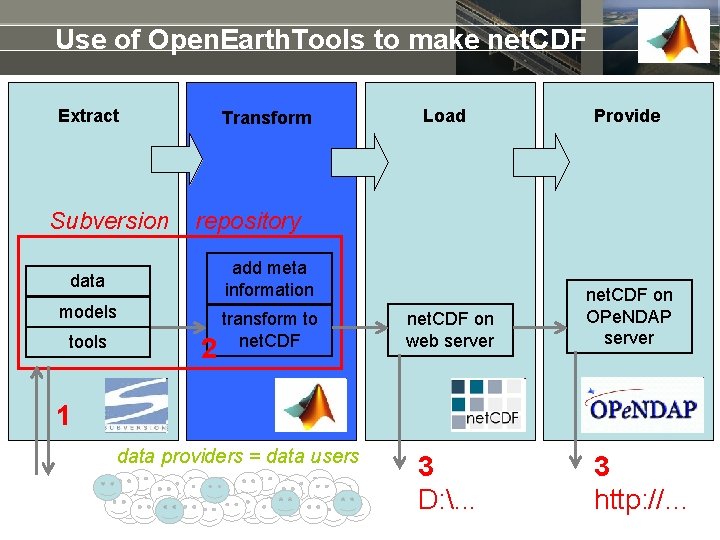
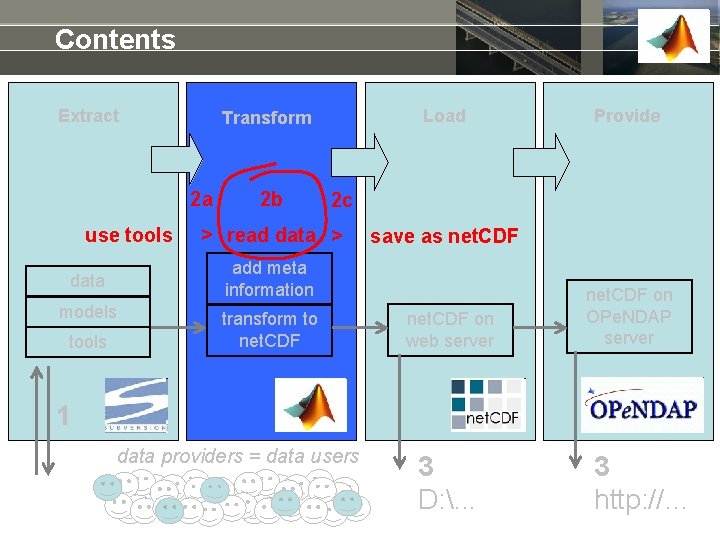
Use of Open. Earth. Tools to make net. CDF Extract Subversion Transform Provide repository add meta information data models tools Load 2 transform to net. CDF on web server net. CDF on OPe. NDAP server 1 data providers = data users 3 D: . . . 3 http: //…

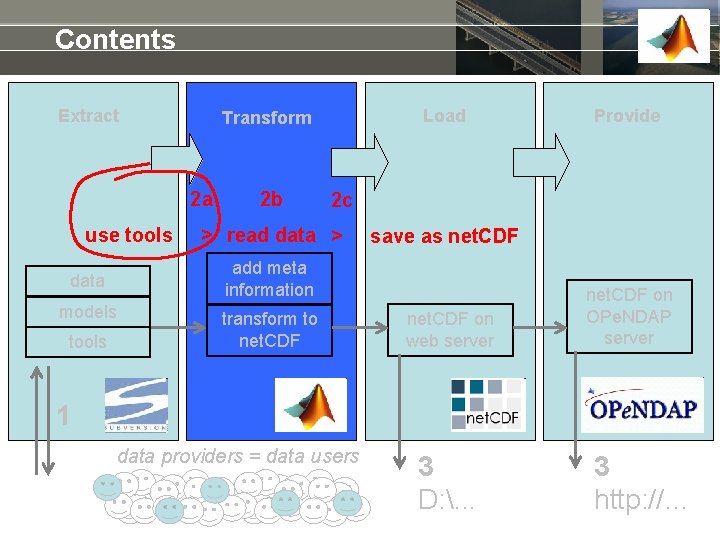
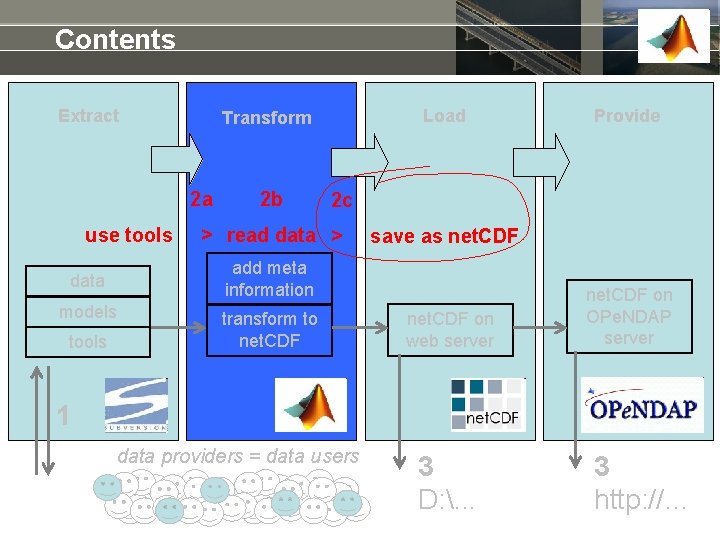
Contents Extract 2 a use tools 2 b models Provide 2 c > read data > save as net. CDF add meta information data tools Load Transform to net. CDF on web server net. CDF on OPe. NDAP server 1 data providers = data users 3 D: . . . 3 http: //…

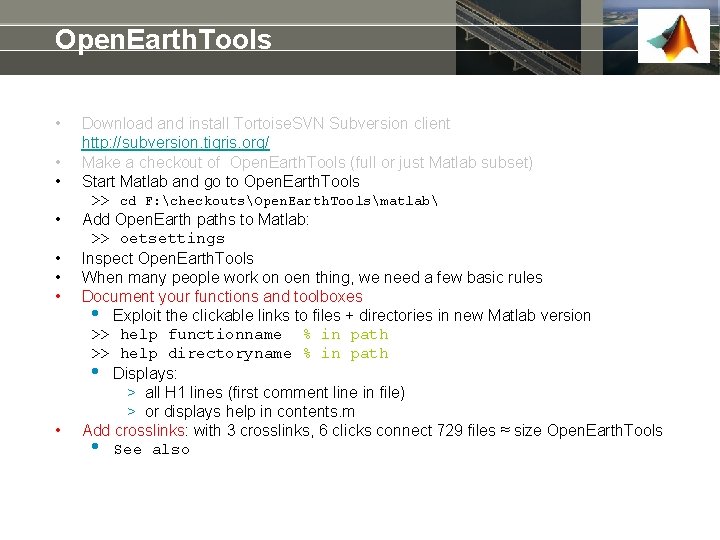
Open. Earth. Tools • • Download and install Tortoise. SVN Subversion client http: //subversion. tigris. org/ Make a checkout of Open. Earth. Tools (full or just Matlab subset) Start Matlab and go to Open. Earth. Tools >> cd F: checkoutsOpen. Earth. Toolsmatlab Add Open. Earth paths to Matlab: >> oetsettings Inspect Open. Earth. Tools When many people work on oen thing, we need a few basic rules Document your functions and toolboxes • Exploit the clickable links to files + directories in new Matlab version >> help functionname % in path >> help directoryname % in path • Displays: > all H 1 lines (first comment line in file) > or displays help in contents. m Add crosslinks: with 3 crosslinks, 6 clicks connect 729 files ≈ size Open. Earth. Tools • See also

