Protein Targeting and Degradation David Shiuan Department of





- Slides: 41

Protein Targeting and Degradation David Shiuan Department of Life Science Institute of Biotechnology Interdisciplinary Program of Bioinformatics National Dong Hwa University 1

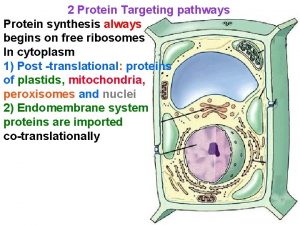
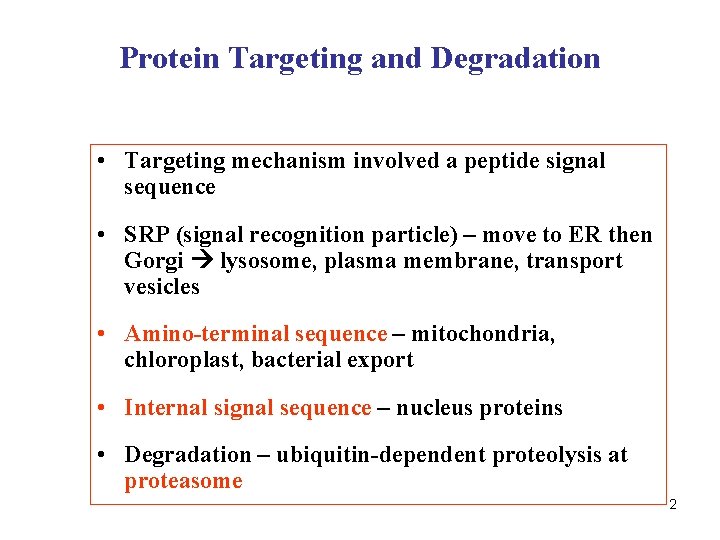
Protein Targeting and Degradation • Targeting mechanism involved a peptide signal sequence • SRP (signal recognition particle) – move to ER then Gorgi lysosome, plasma membrane, transport vesicles • Amino-terminal sequence – mitochondria, chloroplast, bacterial export • Internal signal sequence – nucleus proteins • Degradation – ubiquitin-dependent proteolysis at proteasome 2

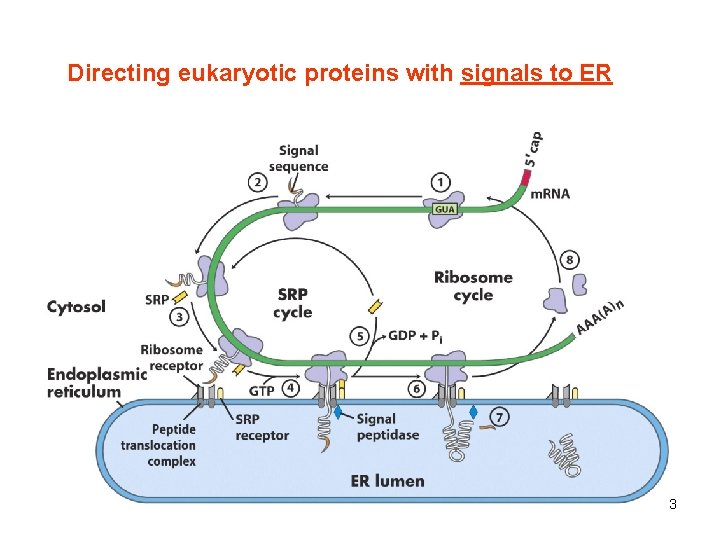
Directing eukaryotic proteins with signals to ER 3

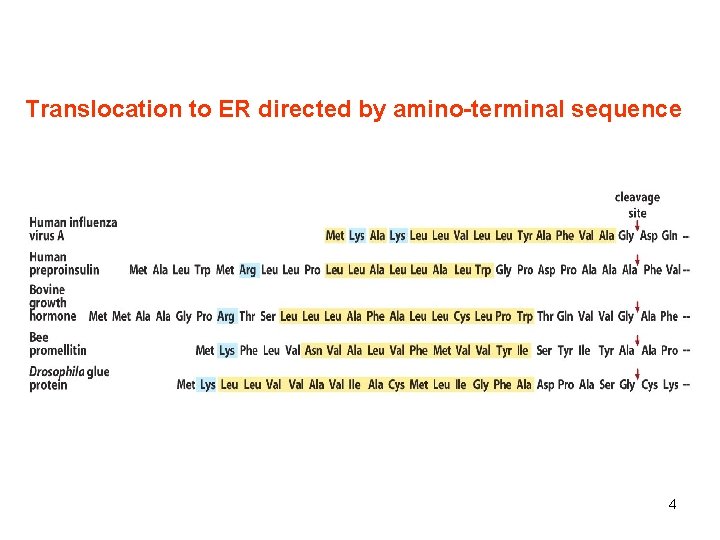
Translocation to ER directed by amino-terminal sequence 4

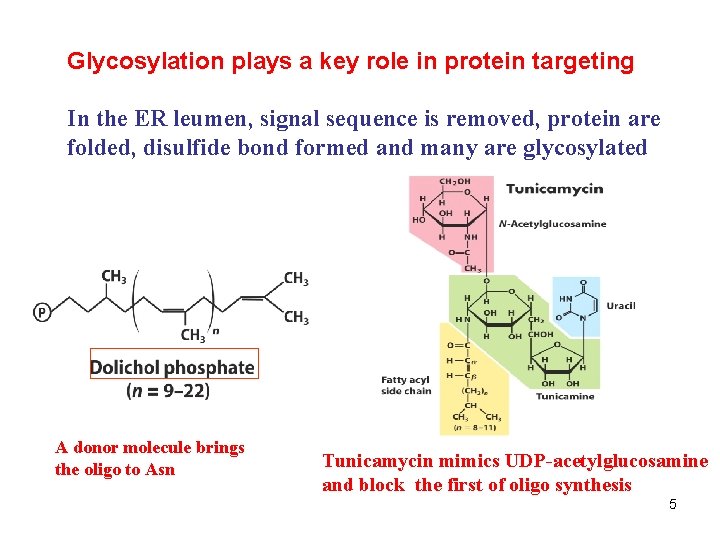
Glycosylation plays a key role in protein targeting In the ER leumen, signal sequence is removed, protein are folded, disulfide bond formed and many are glycosylated A donor molecule brings the oligo to Asn Tunicamycin mimics UDP-acetylglucosamine and block the first of oligo synthesis 5

