Sequence Alignment KunMao Chao Department of Computer Science
























- Slides: 56

Sequence Alignment Kun-Mao Chao (趙坤茂) Department of Computer Science and Information Engineering National Taiwan University, Taiwan E-mail: kmchao@csie. ntu. edu. tw WWW: http: //www. csie. ntu. edu. tw/~kmchao

Bioinformatics 2

Bioinformatics and Computational Biology. Related Journals: • • • Bioinformatics (previously called CABIOS) Bulletin of Mathematical Biology Computers and Biomedical Research Genome Research Genomics Journal of Bioinformatics and Computational Biology Journal of Molecular Biology Nature Nucleic Acid Research Science 3

Bioinformatics and Computational Biology. Related Conferences: • Intelligent Systems for Molecular Biology (ISMB) • Pacific Symposium on Biocomputing (PSB) • The Annual International Conference on Research in Computational Molecular Biology (RECOMB) • The IEEE Computer Society Bioinformatics Conference (CSB) • . . . 4

Bioinformatics and Computational Biology-Related Books: • Calculating the Secrets of Life: Applications of the Mathematical Sciences in Molecular Biology, by Eric S. Lander and Michael S. Waterman (1995) • Introduction to Computational Biology: Maps, Sequences, and Genomes, by Michael S. Waterman (1995) • Introduction to Computational Molecular Biology, by Joao Carlos Setubal and Joao Meidanis (1996) • Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology, by Dan Gusfield (1997) • Computational Molecular Biology: An Algorithmic Approach, by Pavel Pevzner (2000) • Introduction to Bioinformatics, by Arthur M. Lesk (2002) 5

Useful Websites • MIT Biology Hypertextbook – http: //www. mit. edu: 8001/afs/athena/course/other/esgbio/www/ 7001 main. html • The International Society for Computational Biology: – http: //www. iscb. org/ • National Center for Biotechnology Information (NCBI, NIH): – http: //www. ncbi. nlm. nih. gov/ • European Bioinformatics Institute (EBI): – http: //www. ebi. ac. uk/ • DNA Data Bank of Japan (DDBJ): – http: //www. ddbj. nig. ac. jp/ 6

Sequence Alignment 7

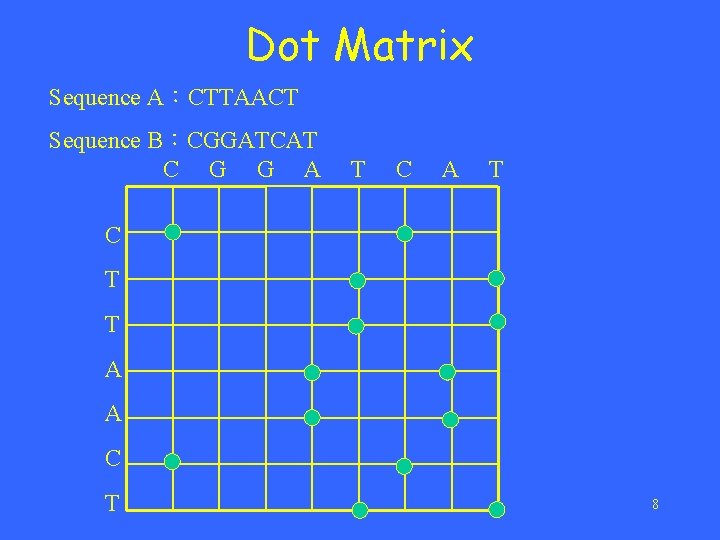
Dot Matrix Sequence A:CTTAACT Sequence B:CGGATCAT C G G A T C T T A A C T 8

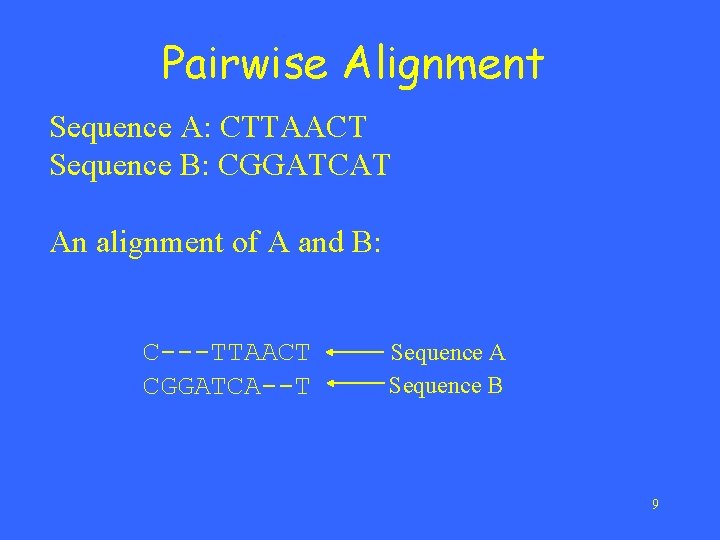
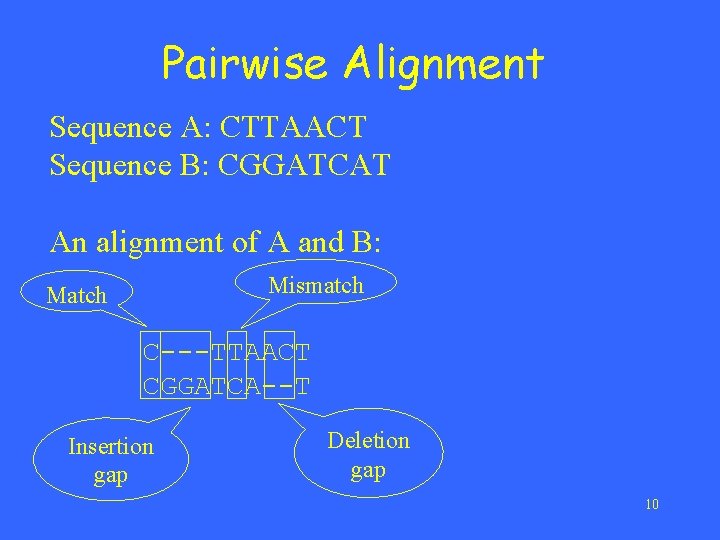
Pairwise Alignment Sequence A: CTTAACT Sequence B: CGGATCAT An alignment of A and B: C---TTAACT CGGATCA--T Sequence A Sequence B 9

Pairwise Alignment Sequence A: CTTAACT Sequence B: CGGATCAT An alignment of A and B: Mismatch Match C---TTAACT CGGATCA--T Insertion gap Deletion gap 10

