Introduction to BPS 4104 Prof Xuhua Xia xxiauottawa

Introduction to BPS 4104 Prof. Xuhua Xia (xxia@uottawa. ca) Course URL: http: //dambe. bio. uottawa. ca/teach/bps 4104. aspx
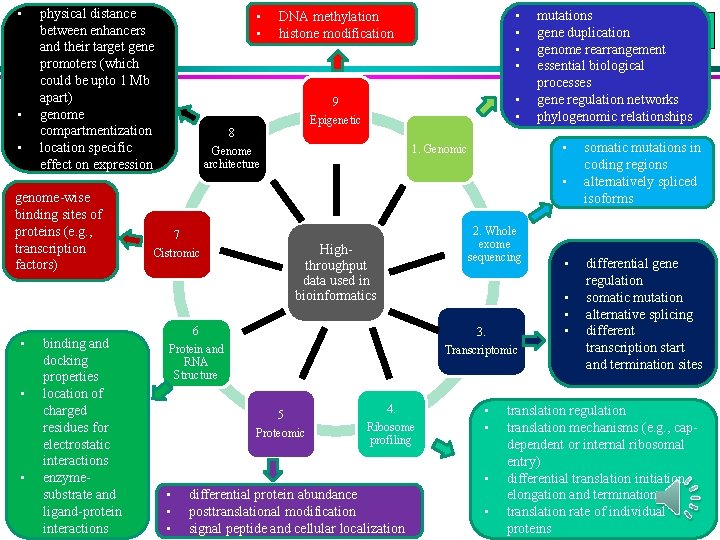
• • • physical distance between enhancers and their target gene promoters (which could be upto 1 Mb apart) genome compartmentization location specific effect on expression • • DNA methylation histone modification • • 9 • • Epigenetic 8 • 1. Genomic Genome architecture mutations gene duplication genome rearrangement essential biological processes gene regulation networks phylogenomic relationships • genome-wise binding sites of proteins (e. g. , transcription factors) • • • binding and docking properties location of charged residues for electrostatic interactions enzymesubstrate and ligand-protein interactions 7 2. Whole exome sequencing Highthroughput data used in bioinformatics Cistromic • • • 6 3. Protein and RNA Structure Transcriptomic 5 Proteomic 4. Ribosome profiling • • • differential protein abundance posttranslational modification signal peptide and cellular localization • • somatic mutations in coding regions alternatively spliced isoforms differential gene regulation somatic mutation alternative splicing different transcription start and termination sites translation regulation translation mechanisms (e. g. , capdependent or internal ribosomal entry) differential translation initiation, elongation and termination translation rate of individual proteins
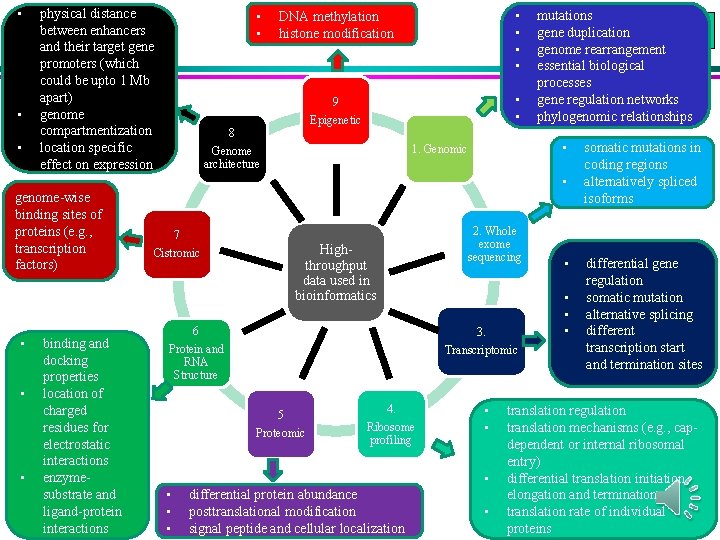
• • • physical distance between enhancers and their target gene promoters (which could be upto 1 Mb apart) genome compartmentization location specific effect on expression • • DNA methylation histone modification • • 9 • • Epigenetic 8 • 1. Genomic Genome architecture mutations gene duplication genome rearrangement essential biological processes gene regulation networks phylogenomic relationships • genome-wise binding sites of proteins (e. g. , transcription factors) • • • binding and docking properties location of charged residues for electrostatic interactions enzymesubstrate and ligand-protein interactions 7 2. Whole exome sequencing Highthroughput data used in bioinformatics Cistromic • • • 6 3. Protein and RNA Structure Transcriptomic 5 Proteomic 4. Ribosome profiling • • • differential protein abundance posttranslational modification signal peptide and cellular localization • • somatic mutations in coding regions alternatively spliced isoforms differential gene regulation somatic mutation alternative splicing different transcription start and termination sites translation regulation translation mechanisms (e. g. , capdependent or internal ribosomal entry) differential translation initiation, elongation and termination translation rate of individual proteins
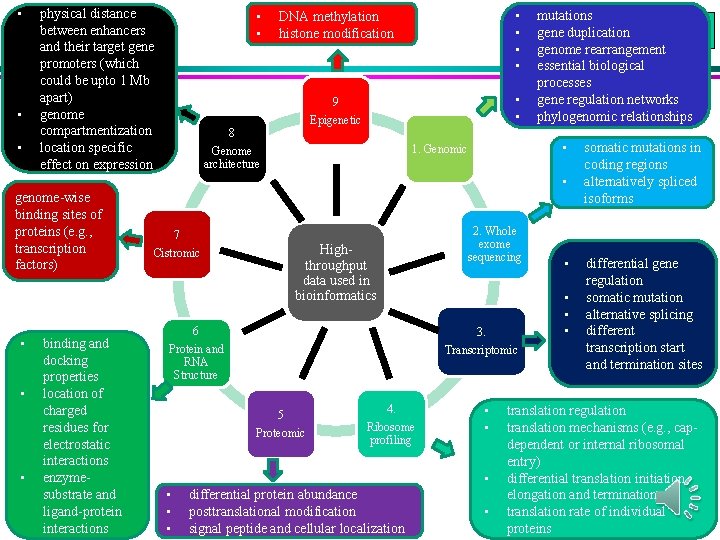
• • • physical distance between enhancers and their target gene promoters (which could be upto 1 Mb apart) genome compartmentization location specific effect on expression • • DNA methylation histone modification • • 9 • • Epigenetic 8 • 1. Genomic Genome architecture mutations gene duplication genome rearrangement essential biological processes gene regulation networks phylogenomic relationships • genome-wise binding sites of proteins (e. g. , transcription factors) • • • binding and docking properties location of charged residues for electrostatic interactions enzymesubstrate and ligand-protein interactions 7 2. Whole exome sequencing Highthroughput data used in bioinformatics Cistromic • • • 6 3. Protein and RNA Structure Transcriptomic 5 Proteomic 4. Ribosome profiling • • • differential protein abundance posttranslational modification signal peptide and cellular localization • • somatic mutations in coding regions alternatively spliced isoforms differential gene regulation somatic mutation alternative splicing different transcription start and termination sites translation regulation translation mechanisms (e. g. , capdependent or internal ribosomal entry) differential translation initiation, elongation and termination translation rate of individual proteins
5 Course objectives • Addressing the central question: What is the most efficient way of converting an ocean of data into knowledge? • Understand fundamental bioinformatic algorithms • Gain experience in using databases and bioinformatics software tools • To learn a set of skills that will enable you to think critically and quantitatively and to address a variety of problems in bioinformatics by hypotheticodeductive methods (a more specific expression for scientific methods)

6 About me • MSc and Ph. D at Western • Postdoctoral fellow at University of Helskinki, University of Toronto and Louisiana State University • Teaching and research at University of Hong Kong and University of Ottawa • Academic interests in: – Parasite-host interaction, with emphasis on pathogenic viruses and bacteria – Microbial genomics and bioinformatics – Molecular biology, phylogenetics and evolution – Software development • My lab web site: http: //dambe. bio. uottawa. ca • I have honours projects for motivated students: COVID-19, Influenza viruses, alternative splicing and diseases, stress response, etc.

Lecture 1 Mathematics and computation behind BLAST and FASTA Xuhua Xia xxia@uottawa. ca http: //dambe. bio. uottawa. ca
Why string matching? • Early applications: Sequence similarity between an oncogene (genes in viruses that cause a cancer-like transformation of the infected cells), v-sis (or p 28 sis or SSV), and the platelet-derived growth factor (PDGF) – M. D. Waterfield et al. 1983. Nature 304: 35 -39 – R. F. Doolittle et al. 1983. Science 221: 275 -277 – Implications: • Cancer can be caused by a constitutively expressed growth factor • Alteration of gene expression can contribute to cancer • Growth factors and the like can be drug targets against cancer

Why string matching? • Fast computational methods in string matching – FASTA – BLAST – Local pair-wise alignment by dynamic programming (Smith and Waterman 1981) • Lipman, D. ; Pearson, W. (1985). "Rapid and sensitive protein similarity searches". Science. 227 (4693): 1435– 1441 • Pearson, WR, DJ Lipman. 1988. Improved tools for biological sequence comparison. PNAS 85: 2444 -2448. • Altschul, SF, W Gish, W Miller, EW Myers, DJ Lipman. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403 -410. • Altschul, SF, DJ Lipman. 1990. Protein database searches for multiple alignments. PNAS 87: 5509 -5513.

Why string matching? • Fast computational methods in string matching – FASTA – BLAST – Local pair-wise alignment by dynamic programming (Smith and Waterman 1981) • Lipman, D. ; Pearson, W. (1985). "Rapid and sensitive protein similarity searches". Science. 227 (4693): 1435– 1441 • Pearson, WR, DJ Lipman. 1988. Improved tools for biological sequence comparison. PNAS 85: 2444 -2448. • Altschul, SF, W Gish, W Miller, EW Myers, DJ Lipman. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403 -410. • Altschul, SF, DJ Lipman. 1990. Protein database searches for multiple alignments. PNAS 87: 5509 -5513.