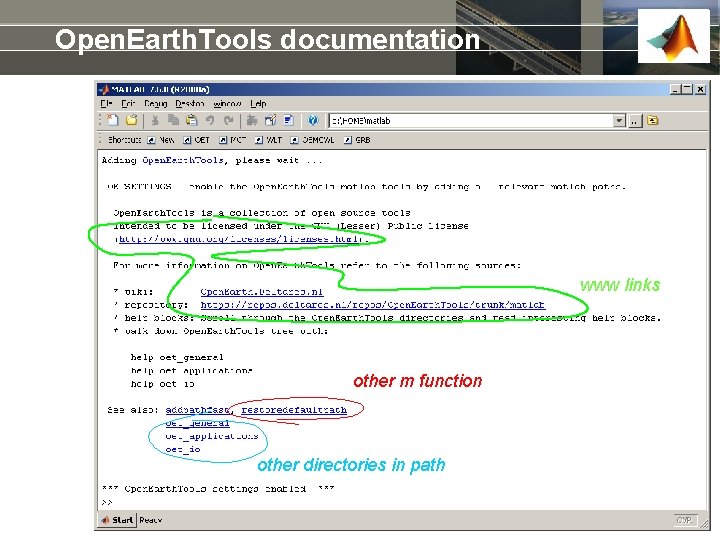
Open. Earth. Tools documentation www links other m function other directories in path

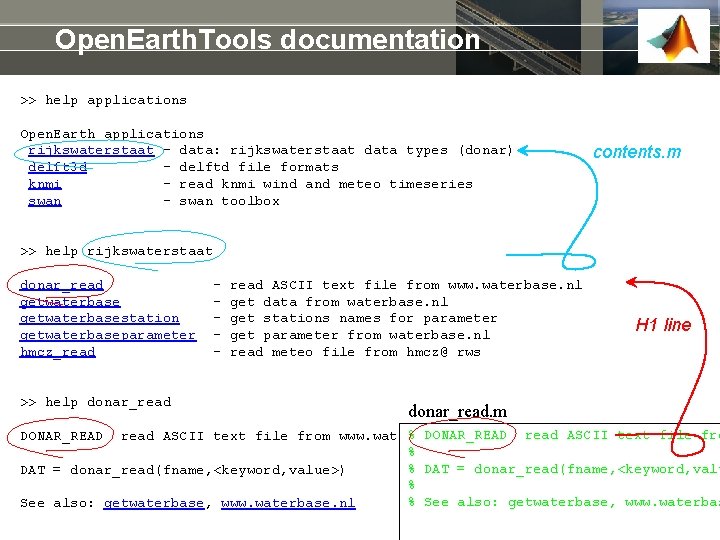
Open. Earth. Tools documentation >> help applications Open. Earth applications rijkswaterstaat - data: rijkswaterstaat data types (donar) delft 3 d - delftd file formats knmi - read knmi wind and meteo timeseries swan - swan toolbox contents. m >> help rijkswaterstaat donar_read getwaterbasestation getwaterbaseparameter hmcz_read >> help donar_read - read ASCII text file from www. waterbase. nl get data from waterbase. nl get stations names for parameter get parameter from waterbase. nl read meteo file from hmcz@ rws H 1 line donar_read. m % DONAR_READ read ASCII text file from www. waterbase. nl % % DAT = donar_read(fname, <keyword, value>) % % See also: getwaterbase, www. waterbase. nl DONAR_READ

Contents Extract 2 a use tools 2 b models Provide 2 c > read data > save as net. CDF add meta information data tools Load Transform to net. CDF on web server net. CDF on OPe. NDAP server 1 data providers = data users 3 D: . . . 3 http: //…

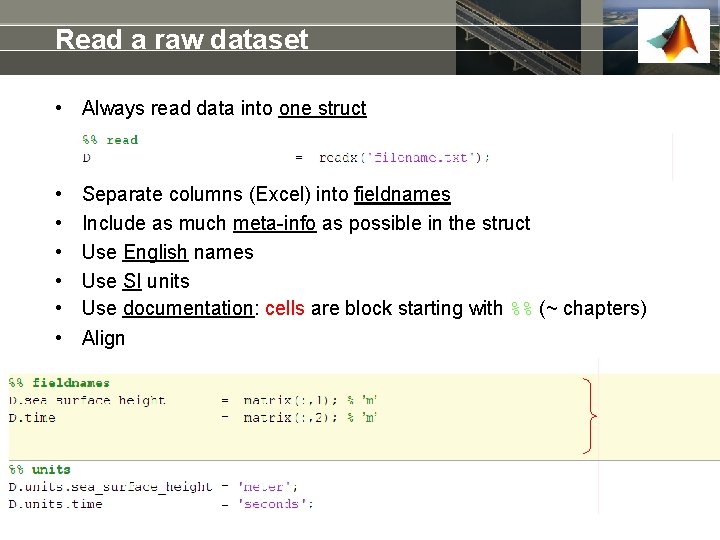
Read a raw dataset • Always read data into one struct • • • Separate columns (Excel) into fieldnames Include as much meta-info as possible in the struct Use English names Use SI units Use documentation: cells are block starting with %% (~ chapters) Align