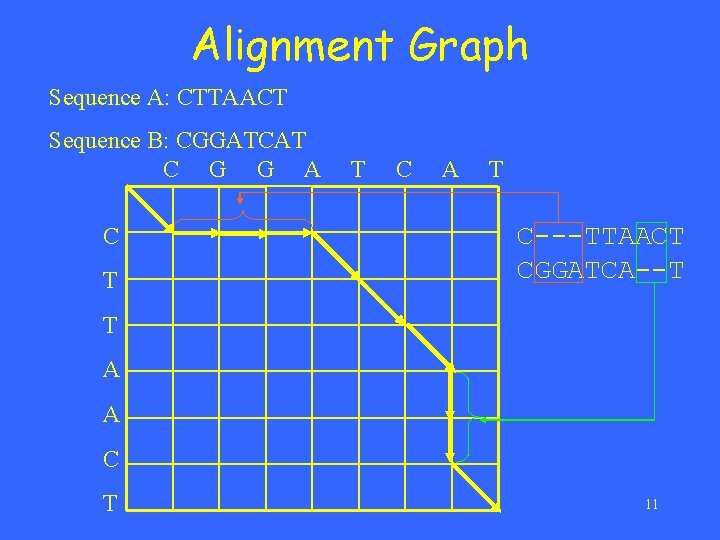
Alignment Graph Sequence A: CTTAACT Sequence B: CGGATCAT C G G A C T T C A T C---TTAACT CGGATCA--T T A A C T 11

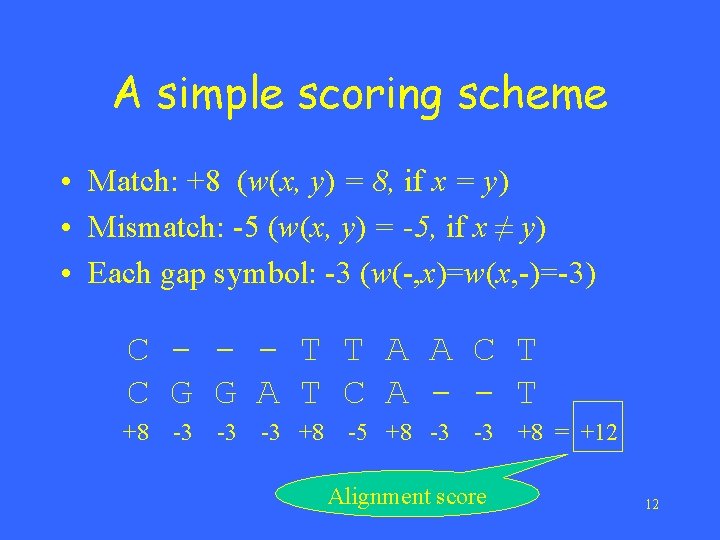
A simple scoring scheme • Match: +8 (w(x, y) = 8, if x = y) • Mismatch: -5 (w(x, y) = -5, if x ≠ y) • Each gap symbol: -3 (w(-, x)=w(x, -)=-3) C - - - T T A A C T C G G A T C A - - T +8 -3 -3 -3 +8 -5 +8 -3 -3 Alignment score +8 = +12 12

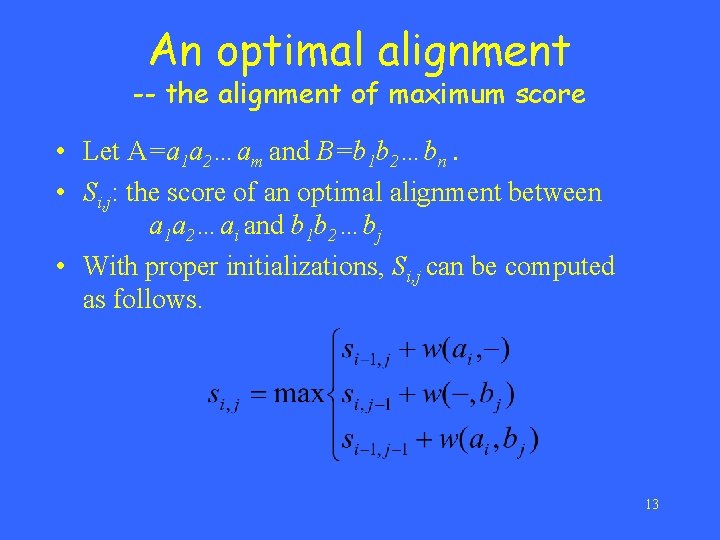
An optimal alignment -- the alignment of maximum score • Let A=a 1 a 2…am and B=b 1 b 2…bn. • Si, j: the score of an optimal alignment between a 1 a 2…ai and b 1 b 2…bj • With proper initializations, Si, j can be computed as follows. 13

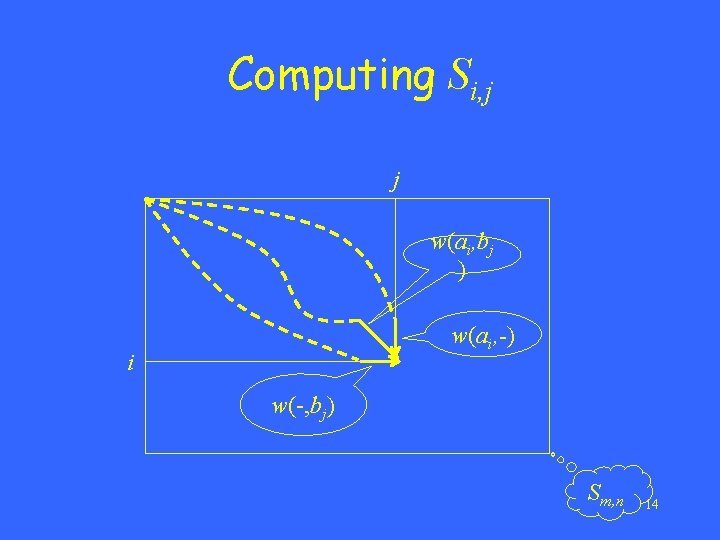
Computing Si, j j w(ai, bj ) w(ai, -) i w(-, bj) Sm, n 14

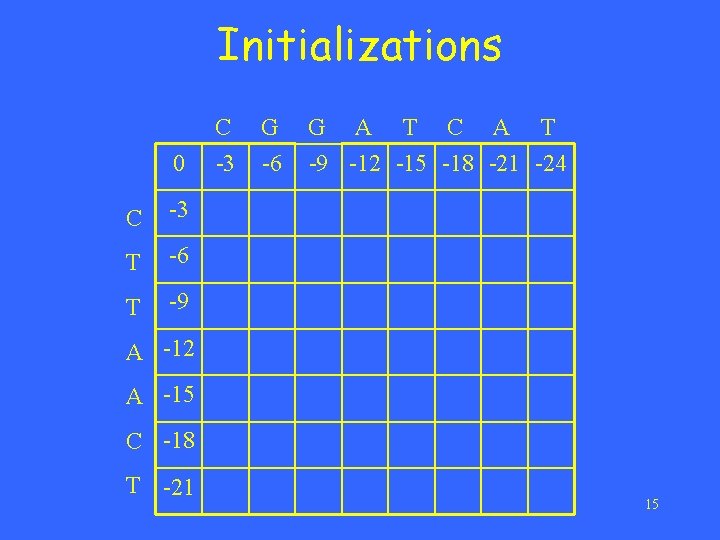
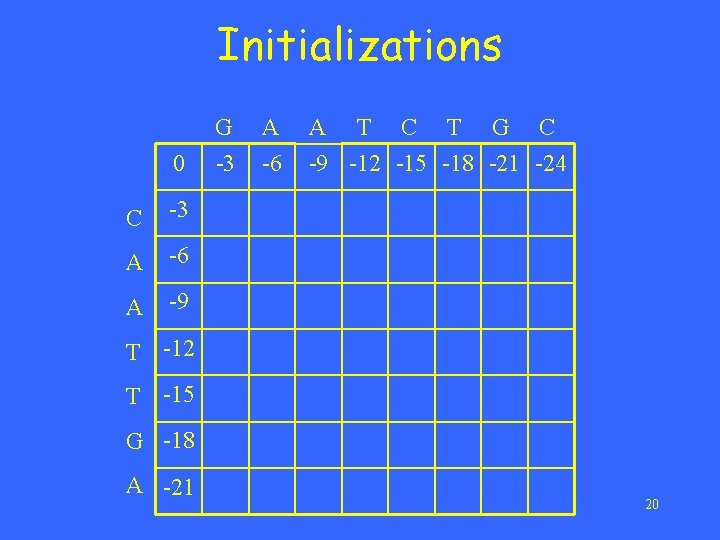
Initializations 0 C -3 T -6 T -9 C -3 G -6 G A T C A T -9 -12 -15 -18 -21 -24 A -12 A -15 C -18 T -21 15