FASTA • Generally considered to be more sensitive than BLAST. • Illustration with two fictitious sequences: Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA Slide 11
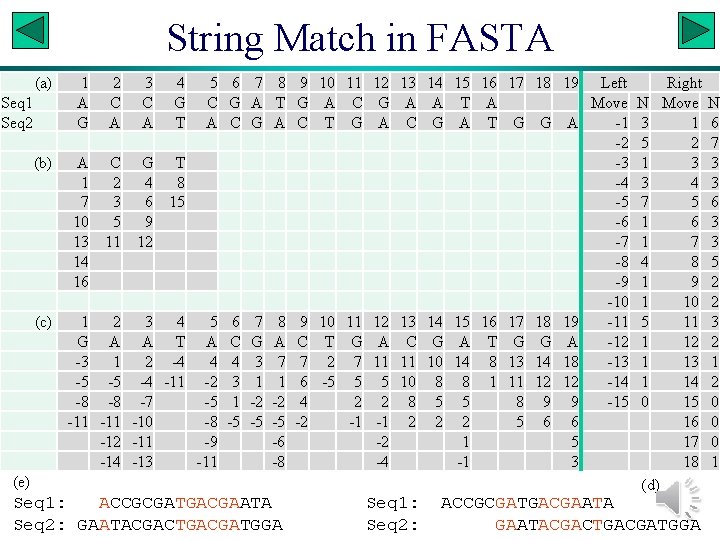
String Match in FASTA (a) Seq 1 Seq 2 (b) (c) 1 2 3 4 5 A C C G A A T A A C G T 1 2 4 8 7 3 6 15 10 5 9 13 11 12 14 16 1 2 3 4 5 G A A T A -3 1 2 -4 4 -5 -5 -4 -11 -2 -8 -8 -7 -5 -11 -10 -8 -12 -11 -9 -14 -13 -11 6 G C 6 C 4 3 1 -5 7 A G 7 G 3 1 -2 -5 8 T A 8 A 7 1 -2 -5 -6 -8 9 G C 9 C 7 6 4 -2 10 A T 10 T 2 -5 11 C G 11 G 7 5 2 -1 12 G A 12 A 11 5 2 -1 -2 -4 13 A C 13 C 11 10 8 2 (e) Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA Seq 1: Seq 2: 14 A G 14 G 10 8 5 2 15 16 17 18 19 Left Right T A Move N Move A T G G A -1 3 1 -2 5 2 -3 1 3 -4 3 4 -5 7 5 -6 1 6 -7 1 7 -8 4 8 -9 1 9 -10 15 16 17 18 19 -11 5 11 A T G G A -12 14 8 13 14 18 -13 1 13 8 1 11 12 12 -14 1 14 5 8 9 9 -15 0 15 2 5 6 6 16 1 5 17 -1 3 18 (d) ACCGCGATGACGAATACGACTGACGATGGA N 6 7 3 3 6 3 3 5 2 2 3 2 1 2 0 0 0 1
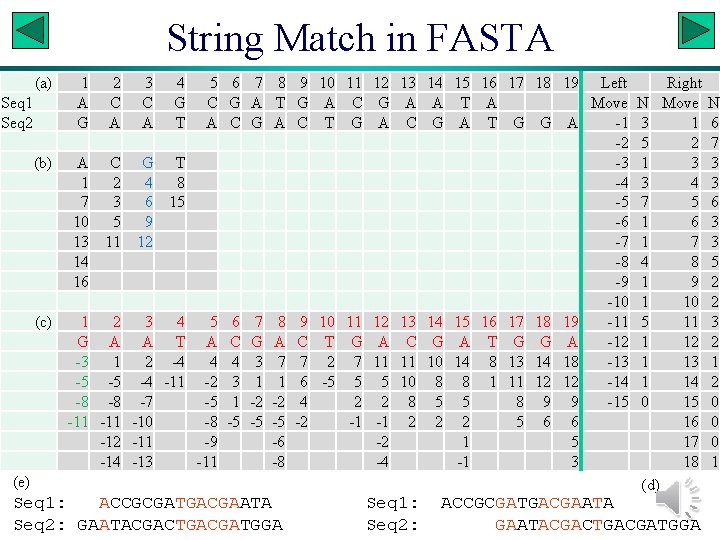
String Match in FASTA (a) Seq 1 Seq 2 (b) (c) 1 2 3 4 5 A C C G A A T A A C G T 1 2 4 8 7 3 6 15 10 5 9 13 11 12 14 16 1 2 3 4 5 G A A T A -3 1 2 -4 4 -5 -5 -4 -11 -2 -8 -8 -7 -5 -11 -10 -8 -12 -11 -9 -14 -13 -11 6 G C 6 C 4 3 1 -5 7 A G 7 G 3 1 -2 -5 8 T A 8 A 7 1 -2 -5 -6 -8 9 G C 9 C 7 6 4 -2 10 A T 10 T 2 -5 11 C G 11 G 7 5 2 -1 12 G A 12 A 11 5 2 -1 -2 -4 13 A C 13 C 11 10 8 2 (e) Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA Seq 1: Seq 2: 14 A G 14 G 10 8 5 2 15 16 17 18 19 Left Right T A Move N Move A T G G A -1 3 1 -2 5 2 -3 1 3 -4 3 4 -5 7 5 -6 1 6 -7 1 7 -8 4 8 -9 1 9 -10 15 16 17 18 19 -11 5 11 A T G G A -12 14 8 13 14 18 -13 1 13 8 1 11 12 12 -14 1 14 5 8 9 9 -15 0 15 2 5 6 6 16 1 5 17 -1 3 18 (d) ACCGCGATGACGAATACGACTGACGATGGA N 6 7 3 3 6 3 3 5 2 2 3 2 1 2 0 0 0 1
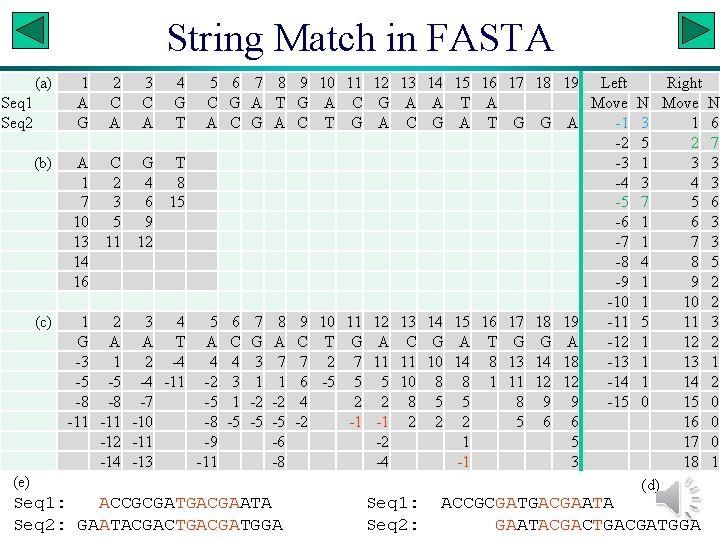