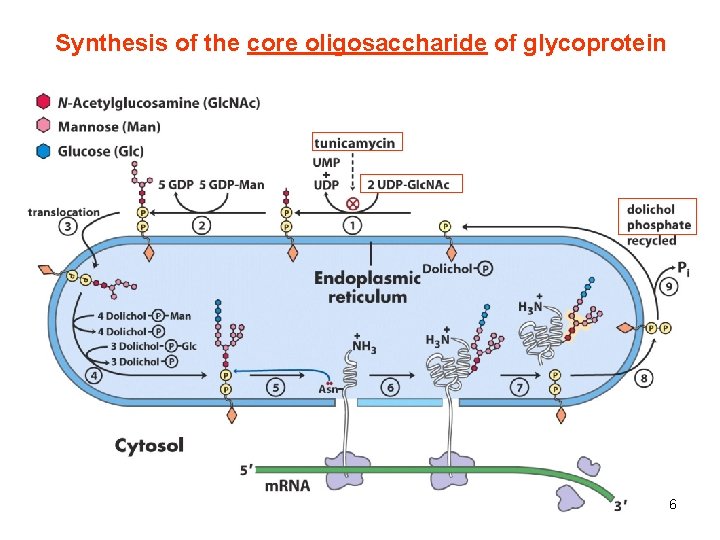
Synthesis of the core oligosaccharide of glycoprotein 6

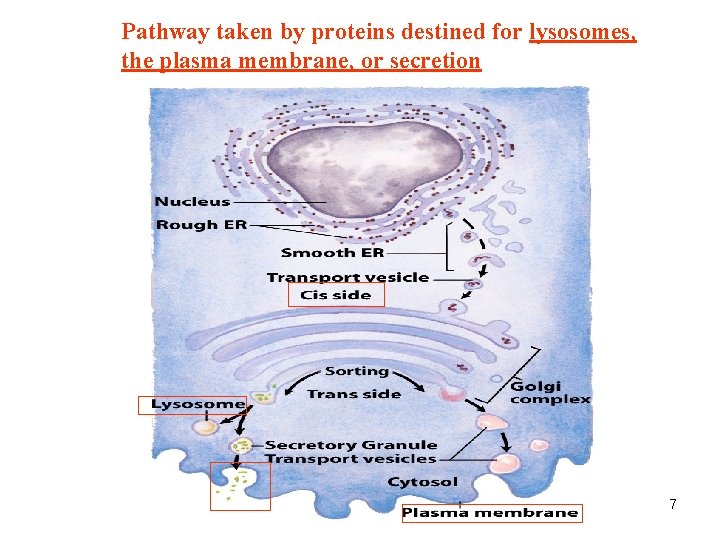
Pathway taken by proteins destined for lysosomes, the plasma membrane, or secretion 7

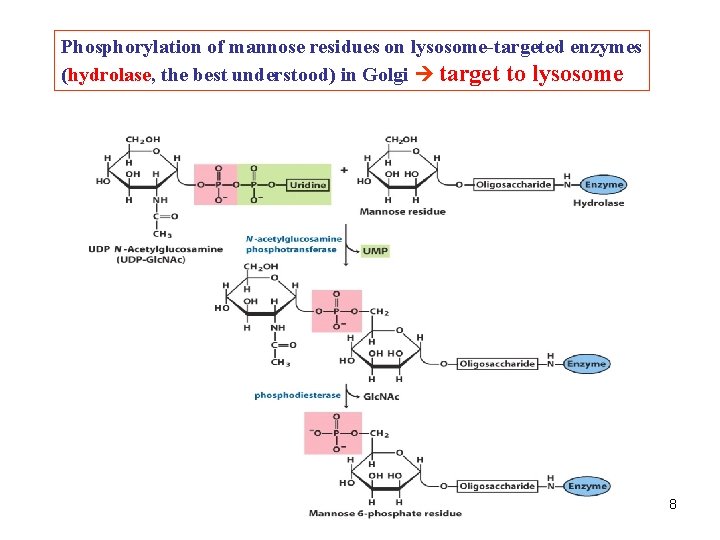
Phosphorylation of mannose residues on lysosome-targeted enzymes (hydrolase, the best understood) in Golgi target to lysosome 8

Targeting of nucleus proteins Ribosomal proteins are imported into nucleus and assembled into 60 s and 40 s. The complete subunits are transported back to cytosol. Nuclear proteins synthesized in cytosol and imported via importin into nucleus. NLS - nucleus localization signal 9

Sanning EM of nucleus surface, showing numerous nuclear pores 10

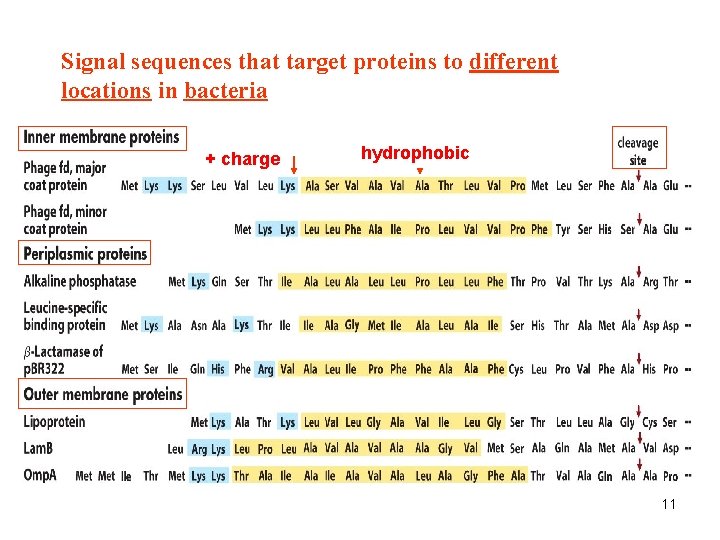
Signal sequences that target proteins to different locations in bacteria + charge hydrophobic 11