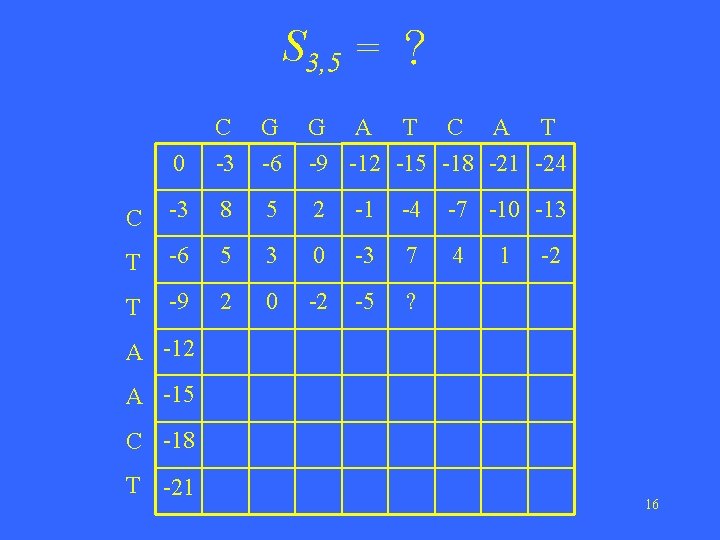
S 3, 5 = ? 0 C -3 G -6 G A T C A T -9 -12 -15 -18 -21 -24 C -3 8 5 2 -1 -4 -7 -10 -13 T -6 5 3 0 -3 7 4 T -9 2 0 -2 -5 ? 1 -2 A -15 C -18 T -21 16

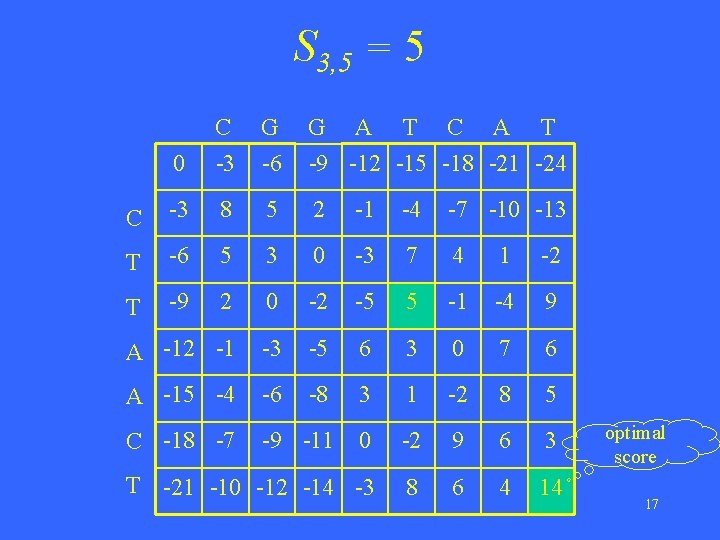
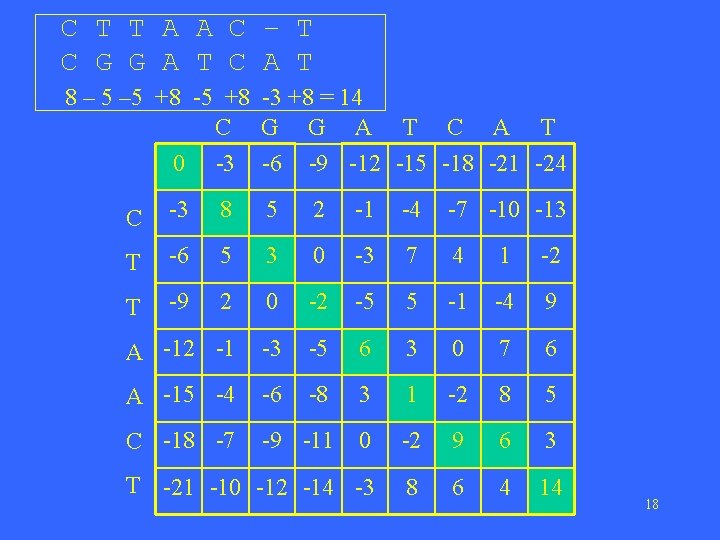
S 3, 5 = 5 0 C -3 G -6 G A T C A T -9 -12 -15 -18 -21 -24 C -3 8 5 2 -1 -4 -7 -10 -13 T -6 5 3 0 -3 7 4 1 -2 T -9 2 0 -2 -5 5 -1 -4 9 A -12 -1 -3 -5 6 3 0 7 6 A -15 -4 -6 -8 3 1 -2 8 5 C -18 -7 -9 -11 0 -2 9 6 3 T -21 -10 -12 -14 -3 8 6 4 14 optimal score 17

C T T A A C – T C G G A T C A T 8 – 5 +8 -5 +8 -3 +8 = 14 C G G A T C A T 0 -3 -6 -9 -12 -15 -18 -21 -24 C -3 8 5 2 -1 -4 -7 -10 -13 T -6 5 3 0 -3 7 4 1 -2 T -9 2 0 -2 -5 5 -1 -4 9 A -12 -1 -3 -5 6 3 0 7 6 A -15 -4 -6 -8 3 1 -2 8 5 C -18 -7 -9 -11 0 -2 9 6 3 T -21 -10 -12 -14 -3 8 6 4 14 18

Now try this example in class Sequence A: CAATTGA Sequence B: GAATCTGC Their optimal alignment? 19

Initializations 0 C -3 A -6 A -9 G -3 A -6 A T C T G C -9 -12 -15 -18 -21 -24 T -12 T -15 G -18 A -21 20