String Match in FASTA (a) Seq 1 Seq 2 (b) (c) 1 2 3 4 5 A C C G A A T A A C G T 1 2 4 8 7 3 6 15 10 5 9 13 11 12 14 16 1 2 3 4 5 G A A T A -3 1 2 -4 4 -5 -5 -4 -11 -2 -8 -8 -7 -5 -11 -10 -8 -12 -11 -9 -14 -13 -11 6 G C 6 C 4 3 1 -5 7 A G 7 G 3 1 -2 -5 8 T A 8 A 7 1 -2 -5 -6 -8 9 G C 9 C 7 6 4 -2 10 A T 10 T 2 -5 11 C G 11 G 7 5 2 -1 12 G A 12 A 11 5 2 -1 -2 -4 13 A C 13 C 11 10 8 2 (e) Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA Seq 1: Seq 2: 14 A G 14 G 10 8 5 2 15 16 17 18 19 Left Right T A Move N Move A T G G A -1 3 1 -2 5 2 -3 1 3 -4 3 4 -5 7 5 -6 1 6 -7 1 7 -8 4 8 -9 1 9 -10 15 16 17 18 19 -11 5 11 A T G G A -12 14 8 13 14 18 -13 1 13 8 1 11 12 12 -14 1 14 5 8 9 9 -15 0 15 2 5 6 6 16 1 5 17 -1 3 18 (d) ACCGCGATGACGAATACGACTGACGATGGA N 6 7 3 3 6 3 3 5 2 2 3 2 1 2 0 0 0 1
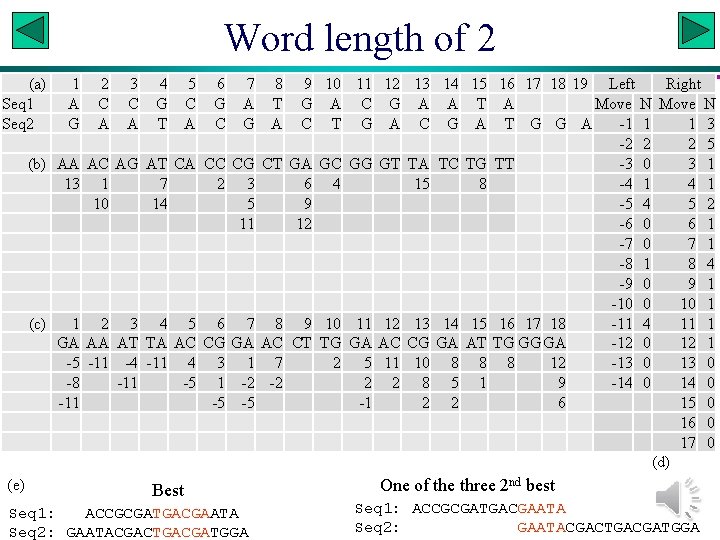
Word length of 2 (a) Seq 1 Seq 2 (b) (c) (e) 1 A G AA 13 1 GA -5 -8 -11 2 C A AC 1 10 2 AA -11 3 C A AG 3 AT -4 -11 4 5 6 7 8 9 10 11 12 G C G A T G A C G T A C G A C T G A AT CA CC CG CT GA GC GG GT 7 2 3 6 4 14 5 9 11 12 4 5 6 7 8 9 10 11 12 TA AC CG GA AC CT TG GA AC -11 4 3 1 7 2 5 11 -5 1 -2 -2 2 2 -5 -5 -1 Best Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA 13 14 15 A A T C G A TA TC TG 15 8 13 14 15 CG GA AT 10 8 8 8 5 1 2 2 16 17 18 19 Left Right A Move N T G G A -1 1 1 3 -2 2 2 5 TT -3 0 3 1 -4 1 -5 4 5 2 -6 0 6 1 -7 0 7 1 -8 1 8 4 -9 0 9 1 -10 0 10 1 16 17 18 -11 4 11 1 TG GG GA -12 0 12 1 8 12 -13 0 9 -14 0 6 15 0 16 0 17 0 (d) One of the three 2 nd best Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA
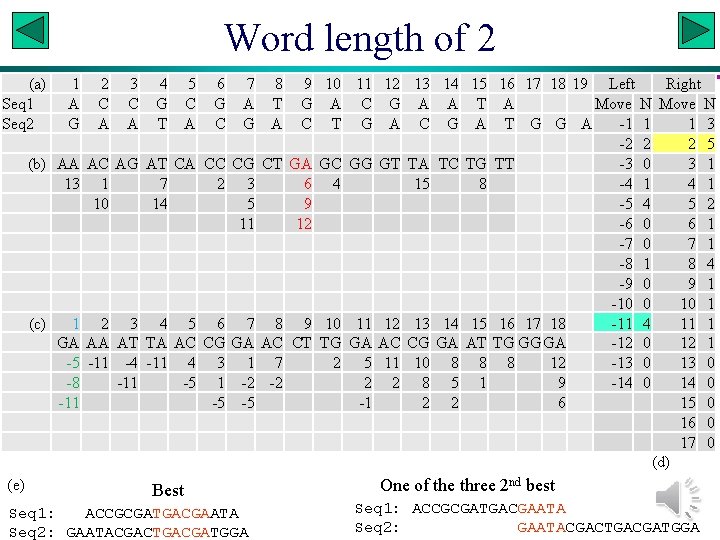
Word length of 2 (a) Seq 1 Seq 2 (b) (c) (e) 1 A G AA 13 1 GA -5 -8 -11 2 C A AC 1 10 2 AA -11 3 C A AG 3 AT -4 -11 4 5 6 7 8 9 10 11 12 G C G A T G A C G T A C G A C T G A AT CA CC CG CT GA GC GG GT 7 2 3 6 4 14 5 9 11 12 4 5 6 7 8 9 10 11 12 TA AC CG GA AC CT TG GA AC -11 4 3 1 7 2 5 11 -5 1 -2 -2 2 2 -5 -5 -1 Best Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA 13 14 15 A A T C G A TA TC TG 15 8 13 14 15 CG GA AT 10 8 8 8 5 1 2 2 16 17 18 19 Left Right A Move N T G G A -1 1 1 3 -2 2 2 5 TT -3 0 3 1 -4 1 -5 4 5 2 -6 0 6 1 -7 0 7 1 -8 1 8 4 -9 0 9 1 -10 0 10 1 16 17 18 -11 4 11 1 TG GG GA -12 0 12 1 8 12 -13 0 9 -14 0 6 15 0 16 0 17 0 (d) One of the three 2 nd best Seq 1: ACCGCGATGACGAATA Seq 2: GAATACGACTGACGATGGA