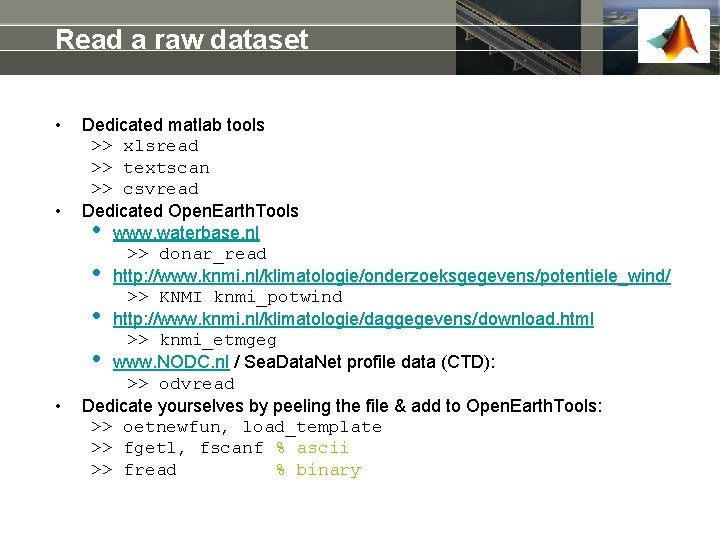
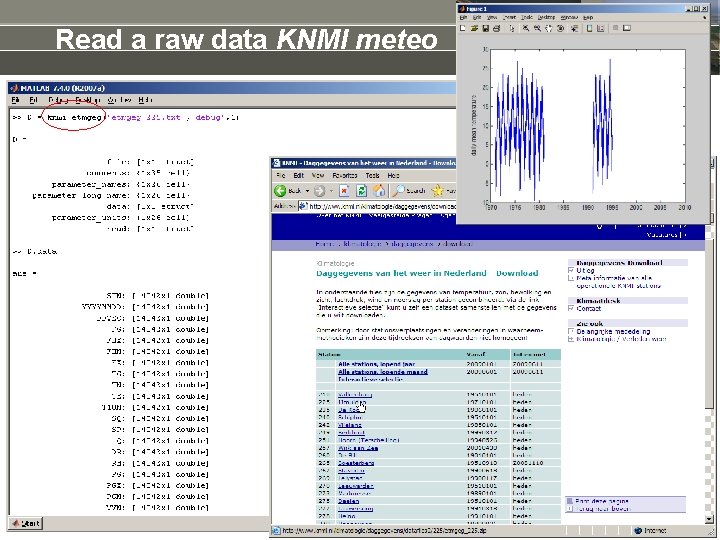
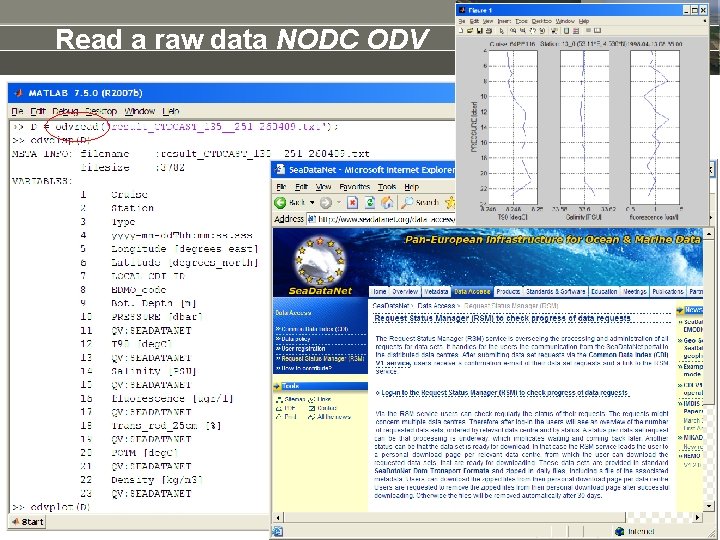
Read a raw dataset • • • Dedicated matlab tools >> xlsread >> textscan >> csvread Dedicated Open. Earth. Tools • www. waterbase. nl >> donar_read • http: //www. knmi. nl/klimatologie/onderzoeksgegevens/potentiele_wind/ >> KNMI knmi_potwind • http: //www. knmi. nl/klimatologie/daggegevens/download. html >> knmi_etmgeg • www. NODC. nl / Sea. Data. Net profile data (CTD): >> odvread Dedicate yourselves by peeling the file & add to Open. Earth. Tools: >> oetnewfun, load_template >> fgetl, fscanf % ascii >> fread % binary

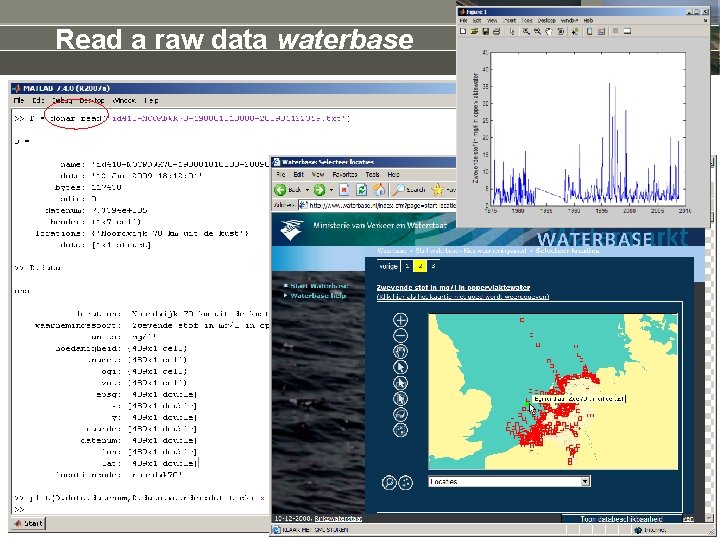
Read a raw data waterbase

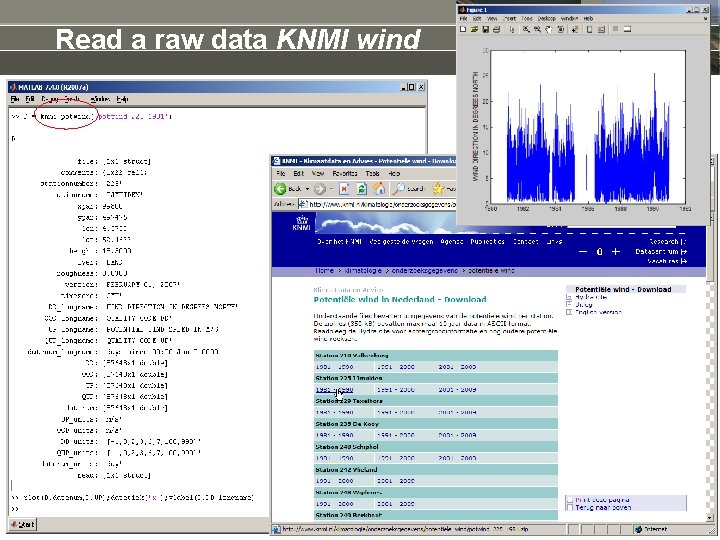
Read a raw data KNMI wind

Read a raw data KNMI meteo

Read a raw data NODC ODV

Test with some raw data Test for yourselve some raw data with dedicated scripts, choose one. Find the associated matlab function using the help documentation. • Rijkswaterstaat www. waterbase. nl • KNMI http: //www. knmi. nl/klimatologie/onderzoeksgegevens/potentiele_wind/ http: //www. knmi. nl/klimatologie/daggegevens/download. html • NODC www. NODC. nl / Sea. Data. Net profile data (CTD):

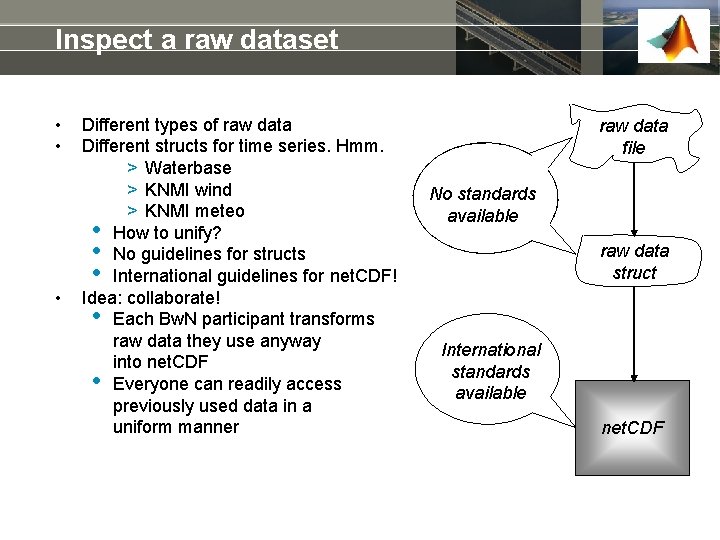
Inspect a raw dataset • • • Different types of raw data Different structs for time series. Hmm. > Waterbase > KNMI wind > KNMI meteo • How to unify? • No guidelines for structs • International guidelines for net. CDF! Idea: collaborate! • Each Bw. N participant transforms raw data they use anyway into net. CDF • Everyone can readily access previously used data in a uniform manner raw data file No standards available raw data struct International standards available net. CDF