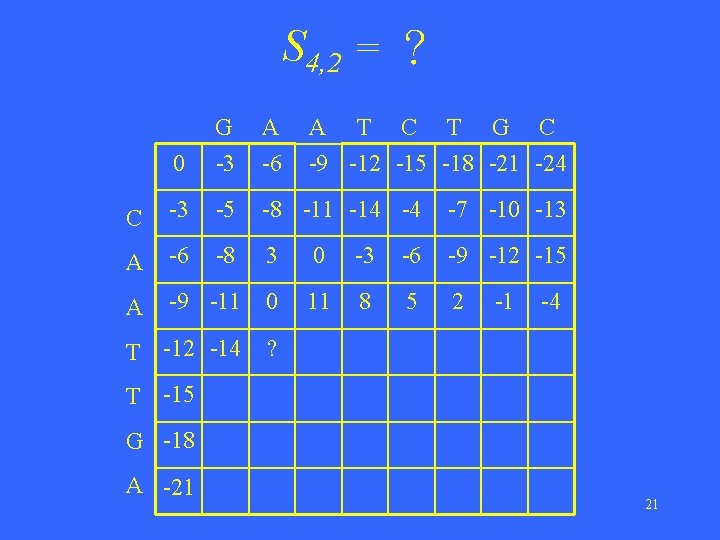
S 4, 2 = ? 0 G -3 A -6 A T C T G C -9 -12 -15 -18 -21 -24 C -3 -5 -8 -11 -14 -4 -7 -10 -13 A -6 -8 3 0 -3 -6 -9 -12 -15 A -9 -11 0 11 8 5 2 T -12 -14 ? -1 -4 T -15 G -18 A -21 21

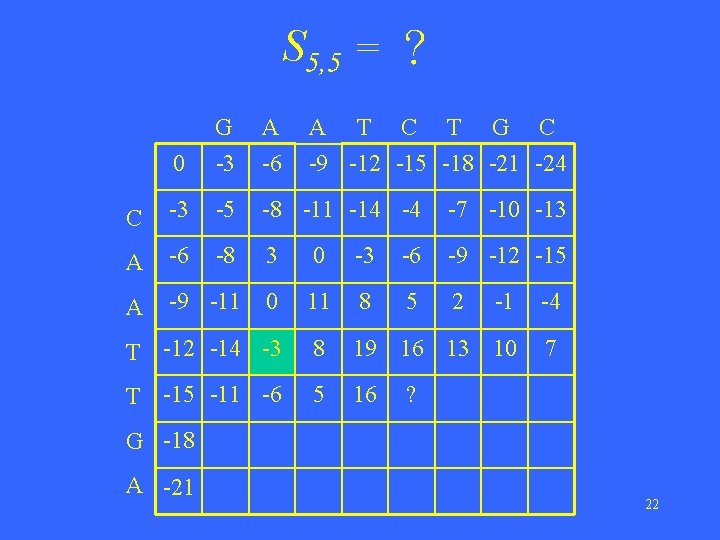
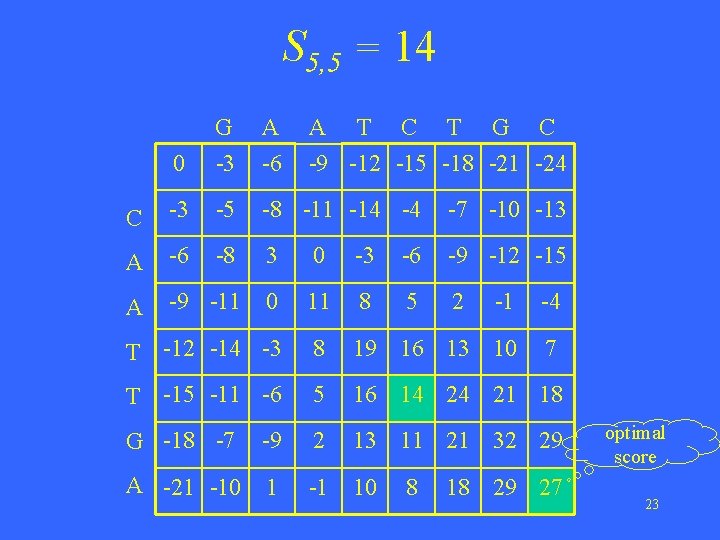
S 5, 5 = ? 0 G -3 A -6 A T C T G C -9 -12 -15 -18 -21 -24 C -3 -5 -8 -11 -14 -4 -7 -10 -13 A -6 -8 3 0 -3 -6 -9 -12 -15 A -9 -11 0 11 8 5 2 -1 -4 T -12 -14 -3 8 19 16 13 10 7 T -15 -11 -6 5 16 ? G -18 A -21 22

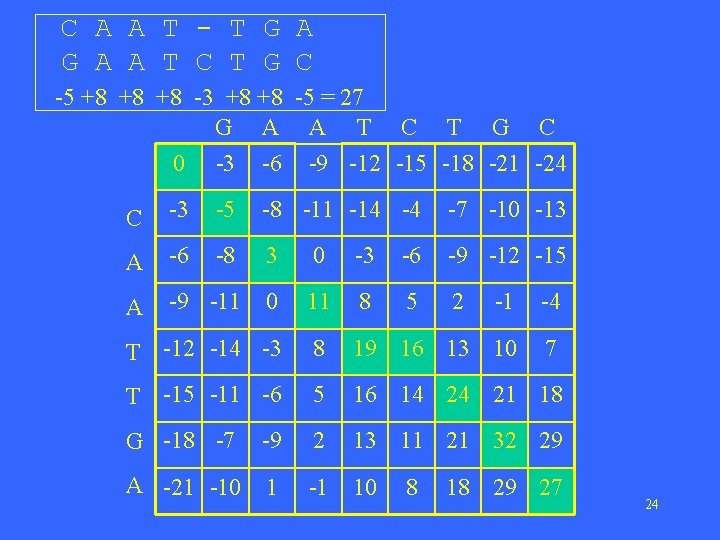
S 5, 5 = 14 0 G -3 A -6 A T C T G C -9 -12 -15 -18 -21 -24 C -3 -5 -8 -11 -14 -4 -7 -10 -13 A -6 -8 3 0 -3 -6 -9 -12 -15 A -9 -11 0 11 8 5 2 -1 -4 T -12 -14 -3 8 19 16 13 10 7 T -15 -11 -6 5 16 14 24 21 18 G -18 -7 -9 2 13 11 21 32 29 A -21 -10 1 -1 10 8 18 29 27 optimal score 23

C A A T - T G A A T C T G C -5 +8 +8 +8 -3 +8 +8 -5 = 27 G A A T C T G C 0 -3 -6 -9 -12 -15 -18 -21 -24 C -3 -5 -8 -11 -14 -4 -7 -10 -13 A -6 -8 3 0 -3 -6 -9 -12 -15 A -9 -11 0 11 8 5 2 -1 -4 T -12 -14 -3 8 19 16 13 10 7 T -15 -11 -6 5 16 14 24 21 18 G -18 -7 -9 2 13 11 21 32 29 A -21 -10 1 -1 10 8 18 29 27 24