Human COX 1 RWLFSTNHKDIGTLYLLFGAWAGVLGTALSLLIRAELGQPGNLLGNDHIYNVIVTAHAF VMI FFMVMPIMIGGFGNWLVPLMIGAPDMAFPRMNNMSFWLLPPSLLLLLASAMVEAGAG TGWTV YPPLAGNYSHPGASVDLTIFSLHLAGVSSILGAINFITTIINMKPPAMTQYQTPLFVWSV LI TAVLLLLSLPVLAAGITMLLTDRNLNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPG FG MISHIVTYYSGKKEPFGYMGMVWAMMSIGFLGFIVWAHHMFTVGMDVDTRAYFTSAT MIIAI PTGVKVFSWLATLHGSNMKWSAAVLWALGFIFLFTVGGLTGIVLANSSLDIVLHDTYY Exact match: VVAH All sequences in the database are pre-indexed. Cys is the rarest in this protein in the database. If a query contains no C or more than one C, then report no exact FHYVLSMGAVFAIMGGFIHWFPLFSGYTLDQTYAKIHFTIMFIGVNLTFFPQHFLGLSG match. MPR If a query sequence contain a C, then go directly to C at site 494 to check

BLAST • Adapted from Crane & Raymer 2003 • Motivation: matching short sequences are faster than matching longer ones • Input sequence: AILVPTVIGCTVPT • Algorithm: – Break the query sequence into words: AILV, ILVP, LVPT, VPTV, PTVI, TVIG, VIGC, IGCT, GCTV, CTVP, TVPT – Discard common words (i. e. , words made entirely of common amino acids) – Search for matches against database sequences, assess significance and decide whether to discard to continue with extension using dynamic programming: AILVPTVIGCTVPT MVQGWALYDFLKCRAILVPTVIACTCVAMLALYDFLKC • Critical decision: Discard or continue? • The E-value as an answer. Slide 18
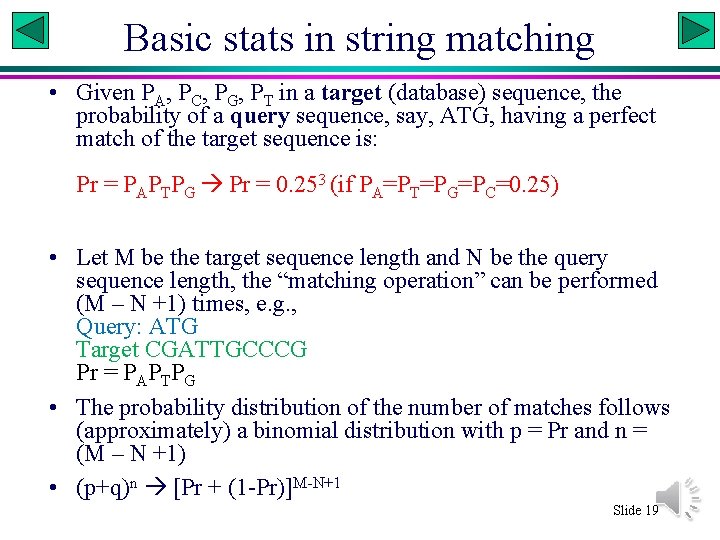
Basic stats in string matching • Given PA, PC, PG, PT in a target (database) sequence, the probability of a query sequence, say, ATG, having a perfect match of the target sequence is: Pr = PAPTPG Pr = 0. 253 (if PA=PT=PG=PC=0. 25) • Let M be the target sequence length and N be the query sequence length, the “matching operation” can be performed (M – N +1) times, e. g. , Query: ATG Target CGATTGCCCG Pr = PAPTPG • The probability distribution of the number of matches follows (approximately) a binomial distribution with p = Pr and n = (M – N +1) • (p+q)n [Pr + (1 -Pr)]M-N+1 Slide 19