Contents Extract 2 a use tools 2 b models Provide 2 c > read data > save as net. CDF add meta information data tools Load Transform to net. CDF on web server net. CDF on OPe. NDAP server 1 data providers = data users 3 D: . . . 3 http: //…

net. CDF • What is net. CDF • How to make net. CDF • Matlab net. CDF interface • Conventions (vocabularies): > Standard meta-info: CF convention + GNU/CC/? > standard quantities: CF convention > Standard units: Unidata UD units > Standard coordinate codes: EPSG • Save script in repository • Open. Earth. Raw. Data: for dedicated script • Open. Earth. Tools: for more general script • Inspect and plot net. CDF

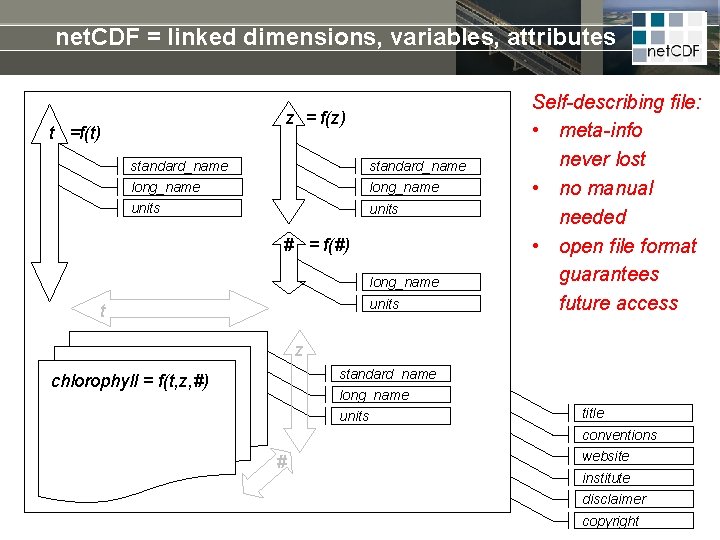
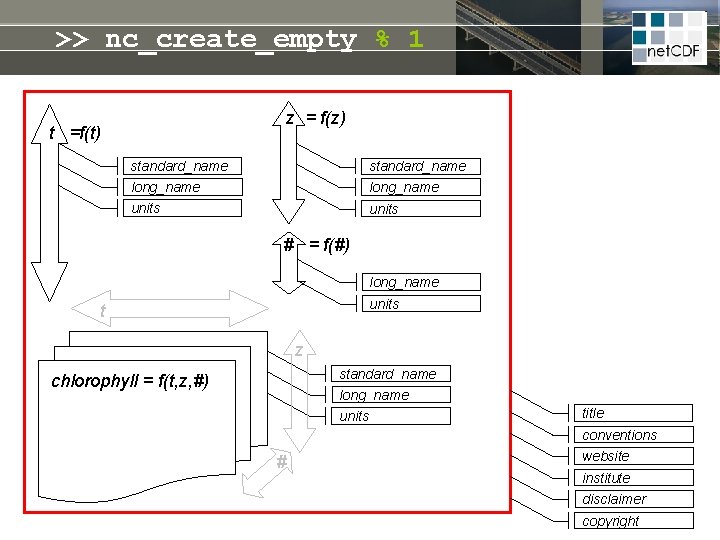
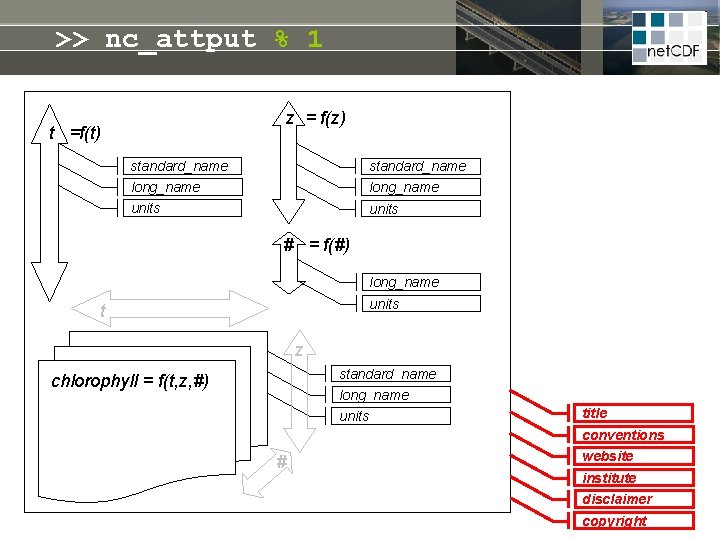
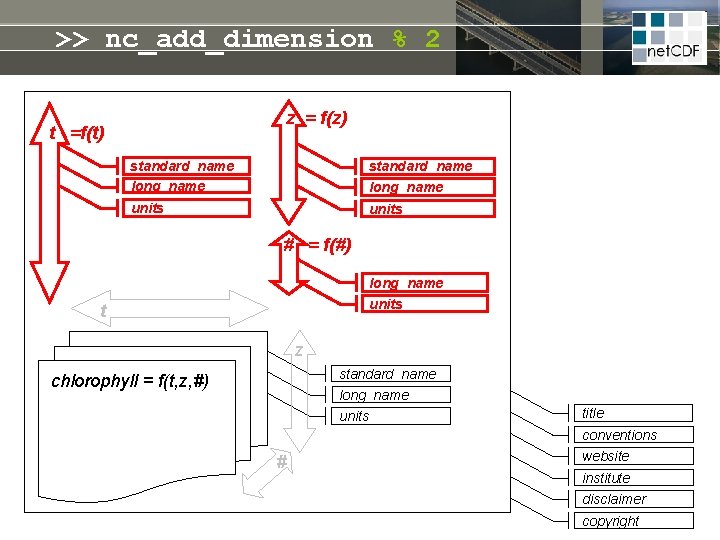
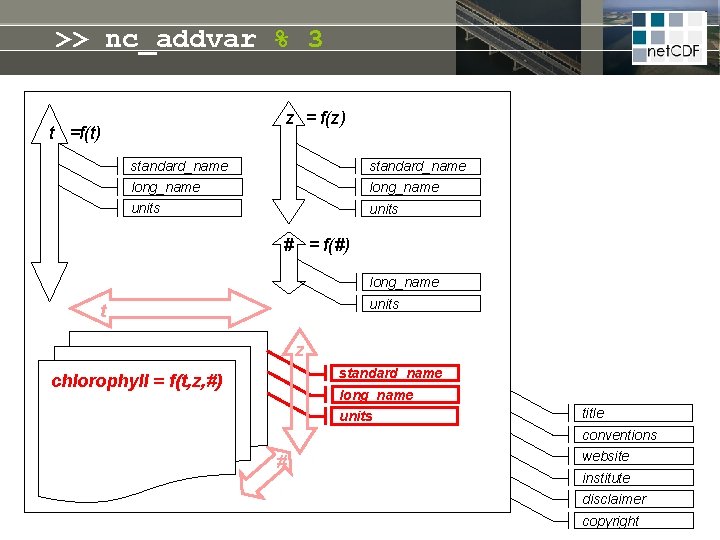
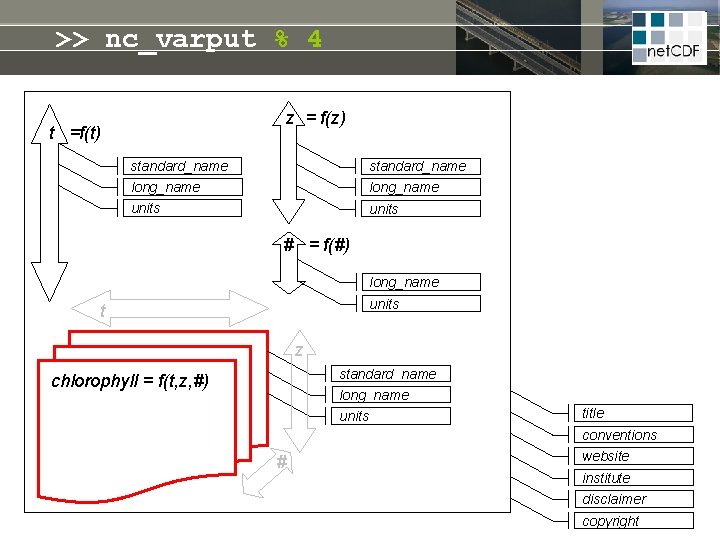
net. CDF = linked dimensions, variables, attributes z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t Self-describing file: • meta-info never lost • no manual needed • open file format guarantees future access z standard_name long_name chlorophyll = f(t, z, #) units title conventions # website institute disclaimer copyright