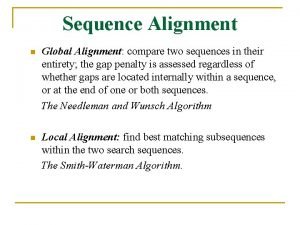
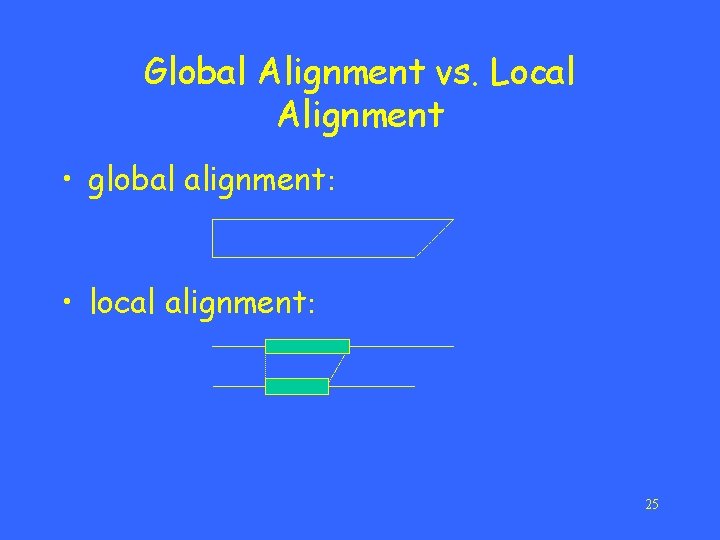
Global Alignment vs. Local Alignment • global alignment: • local alignment: 25

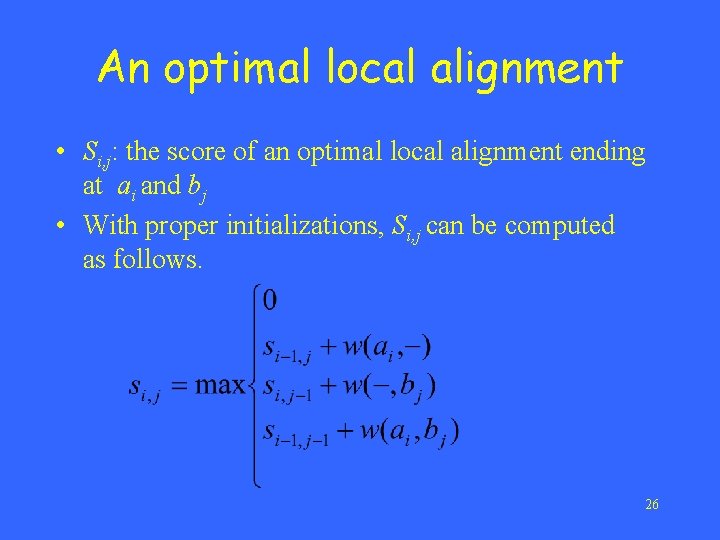
An optimal local alignment • Si, j: the score of an optimal local alignment ending at ai and bj • With proper initializations, Si, j can be computed as follows. 26

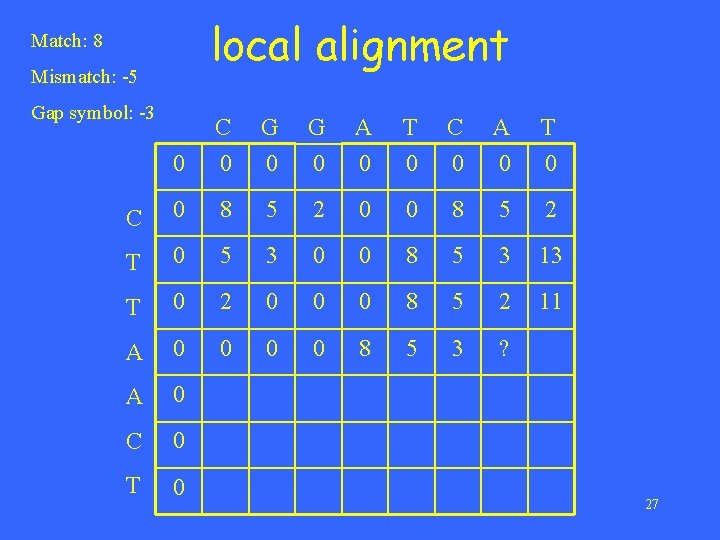
local alignment Match: 8 Mismatch: -5 Gap symbol: -3 0 C 0 G 0 A 0 T 0 C 0 8 5 2 0 0 8 5 2 T 0 5 3 0 0 8 5 3 13 T 0 2 0 0 0 8 5 2 11 A 0 0 8 5 3 ? A 0 C 0 T 0 27

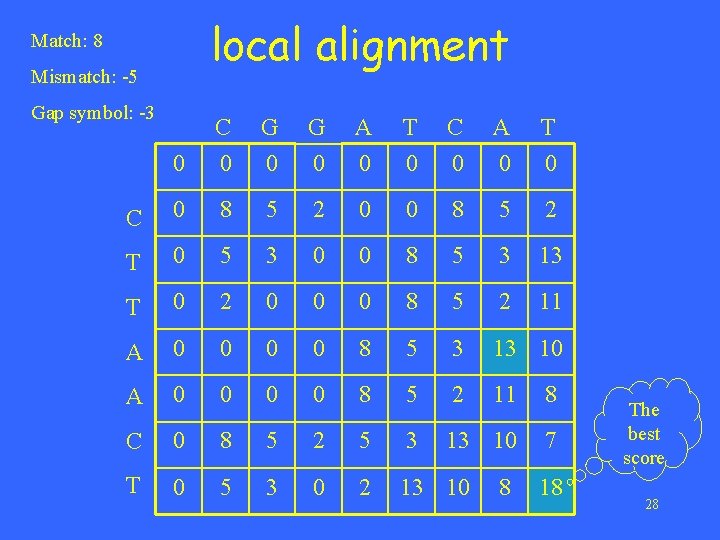
local alignment Match: 8 Mismatch: -5 Gap symbol: -3 0 C 0 G 0 A 0 T 0 C 0 8 5 2 0 0 8 5 2 T 0 5 3 0 0 8 5 3 13 T 0 2 0 0 0 8 5 2 11 A 0 0 8 5 3 13 10 A 0 0 8 5 2 11 8 C 0 8 5 2 5 3 13 10 7 T 0 5 3 0 2 13 10 8 18 The best score 28

A – C - T A T C A T 8 -3+8 = 18 C G 0 0 0 G 0 A 0 T 0 C 0 8 5 2 0 0 8 5 2 T 0 5 3 0 0 8 5 3 13 T 0 2 0 0 0 8 5 2 11 A 0 0 8 5 3 13 10 A 0 0 8 5 2 11 8 C 0 8 5 2 5 3 13 10 7 T 0 5 3 0 2 13 10 8 18 The best score 29