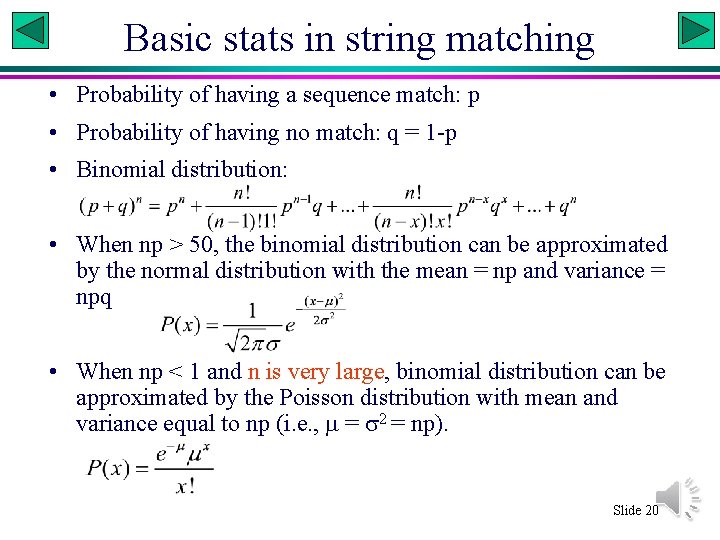
Basic stats in string matching • Probability of having a sequence match: p • Probability of having no match: q = 1 -p • Binomial distribution: • When np > 50, the binomial distribution can be approximated by the normal distribution with the mean = np and variance = npq • When np < 1 and n is very large, binomial distribution can be approximated by the Poisson distribution with mean and variance equal to np (i. e. , = 2 = np). Slide 20
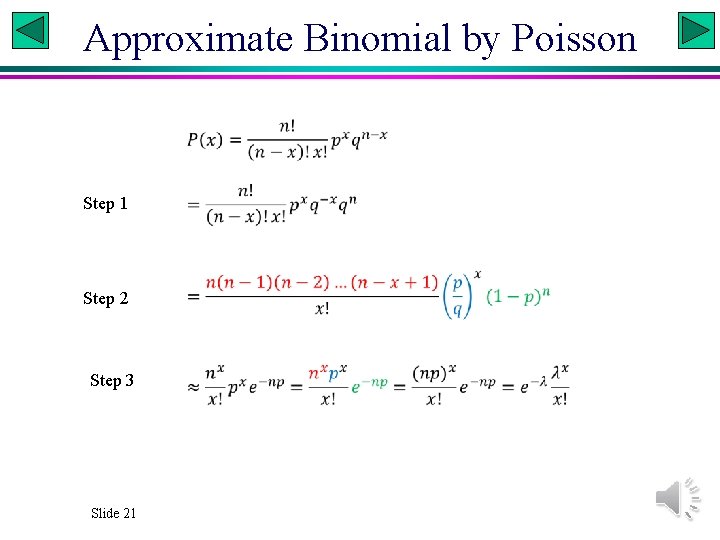
Approximate Binomial by Poisson Step 1 Step 2 Step 3 Slide 21
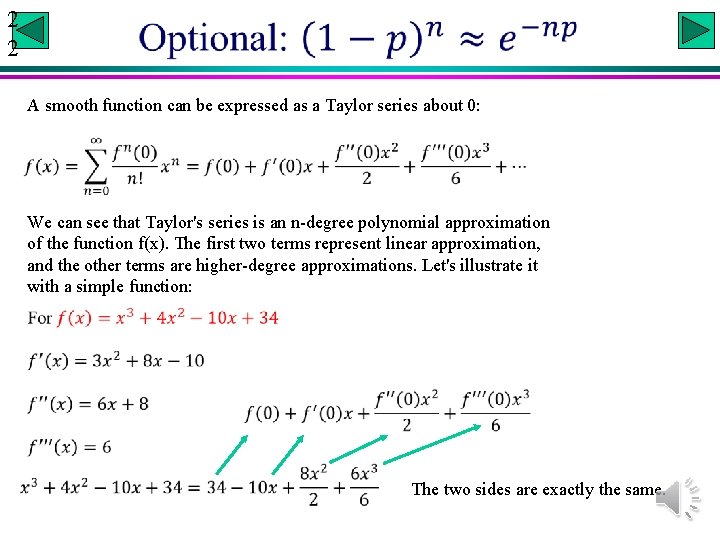
2 2 A smooth function can be expressed as a Taylor series about 0: We can see that Taylor's series is an n-degree polynomial approximation of the function f(x). The first two terms represent linear approximation, and the other terms are higher-degree approximations. Let's illustrate it with a simple function: The two sides are exactly the same.
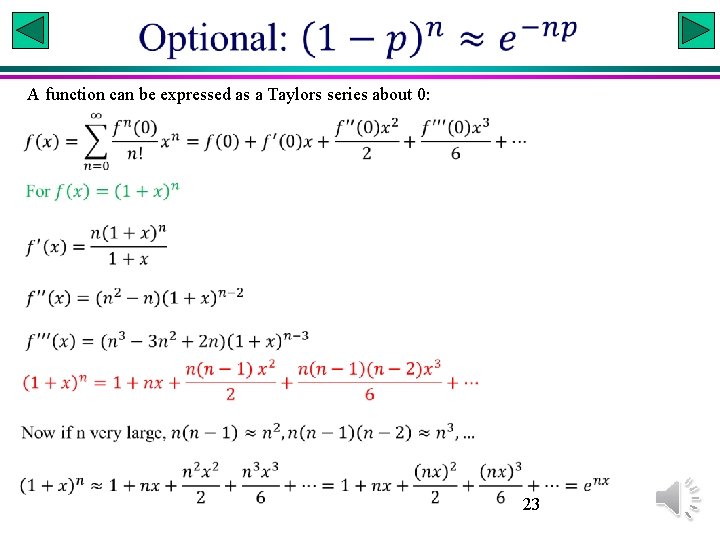
A function can be expressed as a Taylors series about 0: 23
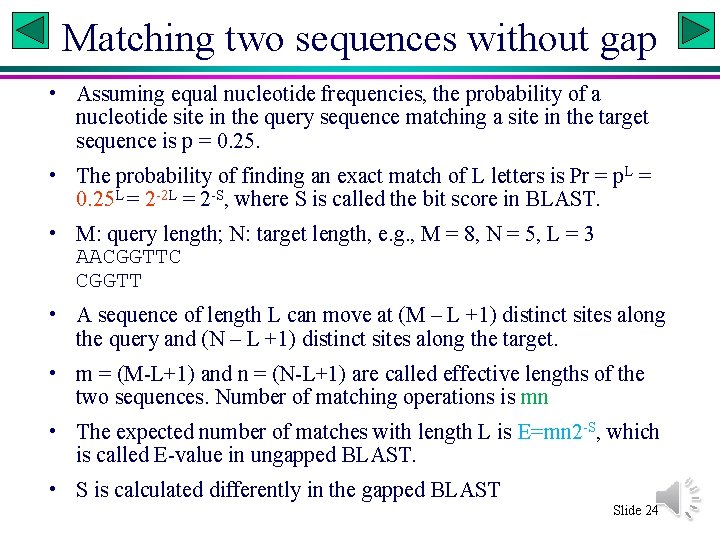
Matching two sequences without gap • Assuming equal nucleotide frequencies, the probability of a nucleotide site in the query sequence matching a site in the target sequence is p = 0. 25. • The probability of finding an exact match of L letters is Pr = p. L = 0. 25 L = 2 -2 L = 2 -S, where S is called the bit score in BLAST. • M: query length; N: target length, e. g. , M = 8, N = 5, L = 3 AACGGTTC CGGTT • A sequence of length L can move at (M – L +1) distinct sites along the query and (N – L +1) distinct sites along the target. • m = (M-L+1) and n = (N-L+1) are called effective lengths of the two sequences. Number of matching operations is mn • The expected number of matches with length L is E=mn 2 -S, which is called E-value in ungapped BLAST. • S is calculated differently in the gapped BLAST Slide 24