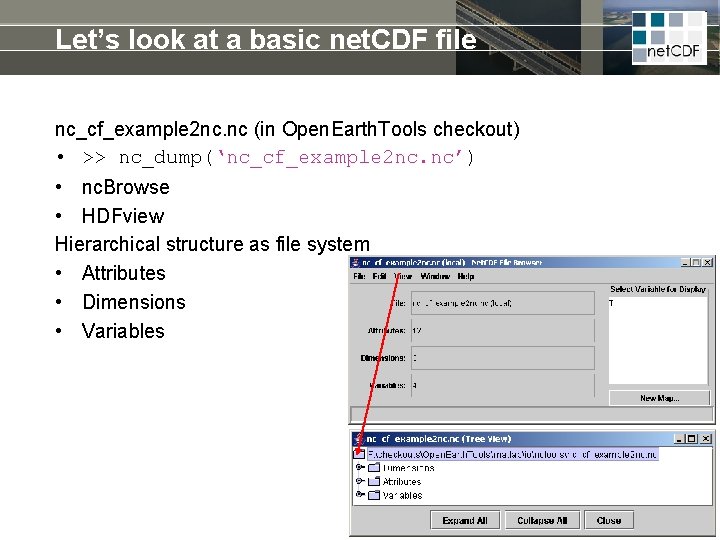
Let’s look at a basic net. CDF file nc_cf_example 2 nc. nc (in Open. Earth. Tools checkout) • >> nc_dump(‘nc_cf_example 2 nc. nc’) • nc. Browse • HDFview Hierarchical structure as file system • Attributes • Dimensions • Variables

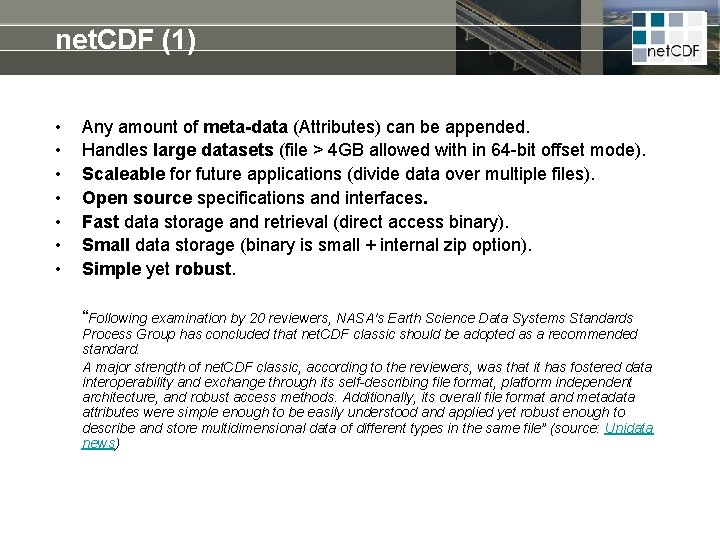
net. CDF (1) • • Any amount of meta-data (Attributes) can be appended. Handles large datasets (file > 4 GB allowed with in 64 -bit offset mode). Scaleable for future applications (divide data over multiple files). Open source specifications and interfaces. Fast data storage and retrieval (direct access binary). Small data storage (binary is small + internal zip option). Simple yet robust. “Following examination by 20 reviewers, NASA's Earth Science Data Systems Standards Process Group has concluded that net. CDF classic should be adopted as a recommended standard. A major strength of net. CDF classic, according to the reviewers, was that it has fostered data interoperability and exchange through its self-describing file format, platform independent architecture, and robust access methods. Additionally, its overall file format and metadata attributes were simple enough to be easily understood and applied yet robust enough to describe and store multidimensional data of different types in the same file” (source: Unidata news)

net. CDF (2) • • • 2 independent implementations of Net. CDF interface (C & Java): • reliable • redundant Generally used, so there’s always someone who can offer help available. net. CDF is de facto standard for coastal ocean circulation models (GOTM, ROMS, ECOM-SED and in the near future also Delft 3 D-FLOW). Lots of simple and free tools on the web that operate on Net. CDF files: • nc. Browse (NOAA) • NCO • Panoply (NASA) • ncview (linux) Implemented in numerous commercial tools too • Quickplot (Delft 3 D)

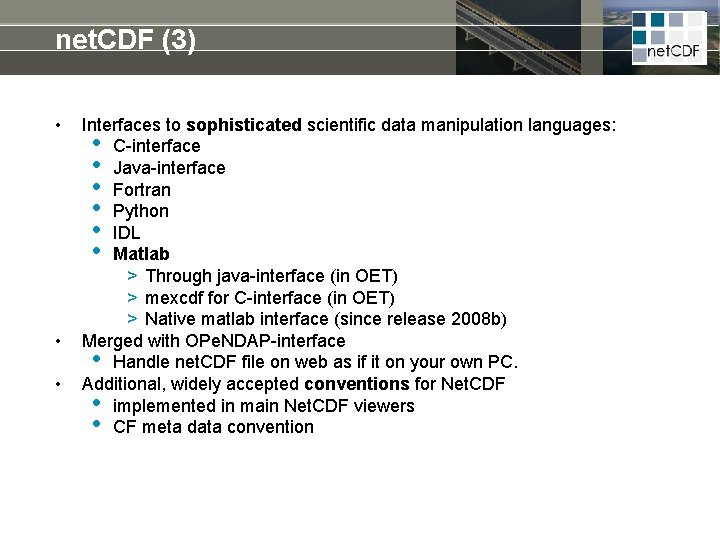
net. CDF (3) • • • Interfaces to sophisticated scientific data manipulation languages: • C-interface • Java-interface • Fortran • Python • IDL • Matlab > Through java-interface (in OET) > mexcdf for C-interface (in OET) > Native matlab interface (since release 2008 b) Merged with OPe. NDAP-interface • Handle net. CDF file on web as if it on your own PC. Additional, widely accepted conventions for Net. CDF • implemented in main Net. CDF viewers • CF meta data convention

Creating a net. CDF file • See : nc_cf_example 2 nc. m • Now step by step walking through • Use nc_cf_example 2 nc. nc as template when doing it yourselves

>> nc_create_empty % 1 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name long_name chlorophyll = f(t, z, #) units title conventions # website institute disclaimer copyright