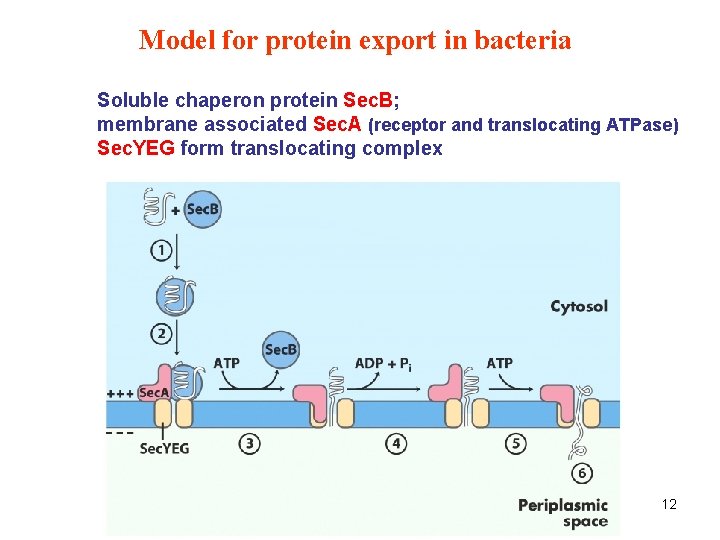
Model for protein export in bacteria Soluble chaperon protein Sec. B; membrane associated Sec. A (receptor and translocating ATPase) Sec. YEG form translocating complex 12

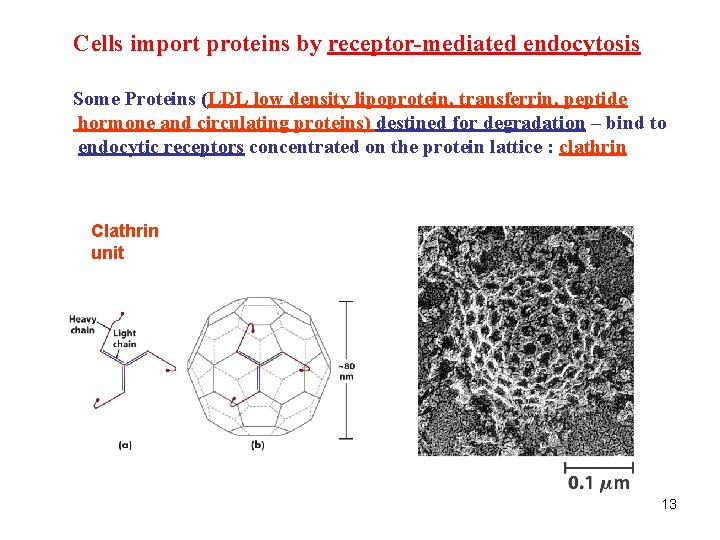
Cells import proteins by receptor-mediated endocytosis Some Proteins (LDL low density lipoprotein, transferrin, peptide hormone and circulating proteins) destined for degradation – bind to endocytic receptors concentrated on the protein lattice : clathrin Clathrin unit 13

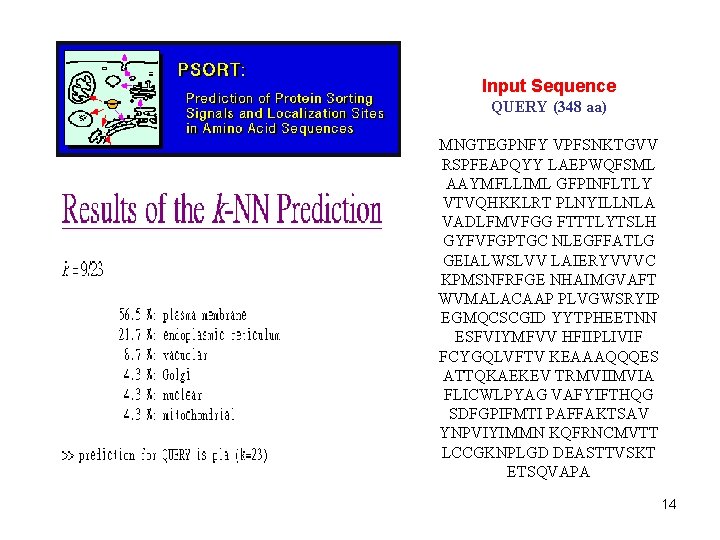
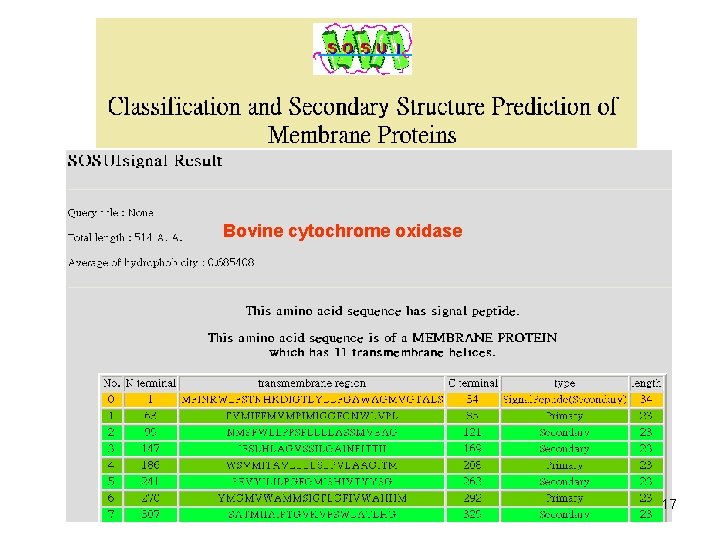
Input Sequence QUERY (348 aa) MNGTEGPNFY VPFSNKTGVV RSPFEAPQYY LAEPWQFSML AAYMFLLIML GFPINFLTLY VTVQHKKLRT PLNYILLNLA VADLFMVFGG FTTTLYTSLH GYFVFGPTGC NLEGFFATLG GEIALWSLVV LAIERYVVVC KPMSNFRFGE NHAIMGVAFT WVMALACAAP PLVGWSRYIP EGMQCSCGID YYTPHEETNN ESFVIYMFVV HFIIPLIVIF FCYGQLVFTV KEAAAQQQES ATTQKAEKEV TRMVIIMVIA FLICWLPYAG VAFYIFTHQG SDFGPIFMTI PAFFAKTSAV YNPVIYIMMN KQFRNCMVTT LCCGKNPLGD DEASTTVSKT ETSQVAPA 14

