Primer Selection Methods for Detection of Genomic Inversions

![Primer Approximation Multiplex PCR (PAMP) p p p Introduced by [Liu&Carson 2007] Experimental technique Primer Approximation Multiplex PCR (PAMP) p p p Introduced by [Liu&Carson 2007] Experimental technique](https://slidetodoc.com/presentation_image_h/132019e01e0be8694674d15986ee6596/image-4.jpg)



- Slides: 31

Primer Selection Methods for Detection of Genomic Inversions and Deletions via PAMP Bhaskar Das. Gupta, University of Illinois at Chicago Jin Jun, and Ion Mandoiu University of Connecticut

Outline p Introduction p Anchored Deletion Detection p Inversion Detection p Conclusions

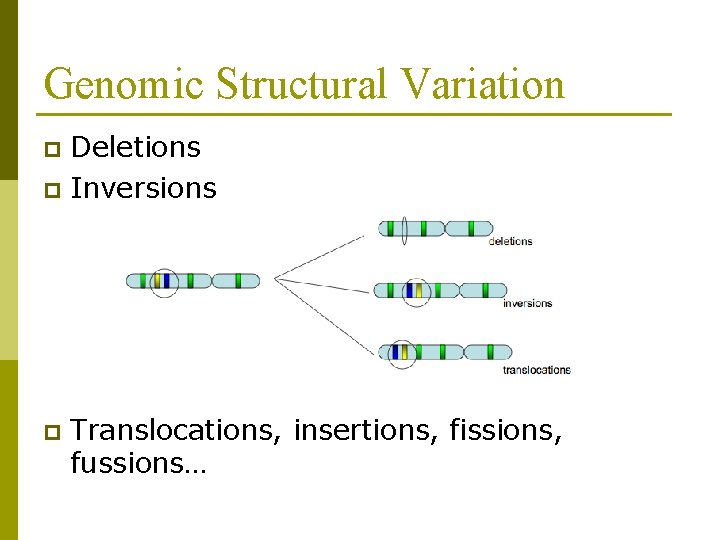
Genomic Structural Variation Deletions p Inversions p p Translocations, insertions, fissions, fussions…
![Primer Approximation Multiplex PCR PAMP p p p Introduced by LiuCarson 2007 Experimental technique Primer Approximation Multiplex PCR (PAMP) p p p Introduced by [Liu&Carson 2007] Experimental technique](https://slidetodoc.com/presentation_image_h/132019e01e0be8694674d15986ee6596/image-4.jpg)
Primer Approximation Multiplex PCR (PAMP) p p p Introduced by [Liu&Carson 2007] Experimental technique for detecting large-scale cancer genome lesions such as inversions and deletions from heterogeneous samples containing a mixture of cancer and normal cells Can be used for n n Tracking how genetic breakpoints are generated during cancer development Monitoring the status of cancer progression with a highly sensitive assays

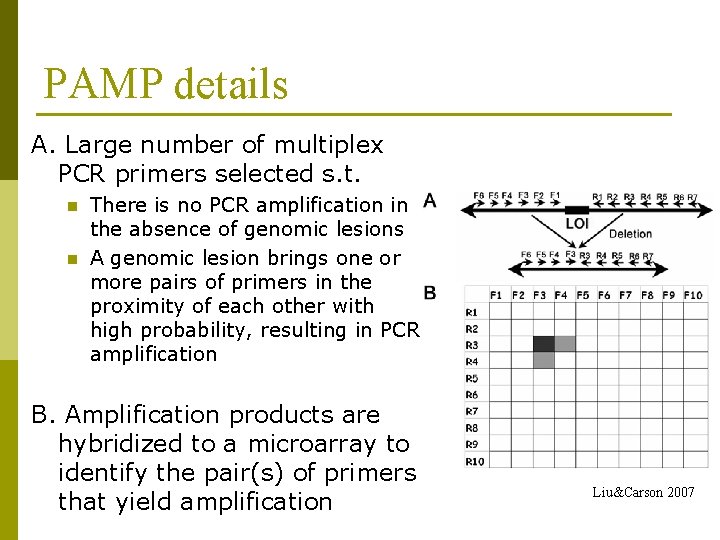
PAMP details A. Large number of multiplex PCR primers selected s. t. n n There is no PCR amplification in the absence of genomic lesions A genomic lesion brings one or more pairs of primers in the proximity of each other with high probability, resulting in PCR amplification B. Amplification products are hybridized to a microarray to identify the pair(s) of primers that yield amplification Liu&Carson 2007

Outline p Introduction p Anchored Deletion Detection p Inversion Detection p Conclusions

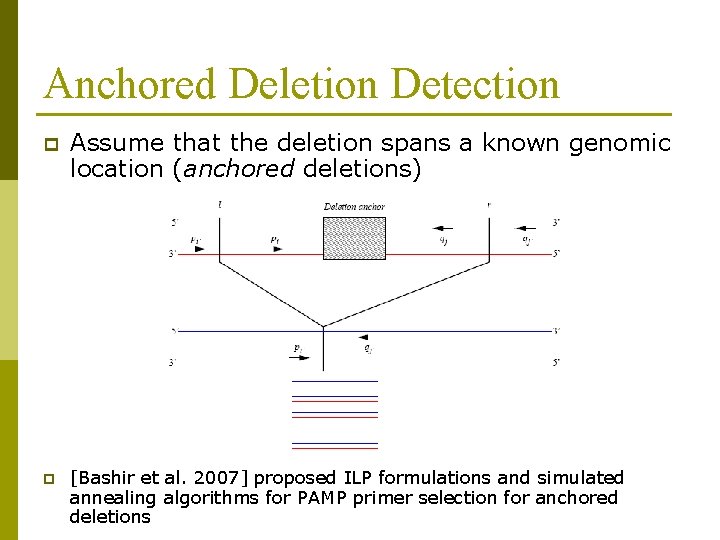
Anchored Deletion Detection p p Assume that the deletion spans a known genomic location (anchored deletions) [Bashir et al. 2007] proposed ILP formulations and simulated annealing algorithms for PAMP primer selection for anchored deletions