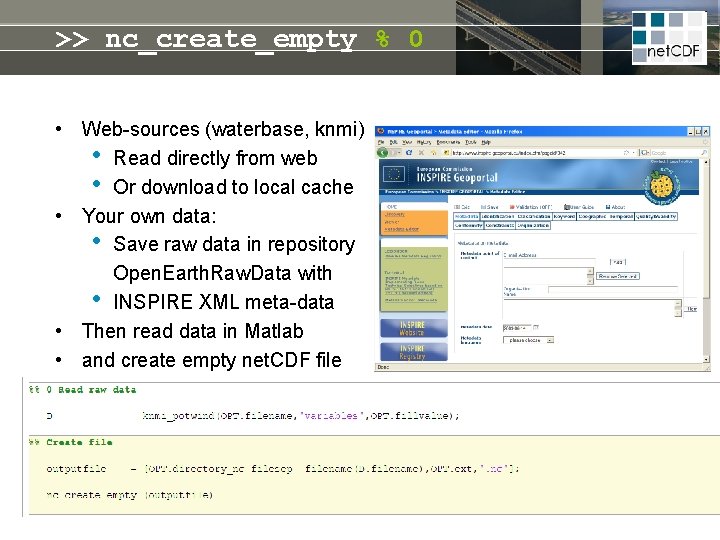
>> nc_create_empty % 0 • Web-sources (waterbase, knmi) • Read directly from web • Or download to local cache • Your own data: • Save raw data in repository Open. Earth. Raw. Data with • INSPIRE XML meta-data • Then read data in Matlab • and create empty net. CDF file

>> nc_attput % 1 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name long_name chlorophyll = f(t, z, #) units title conventions # website institute disclaimer copyright

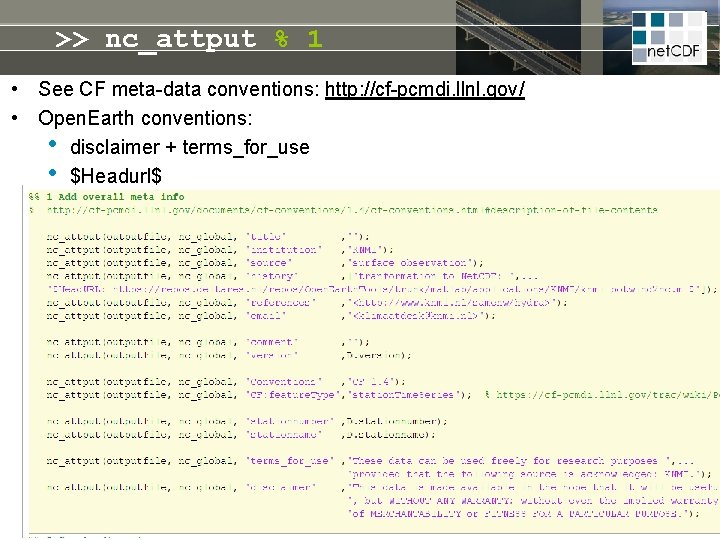
>> nc_attput % 1 • See CF meta-data conventions: http: //cf-pcmdi. llnl. gov/ • Open. Earth conventions: • disclaimer + terms_for_use • $Headurl$

>> nc_add_dimension % 2 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name long_name chlorophyll = f(t, z, #) units title conventions # website institute disclaimer copyright

>> nc_add_dimension % 2

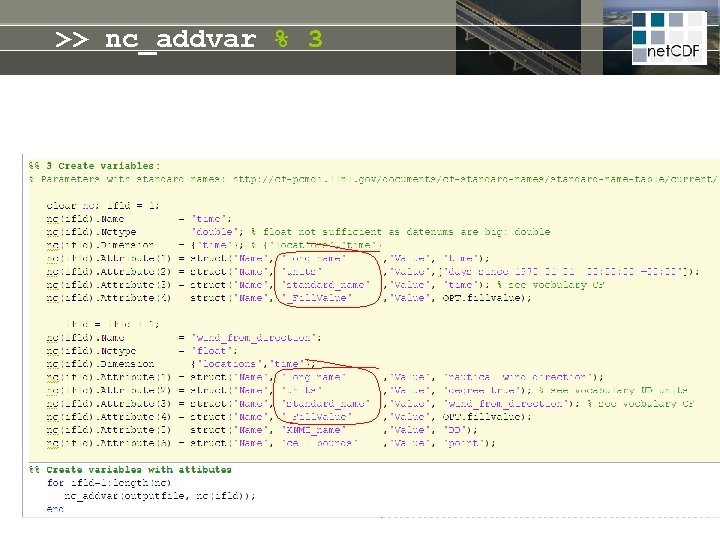
>> nc_addvar % 3 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name chlorophyll = f(t, z, #) long_name units title conventions # website institute disclaimer copyright

>> nc_addvar % 3

>> nc_varput % 4 CF convention vocabulary of quantities

>> nc_varput % 4 UNIDATA UD units package vocabulary of units

>> nc_varput % 4 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name long_name chlorophyll = f(t, z, #) units title conventions # website institute disclaimer copyright