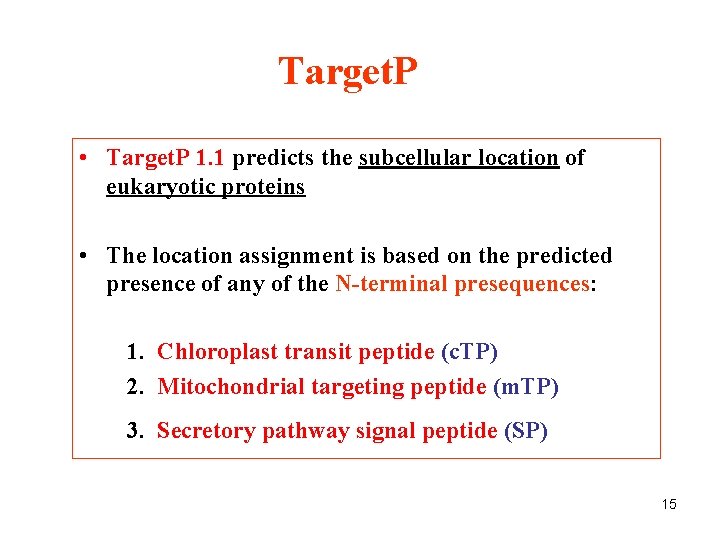
Target. P • Target. P 1. 1 predicts the subcellular location of eukaryotic proteins • The location assignment is based on the predicted presence of any of the N-terminal presequences: 1. Chloroplast transit peptide (c. TP) 2. Mitochondrial targeting peptide (m. TP) 3. Secretory pathway signal peptide (SP) 15

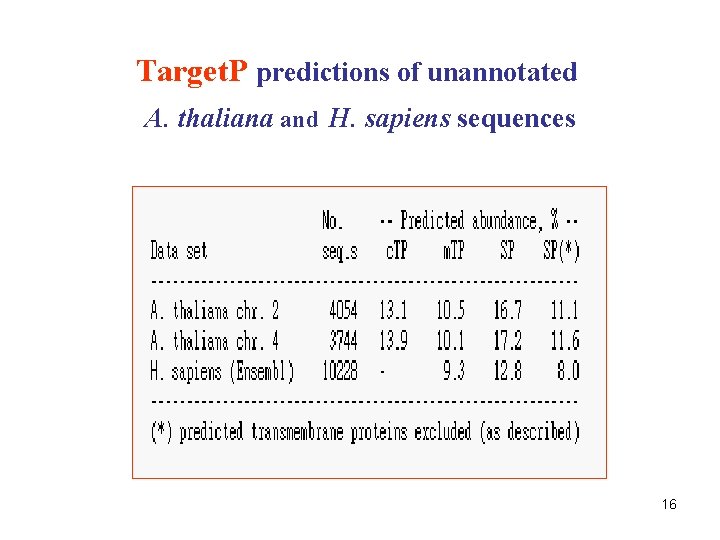
Target. P predictions of unannotated A. thaliana and H. sapiens sequences 16