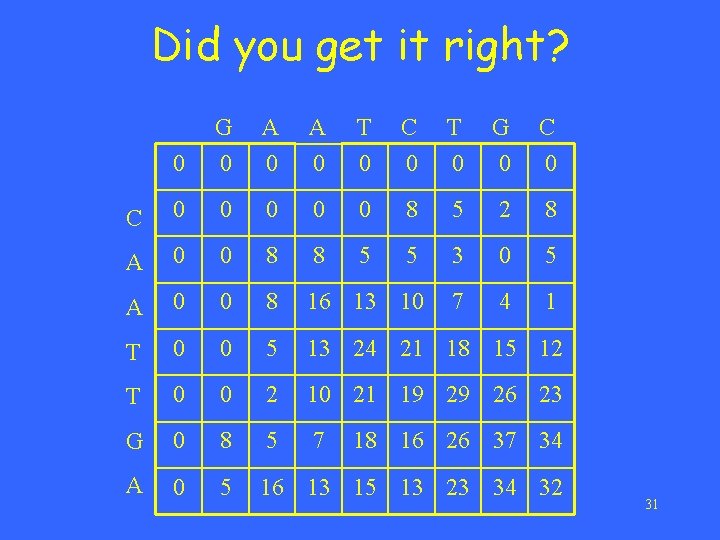
Now try this example in class Sequence A: CAATTGA Sequence B: GAATCTGC Their optimal local alignment? 30

Did you get it right? 0 G 0 A 0 T 0 C 0 T 0 G 0 C 0 0 0 8 5 2 8 A 0 0 8 8 5 5 3 0 5 A 0 0 8 16 13 10 7 4 1 T 0 0 5 13 24 21 18 15 12 T 0 0 2 10 21 19 29 26 23 G 0 8 5 7 A 0 5 16 13 15 13 23 34 32 18 16 26 37 34 31

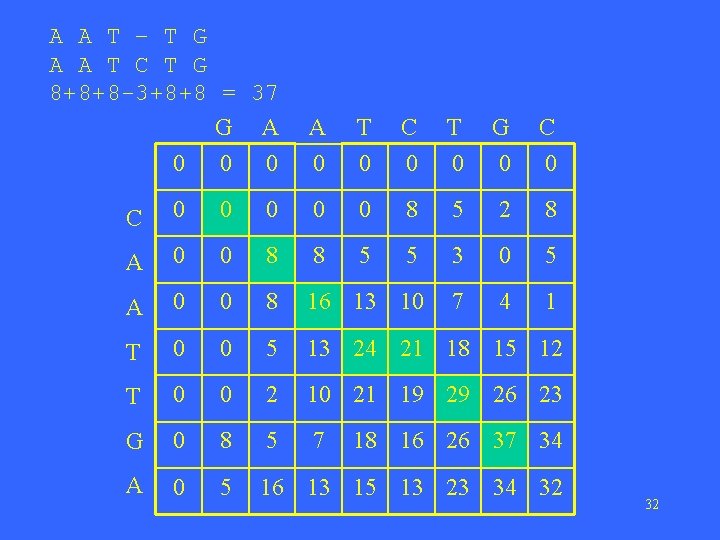
A A T – T G A A T C T G 8+8+8 -3+8+8 = 37 G A 0 0 0 A 0 T 0 C 0 T 0 G 0 C 0 0 0 8 5 2 8 A 0 0 8 8 5 5 3 0 5 A 0 0 8 16 13 10 7 4 1 T 0 0 5 13 24 21 18 15 12 T 0 0 2 10 21 19 29 26 23 G 0 8 5 7 A 0 5 16 13 15 13 23 34 32 18 16 26 37 34 32

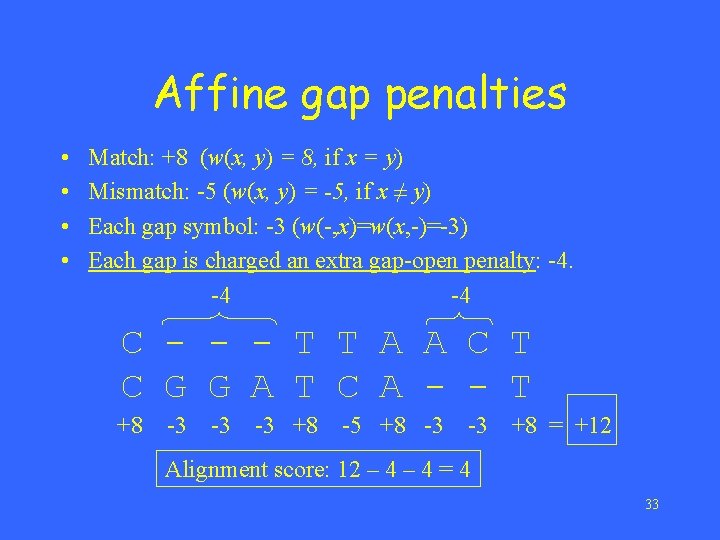
Affine gap penalties • • Match: +8 (w(x, y) = 8, if x = y) Mismatch: -5 (w(x, y) = -5, if x ≠ y) Each gap symbol: -3 (w(-, x)=w(x, -)=-3) Each gap is charged an extra gap-open penalty: -4. -4 -4 C - - - T T A A C T C G G A T C A - - T +8 -3 -3 -3 +8 -5 +8 -3 -3 +8 = +12 Alignment score: 12 – 4 = 4 33

Affine gap panalties • A gap of length k is penalized x + k·y. gap-open penalty Three cases for alignment endings: gap-symbol penalty 1. . x an aligned pair 2. . x. . . - a deletion 3. . . . x an insertion 34

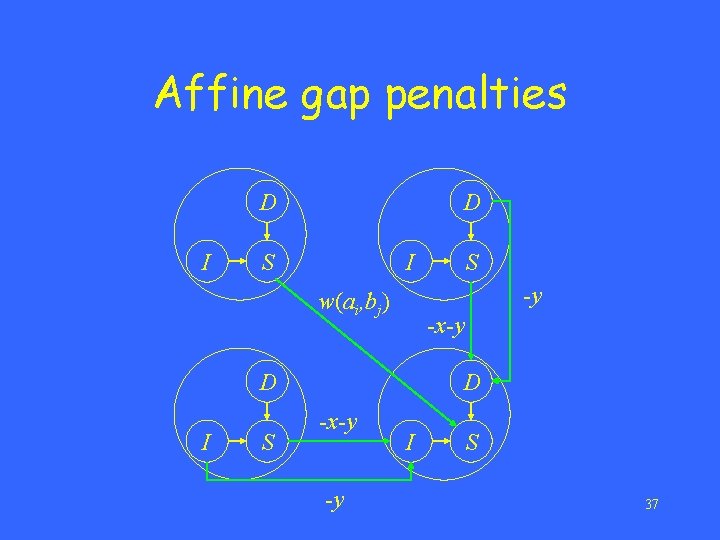
Affine gap penalties • Let D(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj ending with a deletion. • Let I(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj ending with an insertion. • Let S(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj. 35