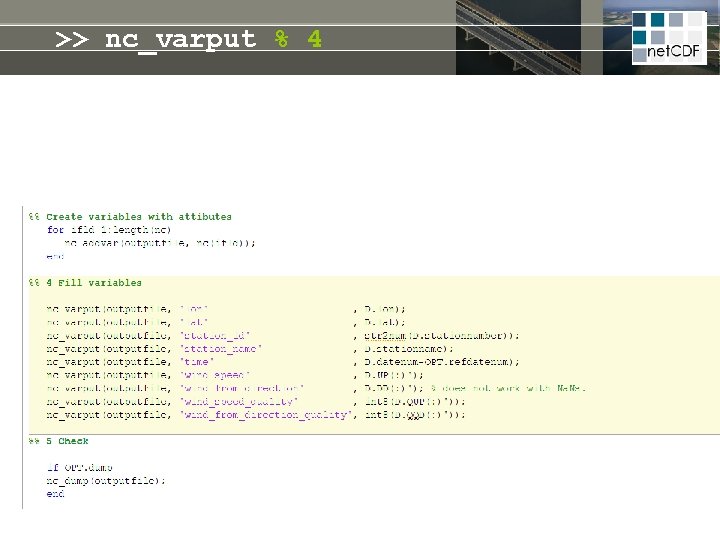
>> nc_varput % 4

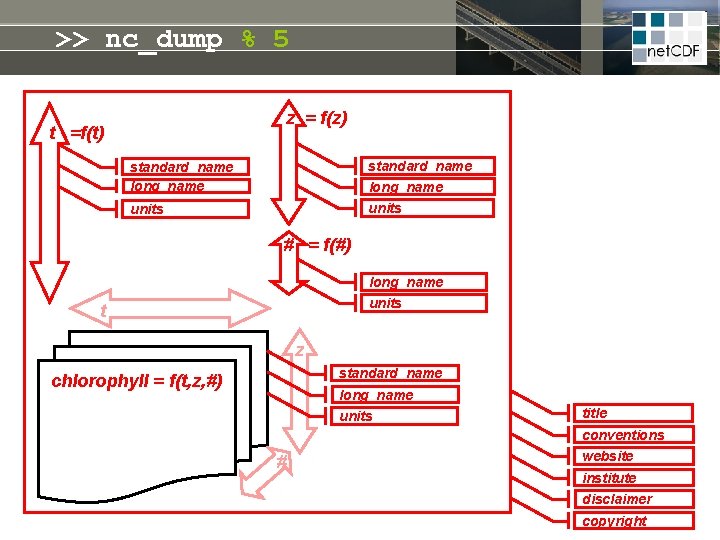
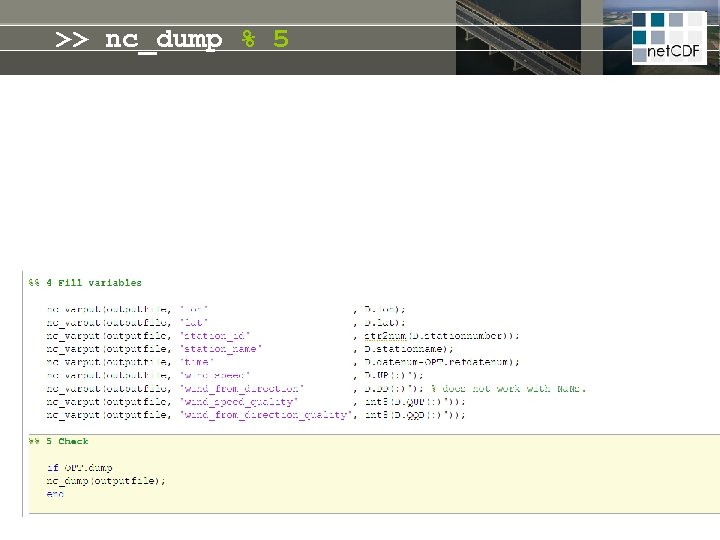
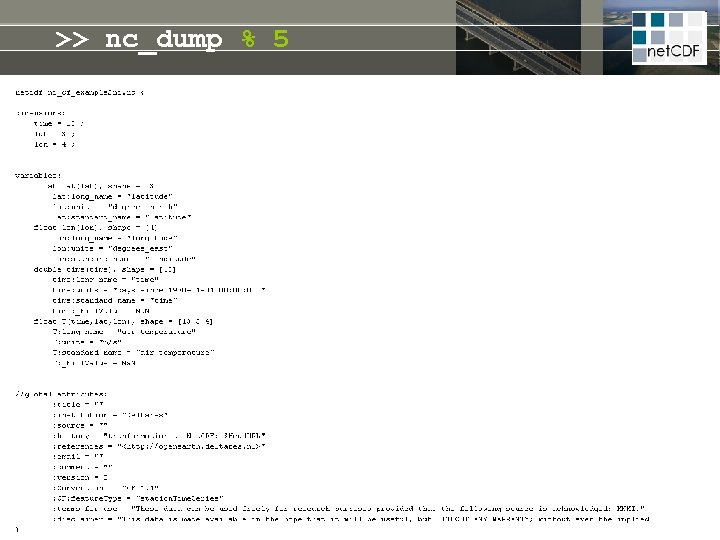
>> nc_dump % 5 z = f(z) t =f(t) standard_name long_name units # = f(#) long_name units t z standard_name chlorophyll = f(t, z, #) long_name units title conventions # website institute disclaimer copyright

>> nc_dump % 5

>> nc_dump % 5

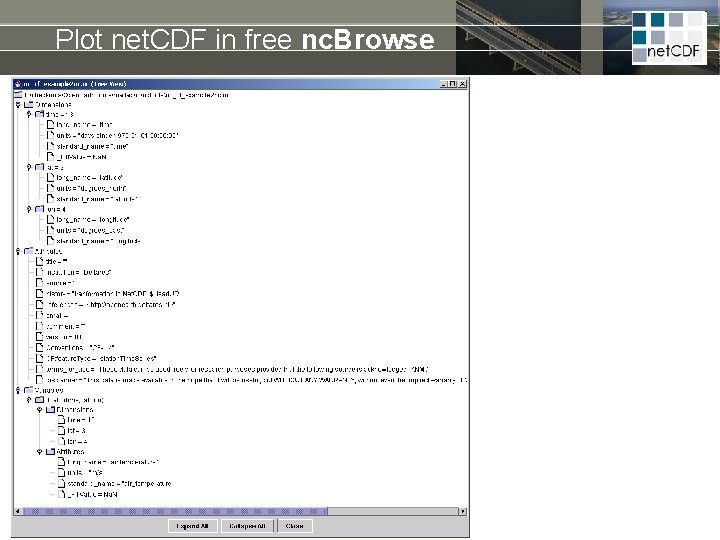
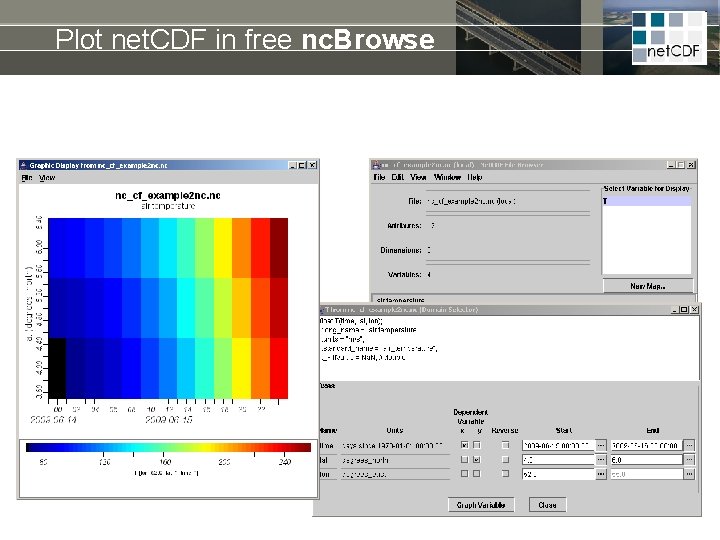
Plot net. CDF in free nc. Browse

Plot net. CDF in free nc. Browse

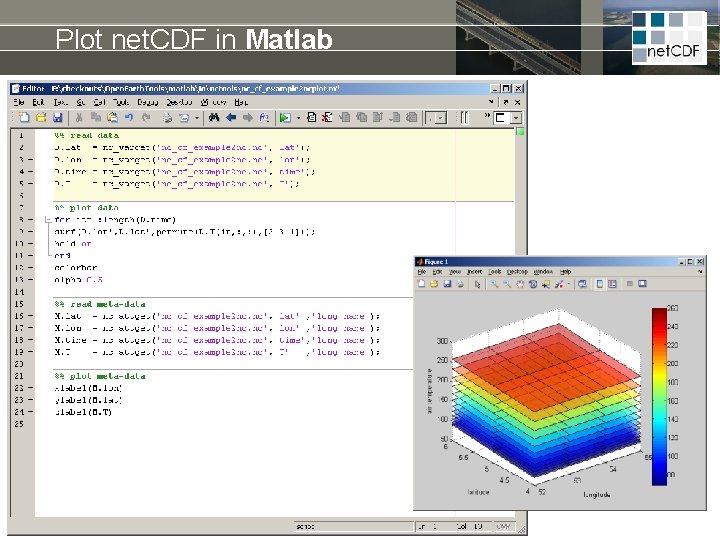
Plot net. CDF in Matlab

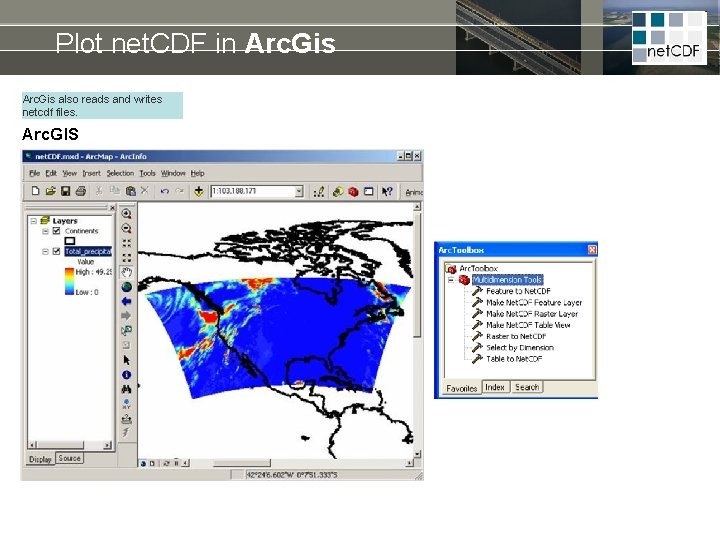
Plot net. CDF in Arc. Gis also reads and writes netcdf files. Arc. GIS