Bovine cytochrome oxidase 17

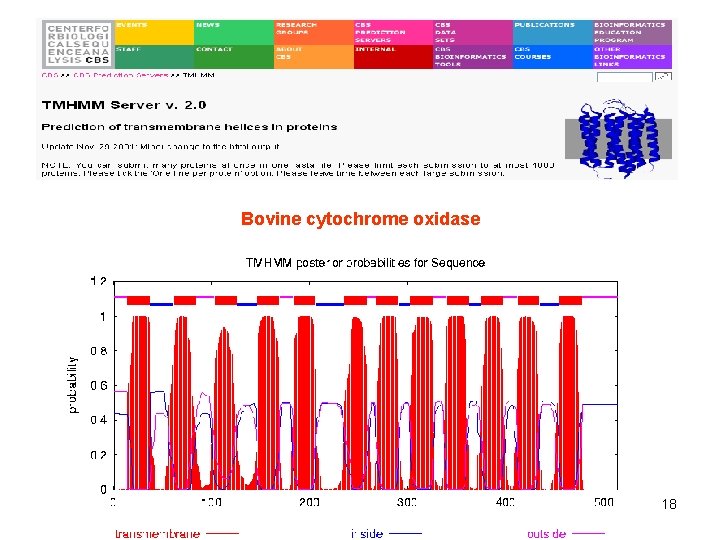
Bovine cytochrome oxidase 18

19

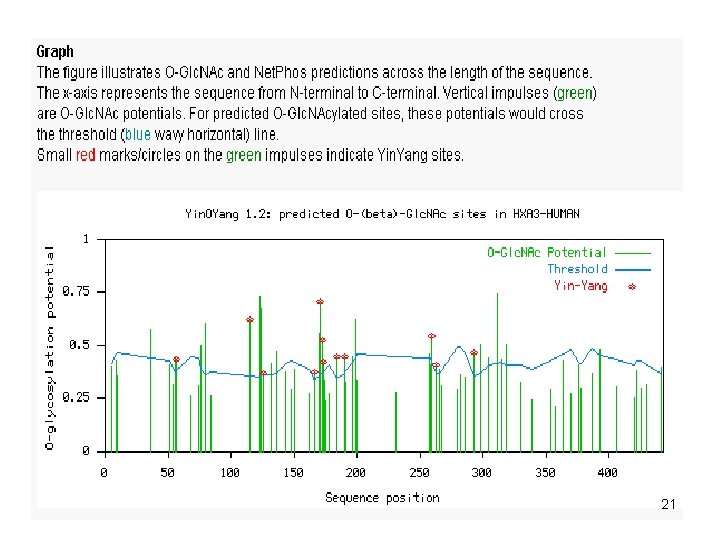
20

21

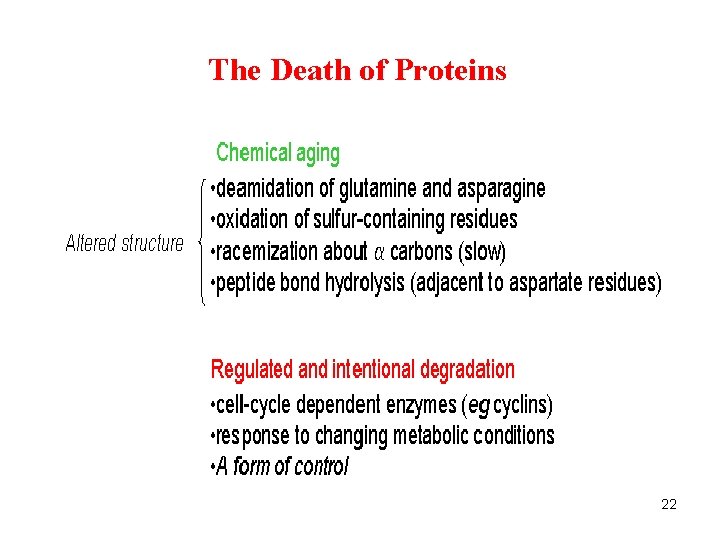
The Death of Proteins 22

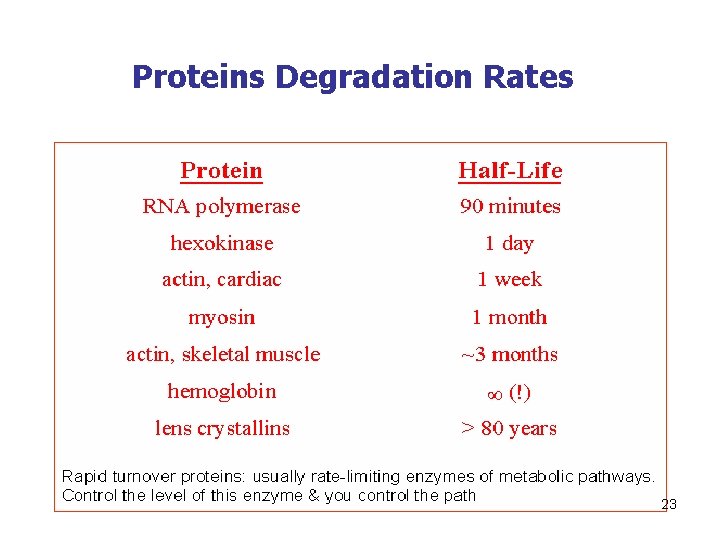
Proteins Degradation Rates 23

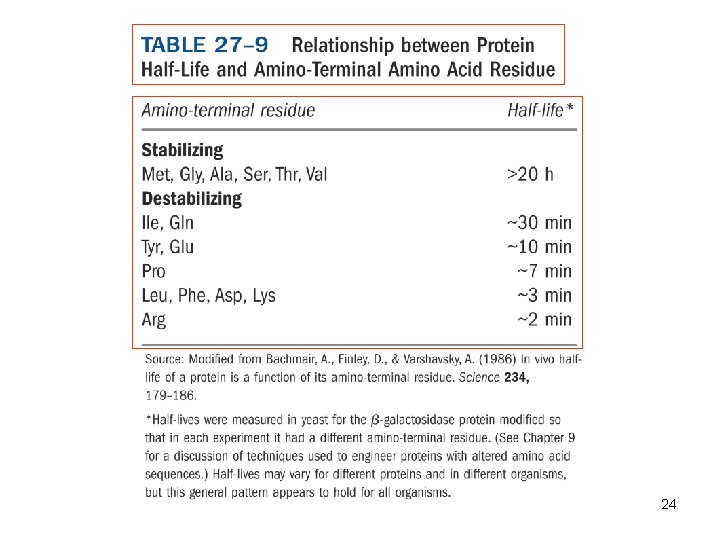
24

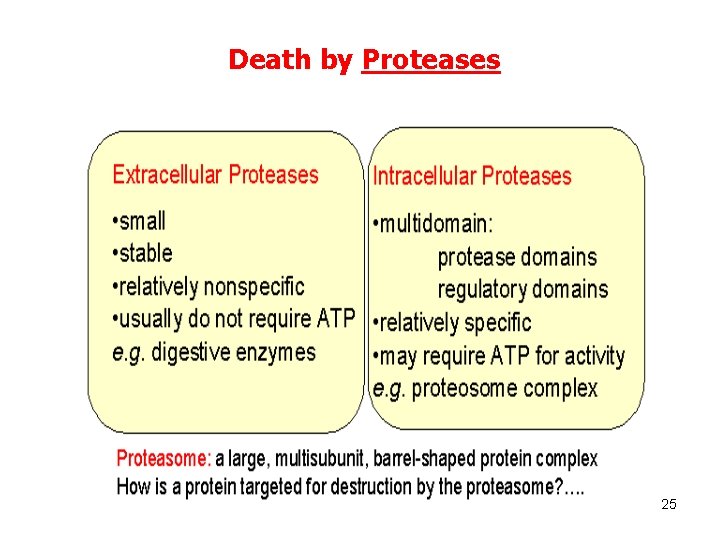
Death by Proteases 25

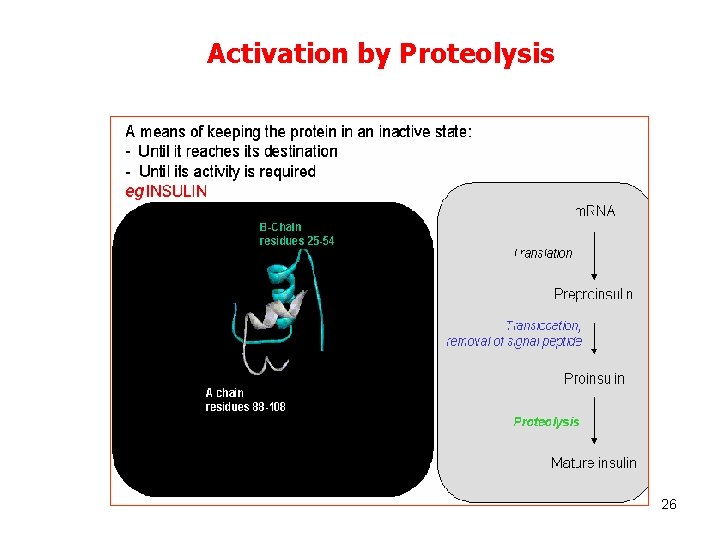
Activation by Proteolysis 26

Ubiquitin 2004 Nobel Prize in Chemistry • Cells are continually building proteins, using them for a single task, and then discarding them. • Signaling or controlling proteins (eg. transcription regulators and the cyclins) - lead very brief lives, carrying their messages and then being thrown away. • Specialized enzymes - built just when they are needed, allowing cells to keep up with their minuteby-minute synthetic needs. • The approach may seem wasteful, but it allows each cell to respond quickly to constantly changing requirements. 27