Affine gap penalties (A gap of length k is penalized x + k·y. ) 36

Affine gap penalties D I D S I -y w(ai, bj) -x-y D I S S D -x-y -y I S 37

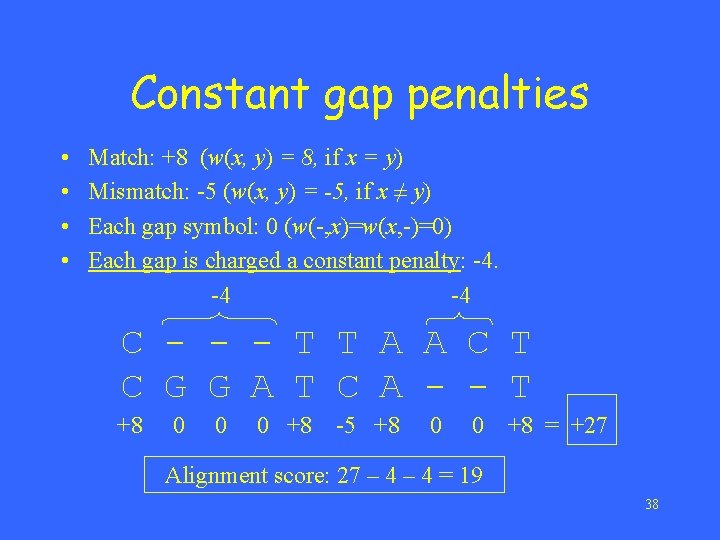
Constant gap penalties • • Match: +8 (w(x, y) = 8, if x = y) Mismatch: -5 (w(x, y) = -5, if x ≠ y) Each gap symbol: 0 (w(-, x)=w(x, -)=0) Each gap is charged a constant penalty: -4. -4 -4 C - - - T T A A C T C G G A T C A - - T +8 0 0 0 +8 -5 +8 0 0 +8 = +27 Alignment score: 27 – 4 = 19 38

Constant gap penalties • Let D(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj ending with a deletion. • Let I(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj ending with an insertion. • Let S(i, j) denote the maximum score of any alignment between a 1 a 2…ai and b 1 b 2…bj. 39

Constant gap penalties 40

Restricted affine gap panalties • A gap of length k is penalized x + f(k)·y. where f(k) = k for k <= c and f(k) = c for k > c Five cases for alignment endings: 1. . x an aligned pair 2. . x. . . - a deletion 3. . . . x an insertion 4. and 5. for long gaps 41

Restricted affine gap penalties 42

D(i, j) vs. D’(i, j) • Case 1: the best alignment ending at (i, j) with a deletion at the end has the last deletion gap of length <= c D(i, j) >= D’(i, j) • Case 2: the best alignment ending at (i, j) with a deletion at the end has the last deletion gap of length >= c D(i, j) <= D’(i, j) 43

k best local alignments • Smith-Waterman (Smith and Waterman, 1981; Waterman and Eggert, 1987) • FASTA (Wilbur and Lipman, 1983; Lipman and Pearson, 1985) • BLAST (Altschul et al. , 1990; Altschul et al. , 1997) 44

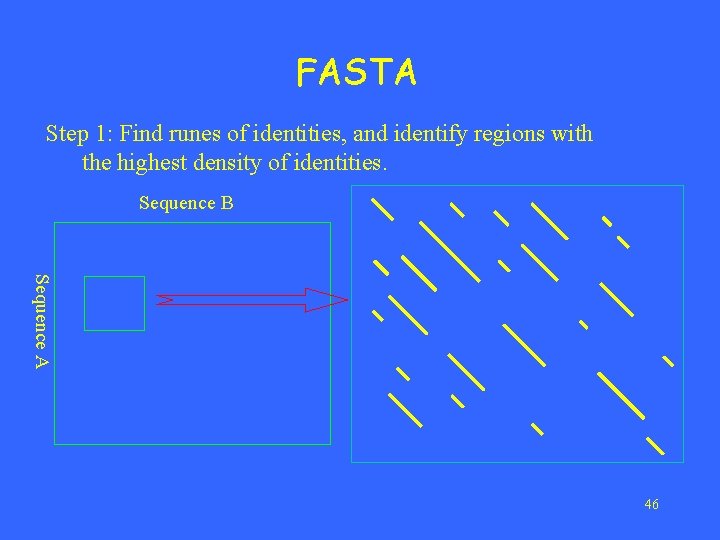
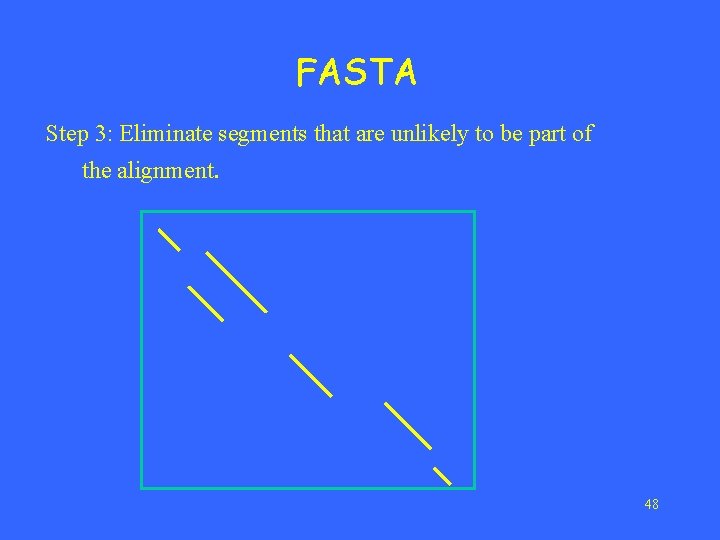
FASTA 1) Find runs of identities, and identify regions with the highest density of identities. 2) Re-score using PAM matrix, and keep top scoring segments. 3) Eliminate segments that are unlikely to be part of the alignment. 4) Optimize the alignment in a band. 45

FASTA Step 1: Find runes of identities, and identify regions with the highest density of identities. Sequence B Sequence A 46

FASTA Step 2: Re-score using PAM matrix, and keep top scoring segments. 47

FASTA Step 3: Eliminate segments that are unlikely to be part of the alignment. 48

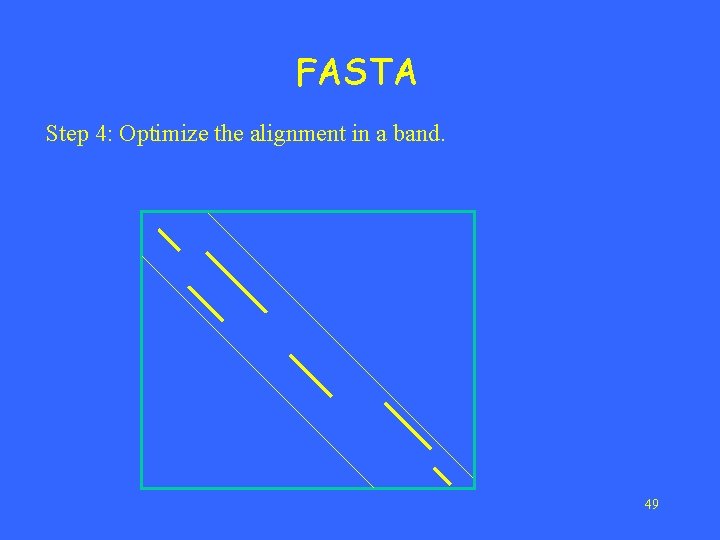
FASTA Step 4: Optimize the alignment in a band. 49

BLAST ü Basic Local Alignment Search Tool (by Altschul, Gish, Miller, Myers and Lipman) ü The central idea of the BLAST algorithm is that a statistically significant alignment is likely to contain a high-scoring pair of aligned words. 50