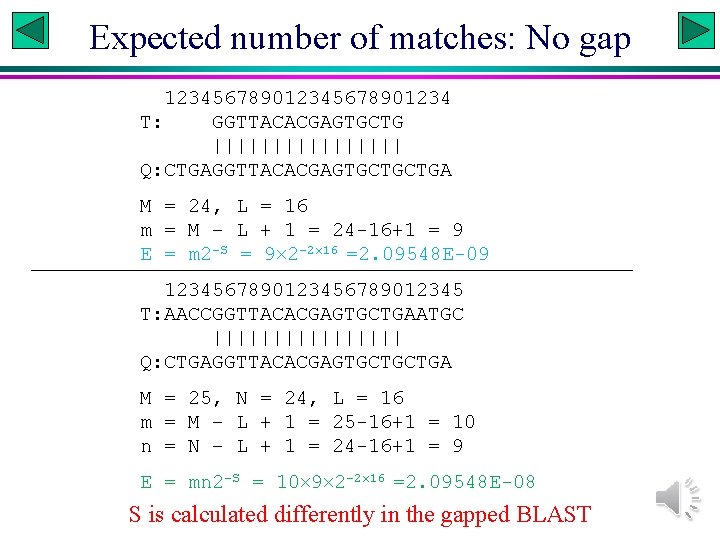
Expected number of matches: No gap 12345678901234 T: GGTTACACGAGTGCTG |||||||| Q: CTGAGGTTACACGAGTGCTGCTGA M = 24, L = 16 m = M – L + 1 = 24 -16+1 = 9 E = m 2 -S = 9 2 -2 16 =2. 09548 E-09 123456789012345 T: AACCGGTTACACGAGTGCTGAATGC |||||||| Q: CTGAGGTTACACGAGTGCTGCTGA M = 25, N = 24, L = 16 m = M – L + 1 = 25 -16+1 = 10 n = N – L + 1 = 24 -16+1 = 9 E = mn 2 -S = 10 9 2 -2 16 =2. 09548 E-08 S is calculated differently in the gapped BLAST
![Blast Output (Nuc. Seq. ) BLASTN 2. 2. 4 [Aug-26 -2002]. . . Query= Blast Output (Nuc. Seq. ) BLASTN 2. 2. 4 [Aug-26 -2002]. . . Query=](http://slidetodoc.com/presentation_image_h/31fc1266f1983e092666a898ce31944c/image-26.jpg)
Blast Output (Nuc. Seq. ) BLASTN 2. 2. 4 [Aug-26 -2002]. . . Query= Seq 1 38 Database: Mg. CDS 480 sequences; 526, 317 total letters Sequences producing significant alignments: MG 001 1095 bases Score = 34. 2 bits (17), Expect = 7 e-004 Identities = 35/40 (87%), Gaps = 2/40 (5%) Query: 1 Sbjct: 1 Constant gap penalty vs affine function penalty Score E (bits) Value 34 7 e-004 atgaataacg--attatttccaacgacaaaaccac 38 ||||||||||| atgaataacgttattatttccaataacaaaataaaaccac 40 Lambda K H 1. 37 0. 711 1. 31 Matrix: blastn matrix: 1 -3 Gap Penalties: Existence: 5, Extension: 2 … effective length of query: 26 effective length of database: 520, 557 Typically one would count only 1 GE here. Matches: 35*1 = 35 Mismatches: 3*(-3) = -9 Gap Open: 1*5 = 5 Gap extension: 2*2 =4 R = 35 - 9 - 5 - 4 = 17 S = [λR – ln(K)]/ln(2) =[1. 37*17 -ln(0. 711)]/ln(2) = 34 E = mn 2 -S = 26 * 520557 * 2 -34 = 7. 878 E-04 x p(x) 0 0. 999265217 1 0. 000734513 … Alternatively, E = Kmn. Exp(-lambda*R)

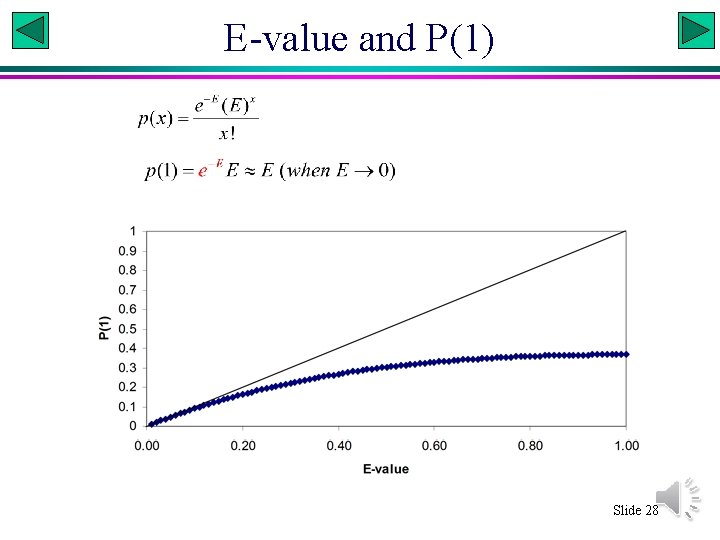