Ubiquitin 2004 Nobel Prize in Chemistry • The small protein ubiquitin plays a central role. Ubiquitin attached to obsolete proteins to dissemble. • Ubiquitin is found in all eukaryotic cells and in cells throughout your body. • The Nobel Prize in Chemistry 2004 was awarded to the Aaron Ciechanover, Avram Hershko , Irwin Rose who discovered its essential function in 1980. 28

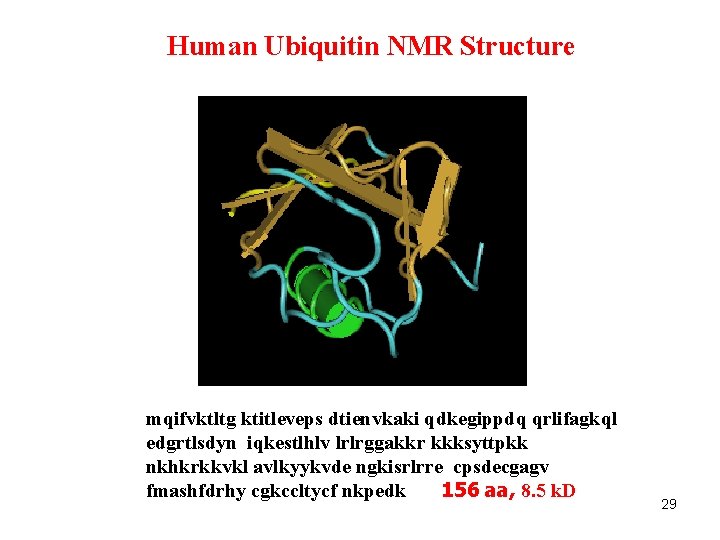
Human Ubiquitin NMR Structure mqifvktltg ktitleveps dtienvkaki qdkegippdq qrlifagkql edgrtlsdyn iqkestlhlv lrlrggakkr kkksyttpkk nkhkrkkvkl avlkyykvde ngkisrlrre cpsdecgagv fmashfdrhy cgkccltycf nkpedk 156 aa, 8. 5 k. D 29

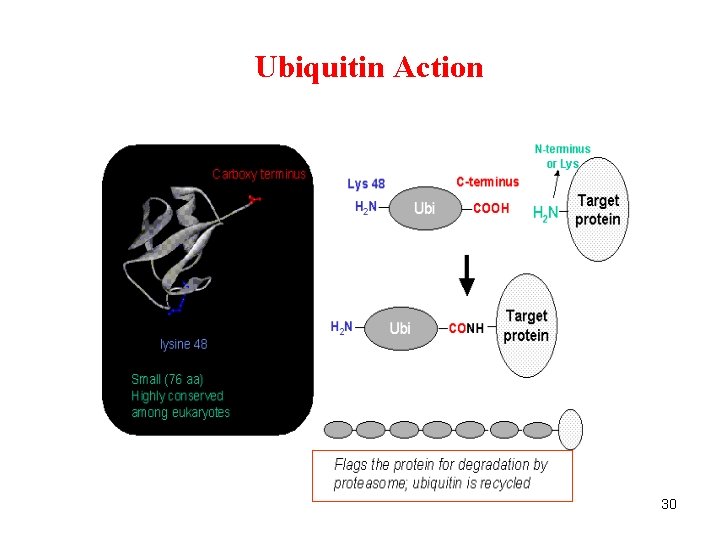
Ubiquitin Action 30

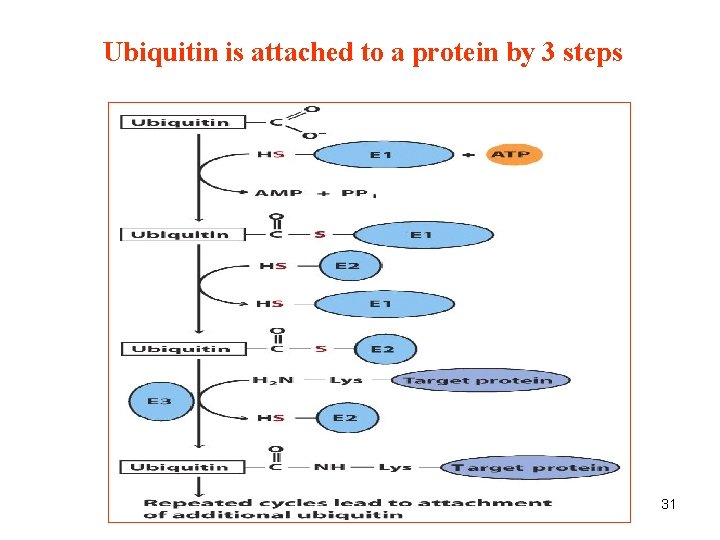
Ubiquitin is attached to a protein by 3 steps 31