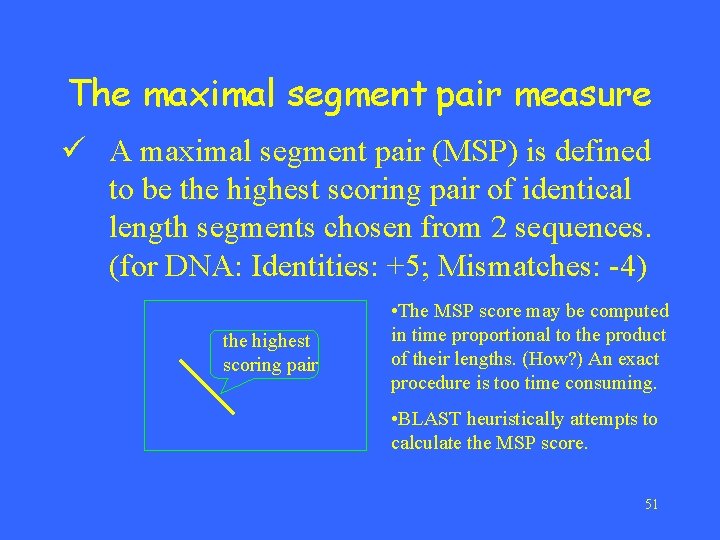
The maximal segment pair measure ü A maximal segment pair (MSP) is defined to be the highest scoring pair of identical length segments chosen from 2 sequences. (for DNA: Identities: +5; Mismatches: -4) the highest scoring pair • The MSP score may be computed in time proportional to the product of their lengths. (How? ) An exact procedure is too time consuming. • BLAST heuristically attempts to calculate the MSP score. 51

BLAST 1) Build the hash table for Sequence A. 2) Scan Sequence B for hits. 3) Extend hits. 52

BLAST Step 1: Build the hash table for Sequence A. (3 -tuple example) For protein sequences: For DNA sequences: Seq. A = AGATCGAT 12345678 AAA AAC. . AGA. . ATC. . CGA. . GAT. . TCG. . TTT 1 3 5 2 Seq. A = ELVIS Add xyz to the hash table if Score(xyz, ELV) ≧ T; Add xyz to the hash table if Score(xyz, LVI) ≧ T; Add xyz to the hash table if Score(xyz, VIS) ≧ T; 6 4 53

BLAST Step 2: Scan sequence B for hits. 54

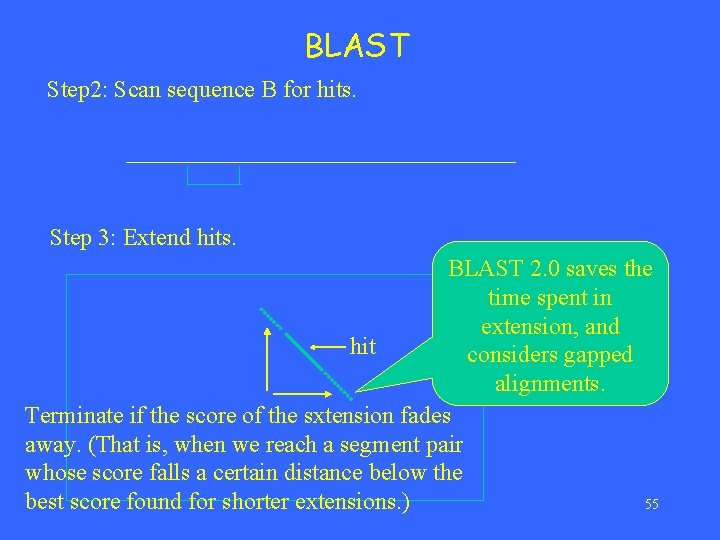
BLAST Step 2: Scan sequence B for hits. Step 3: Extend hits. BLAST 2. 0 saves the time spent in extension, and hit considers gapped alignments. Terminate if the score of the sxtension fades away. (That is, when we reach a segment pair whose score falls a certain distance below the 55 best score found for shorter extensions. )

Remarks • Filtering is based on the observation that a good alignment usually includes short identical or very similar fragments. • The idea of filtration was used in both FASTA and BLAST. 56
 Massimo ferrario infn
Massimo ferrario infn Kunmao
Kunmao Global vs local alignment
Global vs local alignment Global alignment vs local alignment
Global alignment vs local alignment Sequence alignment
Sequence alignment Actcg
Actcg Global alignment vs local alignment
Global alignment vs local alignment Ucl ridgmount practice
Ucl ridgmount practice Northwestern computer science department
Northwestern computer science department Computer science department rutgers
Computer science department rutgers Meredith hutchin stanford
Meredith hutchin stanford Florida state university computer science
Florida state university computer science Trimentoring
Trimentoring Department of computer science christ
Department of computer science christ Computer science department columbia
Computer science department columbia Tcoffee alignment
Tcoffee alignment Pasta multiple sequence alignment
Pasta multiple sequence alignment Bioedit download
Bioedit download Dot plot sequence alignment
Dot plot sequence alignment Tcoffee multiple sequence alignment
Tcoffee multiple sequence alignment Kkllkk profile
Kkllkk profile Clustal omega alignment
Clustal omega alignment Python multiple sequence alignment
Python multiple sequence alignment Sequence alignment
Sequence alignment Sequence alignment
Sequence alignment Sequence alignment
Sequence alignment What is gap penalty in bioinformatics
What is gap penalty in bioinformatics Praline multiple sequence alignment
Praline multiple sequence alignment My favorite subject is art because
My favorite subject is art because Dysplastic obesity
Dysplastic obesity Paralelos chão
Paralelos chão Quy trình sản xuất chao
Quy trình sản xuất chao Jackson chao
Jackson chao Cha cha cha con el jaleo del tren letra
Cha cha cha con el jaleo del tren letra Shih chao-hwei
Shih chao-hwei Enem 2016 para reciclar um motor
Enem 2016 para reciclar um motor Batatinha quando nasce espalha a rama pelo chão
Batatinha quando nasce espalha a rama pelo chão Chao seader method
Chao seader method Chao-hsien chu
Chao-hsien chu Nucleotides of rna
Nucleotides of rna Pseudocode repetition
Pseudocode repetition Differentiate finite sequence and infinite sequence
Differentiate finite sequence and infinite sequence Convolutional sequence to sequence learning
Convolutional sequence to sequence learning Department of forensic science dc
Department of forensic science dc Ohio tpes
Ohio tpes Eacademics.iitd
Eacademics.iitd Tum department of electrical and computer engineering
Tum department of electrical and computer engineering Computer engineering department
Computer engineering department Bps4104
Bps4104 Victorian curriculum science scope and sequence
Victorian curriculum science scope and sequence Design of animation sequence in computer graphics
Design of animation sequence in computer graphics Social science vs natural science
Social science vs natural science Branches of biology
Branches of biology Natural and physical science
Natural and physical science Applied science vs pure science
Applied science vs pure science Anthropology vs sociology
Anthropology vs sociology Sciencefusion think central
Sciencefusion think central