E-Value in BLAST • The e-value is the expected number of random matches that is equally good or better than the reported match. It can be a number near zero or much larger than 1. • It is NOT the probability of finding the reported match. • Only when the e-value is extremely small can it be interpreted as the probability of finding 1 match that is as good as the reported one (see next slide). Slide 27
E-value and P(1) Slide 28
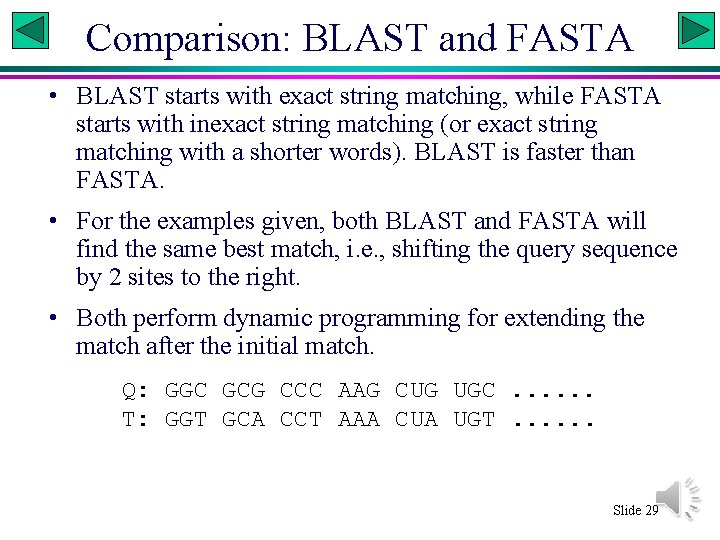
Comparison: BLAST and FASTA • BLAST starts with exact string matching, while FASTA starts with inexact string matching (or exact string matching with a shorter words). BLAST is faster than FASTA. • For the examples given, both BLAST and FASTA will find the same best match, i. e. , shifting the query sequence by 2 sites to the right. • Both perform dynamic programming for extending the match after the initial match. Q: GGC GCG CCC AAG CUG UGC. . . T: GGT GCA CCT AAA CUA UGT. . . Slide 29
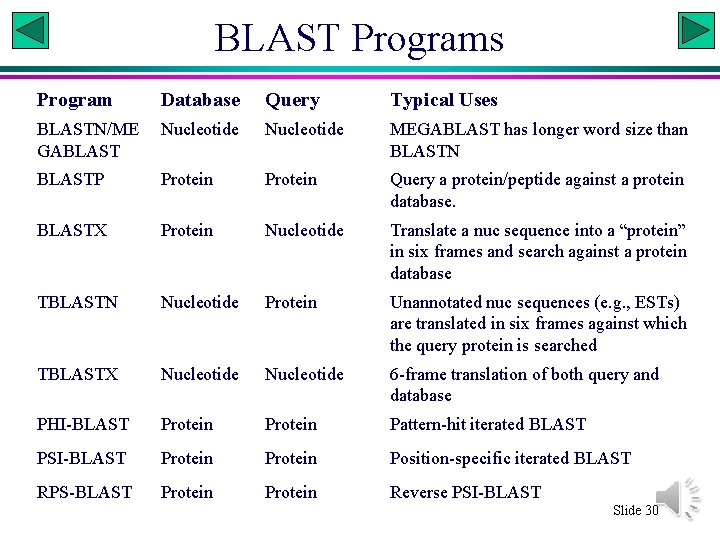
BLAST Programs Program Database Query Typical Uses BLASTN/ME GABLAST Nucleotide MEGABLAST has longer word size than BLASTN BLASTP Protein Query a protein/peptide against a protein database. BLASTX Protein Nucleotide Translate a nuc sequence into a “protein” in six frames and search against a protein database TBLASTN Nucleotide Protein Unannotated nuc sequences (e. g. , ESTs) are translated in six frames against which the query protein is searched TBLASTX Nucleotide 6 -frame translation of both query and database PHI-BLAST Protein Pattern-hit iterated BLAST PSI-BLAST Protein Position-specific iterated BLAST RPS-BLAST Protein Reverse PSI-BLAST Slide 30

Local BLAST • Most biopharmaceutical companies have their own BLAST database and would do local BLAST • Steps for local BLAST: – Create a file in FASTA format – Create a BLAST database – BLAST query sequences against the locally created BLAST database instead of against remote databases such as Gen. Bank hosted in NCBI. • Advantage of local BLAST – Fast – Keep local BLAST database private • You will learn this in the lab. Slide 31
- Slides: 31