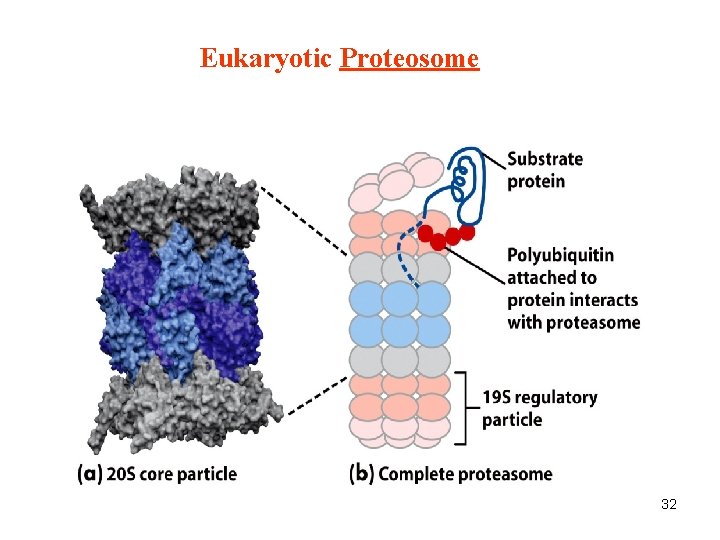
Eukaryotic Proteosome 32

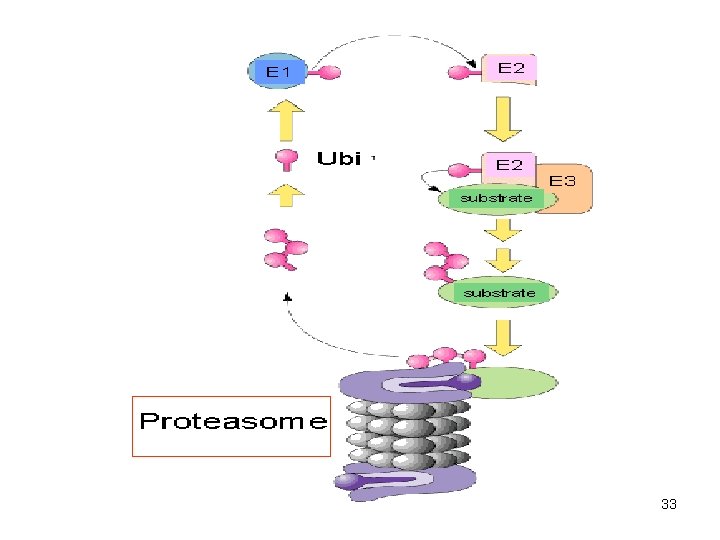
33

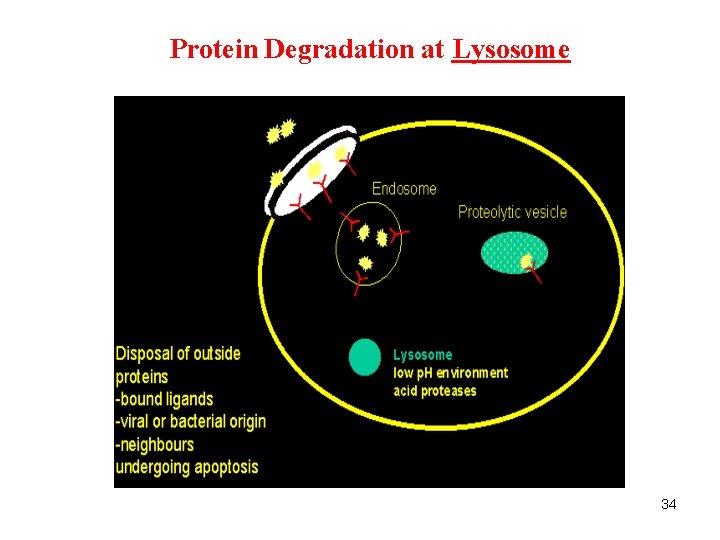
Protein Degradation at Lysosome 34

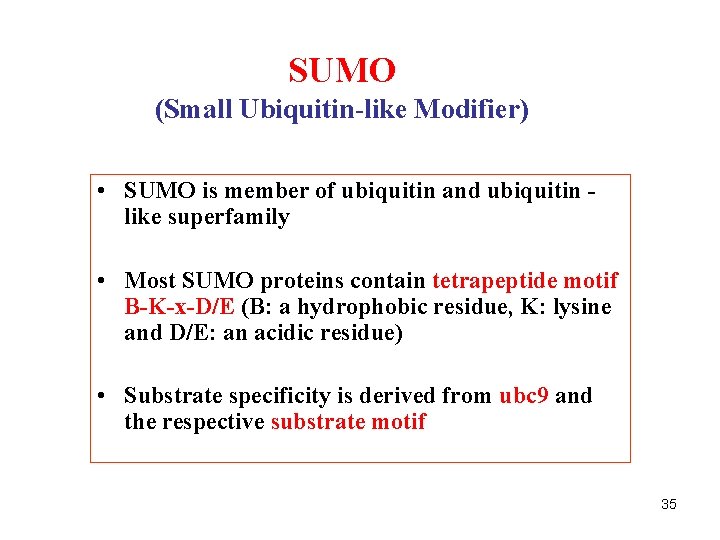
SUMO (Small Ubiquitin-like Modifier) • SUMO is member of ubiquitin and ubiquitin like superfamily • Most SUMO proteins contain tetrapeptide motif B-K-x-D/E (B: a hydrophobic residue, K: lysine and D/E: an acidic residue) • Substrate specificity is derived from ubc 9 and the respective substrate motif 35

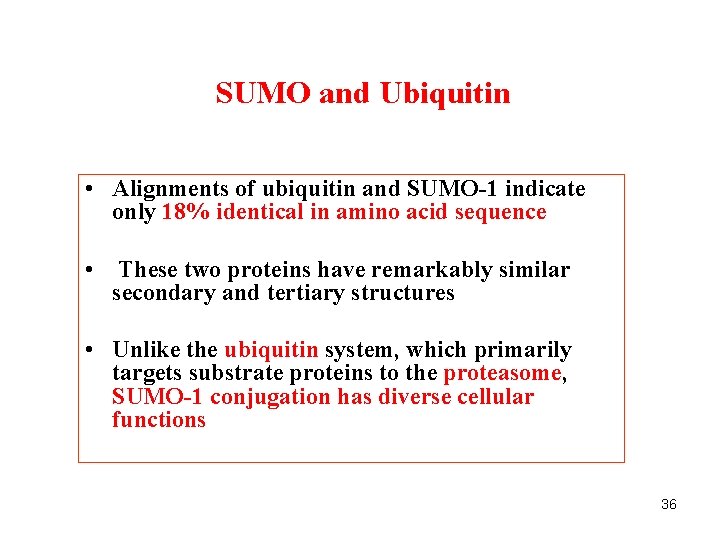
SUMO and Ubiquitin • Alignments of ubiquitin and SUMO-1 indicate only 18% identical in amino acid sequence • These two proteins have remarkably similar secondary and tertiary structures • Unlike the ubiquitin system, which primarily targets substrate proteins to the proteasome, SUMO-1 conjugation has diverse cellular functions 36