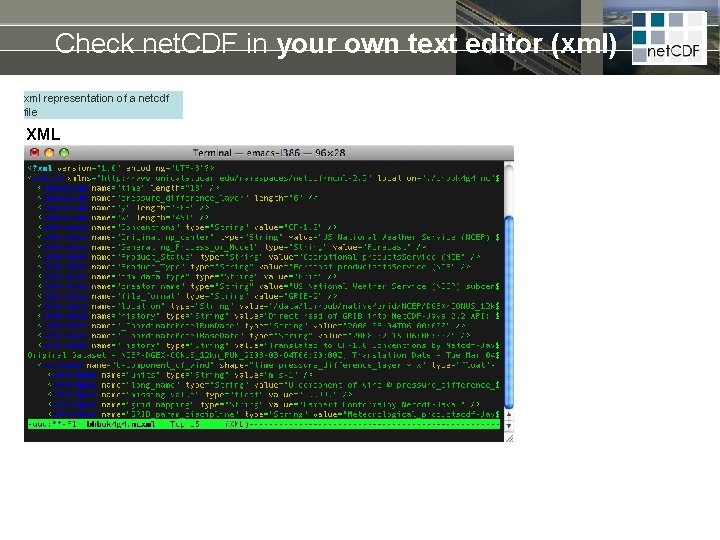
Check net. CDF in your own text editor (xml) xml representation of a netcdf file XML

Check net. CDF in … Very useful NCO #diff ncdiff -v time file 1. nc file 2. nc #compression & packingncpdq -4 -L 9 in. nc out. nc # Deflated packing (~80% lossy compression) #selecting variables by regex ncks -v '^Q. . ' in. nc # Q 01 --Q 99, QAA--QZZ, etc. Useful JAVA net. CDF tools Web hyperslabs, cool! Not so stable. IDV

Summary current session Extract 2 a use tools 2 b models Provide 2 c > read data > save as net. CDF add meta information data tools Load Transform to net. CDF on web server net. CDF on OPe. NDAP server 1 data providers = data users 3 D: . . . 3 http: //…

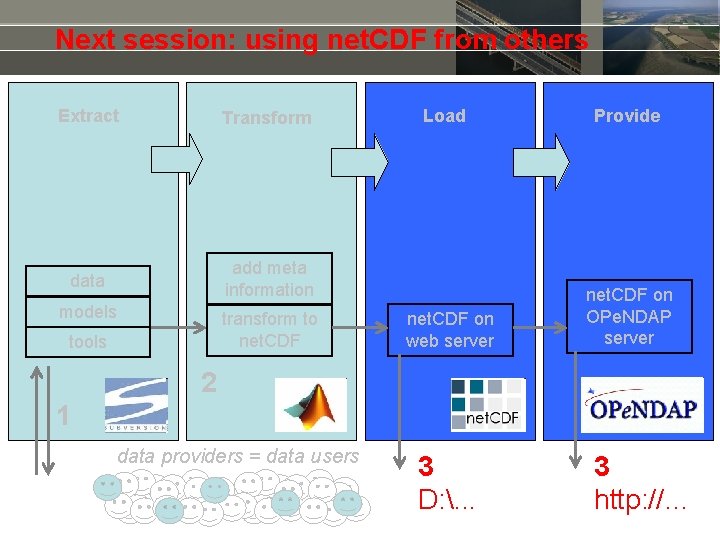
Next session: using net. CDF from others Extract Transform data add meta information models transform to net. CDF tools Load net. CDF on web server Provide net. CDF on OPe. NDAP server 2 1 data providers = data users 3 D: . . . 3 http: //…
 Open earth tools
Open earth tools 영국 beis
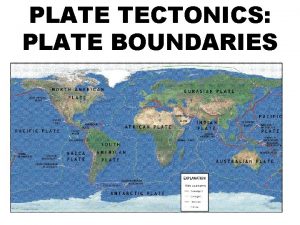
영국 beis What layers of earth make up the lithosphere
What layers of earth make up the lithosphere Make the lie big
Make the lie big Go make a difference we can make a difference
Go make a difference we can make a difference Make the lie big, make it simple
Make the lie big, make it simple Classification of tools used to assemble wood.
Classification of tools used to assemble wood. Bigearthnet
Bigearthnet Open system closed system and isolated system
Open system closed system and isolated system The appropriate cutting tool used in cutting fabrics
The appropriate cutting tool used in cutting fabrics Arcade game generator
Arcade game generator Open health tools
Open health tools Nsm network security monitoring open systems
Nsm network security monitoring open systems Ocr tools open source
Ocr tools open source Open health tools
Open health tools Google fair use images
Google fair use images Achmed lach net ich krieg mein tach net
Achmed lach net ich krieg mein tach net Ado.net vb.net
Ado.net vb.net Net open position adalah
Net open position adalah On delay timer coil symbol
On delay timer coil symbol Open hearts open hands
Open hearts open hands Make zero conditional sentences use cues
Make zero conditional sentences use cues Chapter 11 study guide business and technology
Chapter 11 study guide business and technology Contrived experiences activities
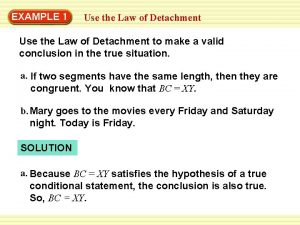
Contrived experiences activities Examples of law of detachment
Examples of law of detachment Use it up wear it out make it do
Use it up wear it out make it do Combine the sentences using non defining relative clauses
Combine the sentences using non defining relative clauses Will may and might for predictions
Will may and might for predictions Tools use in sewing
Tools use in sewing What is eraser tool in ms paint
What is eraser tool in ms paint Common business tools and technologies
Common business tools and technologies Use appropriate tools strategically
Use appropriate tools strategically What tools did the neolithic use
What tools did the neolithic use Use developer tools to create custom visuals power bi
Use developer tools to create custom visuals power bi Draw segment sr the bisector of the vertex angle prq
Draw segment sr the bisector of the vertex angle prq How you use ict today and how you will use it tomorrow
How you use ict today and how you will use it tomorrow World geography chapter 3 climates of the earth answers
World geography chapter 3 climates of the earth answers Which planet is called the blue planet
Which planet is called the blue planet Distribution of earth's water
Distribution of earth's water Moonfall msv
Moonfall msv Accurate representation meaning
Accurate representation meaning Earth layers physical and chemical
Earth layers physical and chemical Igneous rock formation
Igneous rock formation Goole doc
Goole doc A pattern of meteorological symbols that represent weather
A pattern of meteorological symbols that represent weather Different zones of the earth
Different zones of the earth