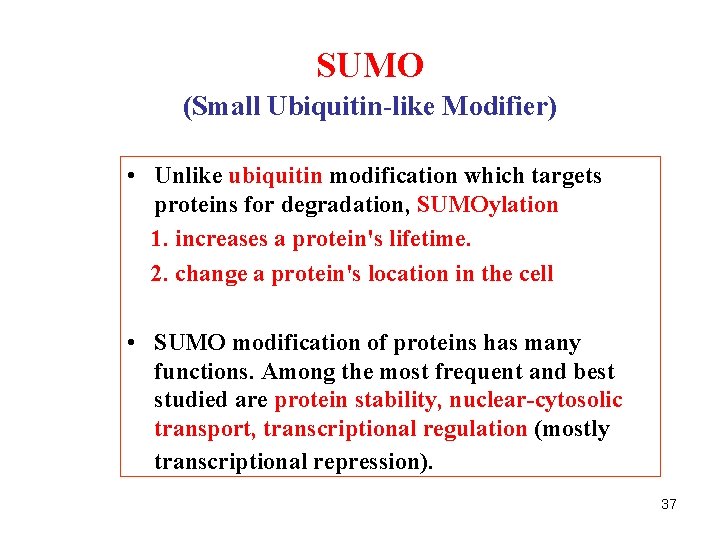
SUMO (Small Ubiquitin-like Modifier) • Unlike ubiquitin modification which targets proteins for degradation, SUMOylation 1. increases a protein's lifetime. 2. change a protein's location in the cell • SUMO modification of proteins has many functions. Among the most frequent and best studied are protein stability, nuclear-cytosolic transport, transcriptional regulation (mostly transcriptional repression). 37

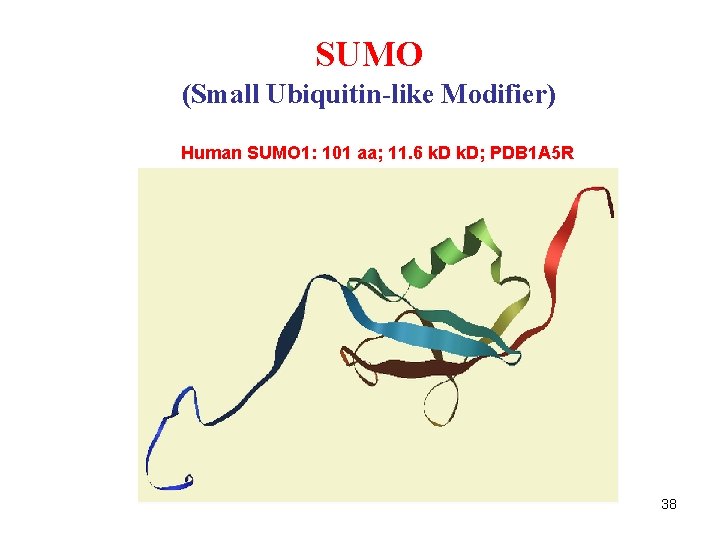
SUMO (Small Ubiquitin-like Modifier) Human SUMO 1: 101 aa; 11. 6 k. D; PDB 1 A 5 R 38

李 水 龍 (Steven Shoei-Lung Li) 高雄醫學大學醫學研究所 講座教授 Daxx is a shuttle protein participating in biological functions on various subcellular localizations. Human Fas death domainassociated protein (Daxx) is a 740 -amino acids protein mainly localized in nucleus. It functioned as a transcriptional repressor when associated with chromatin in nucleus. 39

40

41
 Protein targeting pathways
Protein targeting pathways Carrier vs channel proteins
Carrier vs channel proteins Protein-protein docking
Protein-protein docking Estimation of degradation function
Estimation of degradation function Edman degradation steps
Edman degradation steps Prdp biochemistry
Prdp biochemistry Mechanical degradation
Mechanical degradation Degradation of ketone bodies
Degradation of ketone bodies Importance of environmental degradation
Importance of environmental degradation Noise
Noise Light induced degradation
Light induced degradation Edman degradation
Edman degradation Salting out proteins
Salting out proteins Conclusion of environmental degradation
Conclusion of environmental degradation How environmental degradation occurs
How environmental degradation occurs Land degradation definition
Land degradation definition Sucrose synthesis
Sucrose synthesis Potential induced degradation
Potential induced degradation Abnormal degradation of disaccharides
Abnormal degradation of disaccharides Edman degradation
Edman degradation Tag degradation
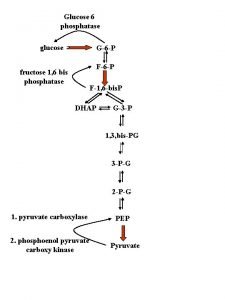
Tag degradation Glucose 6 phosphatase
Glucose 6 phosphatase Linear position invariant degradation
Linear position invariant degradation Market segmentation and targeting ppt
Market segmentation and targeting ppt Avon market segmentation
Avon market segmentation Requirements for effective segmentation
Requirements for effective segmentation Targeting
Targeting Sony market segmentation, targeting and positioning
Sony market segmentation, targeting and positioning Segmentation targeting differentiation and positioning
Segmentation targeting differentiation and positioning Https://www.census.gov/popclock/
Https://www.census.gov/popclock/ Hcp segment
Hcp segment Case study segmentation targeting positioning
Case study segmentation targeting positioning Nivea segmentation
Nivea segmentation Chapter 7 segmentation targeting and positioning
Chapter 7 segmentation targeting and positioning Geographic segmentation examples pakistan
Geographic segmentation examples pakistan Principles of market targeting
Principles of market targeting Market segmentation, targeting and positioning
Market segmentation, targeting and positioning Rural market segmentation
Rural market segmentation Segmentation targeting and positioning of colgate
Segmentation targeting and positioning of colgate Benefits of market segmentation targeting and positioning
Benefits of market segmentation targeting and positioning Segmentation
Segmentation Concentrated marketing
Concentrated marketing