Criteria for Primer Selection p Standard criteria for multiplex PCR primer selection n Melting temperature, Tm Lack of hairpin secondary structure, and No dimerization between pairs of primers p Single pair of dimerizing primers is sufficient to negate the amplification [Bashir et al. 2007]

Optimization Objective p Multiplex PCR primer set selection n p Minimize number of primers and/or multiplex PCR reactions needed to amplify a given set of discrete amplification targets PAMP primer set selection n Minimize the probability that an unknown genomic lesion fails to be detected by the assay

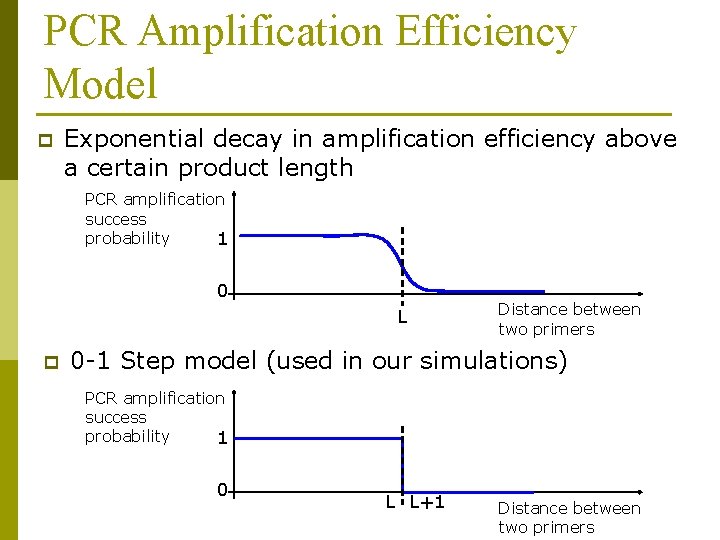
PCR Amplification Efficiency Model p Exponential decay in amplification efficiency above a certain product length PCR amplification success probability 1 0 L p Distance between two primers 0 -1 Step model (used in our simulations) PCR amplification success probability 1 0 L L+1 Distance between two primers

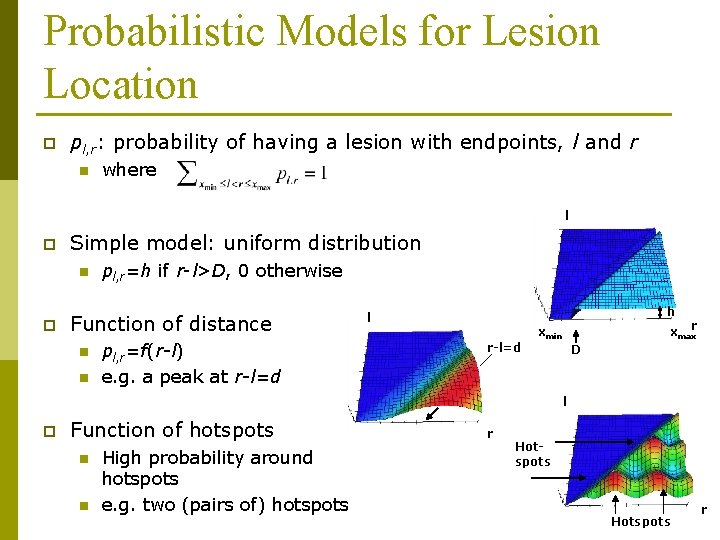
Probabilistic Models for Lesion Location p pl, r: probability of having a lesion with endpoints, l and r n where l p Simple model: uniform distribution n p pl, r=h if r-l>D, 0 otherwise Function of distance n n pl, r=f(r-l) e. g. a peak at r-l=d h l r-l=d xmaxr xmin D l p Function of hotspots n n High probability around hotspots e. g. two (pairs of) hotspots r Hotspots r

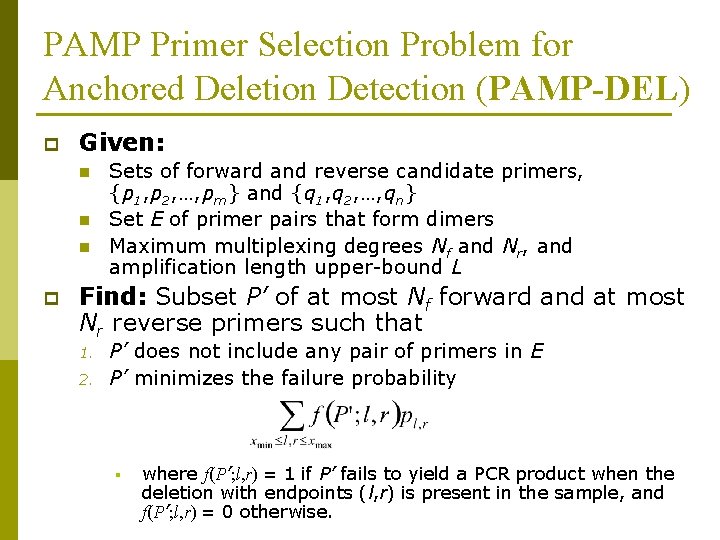
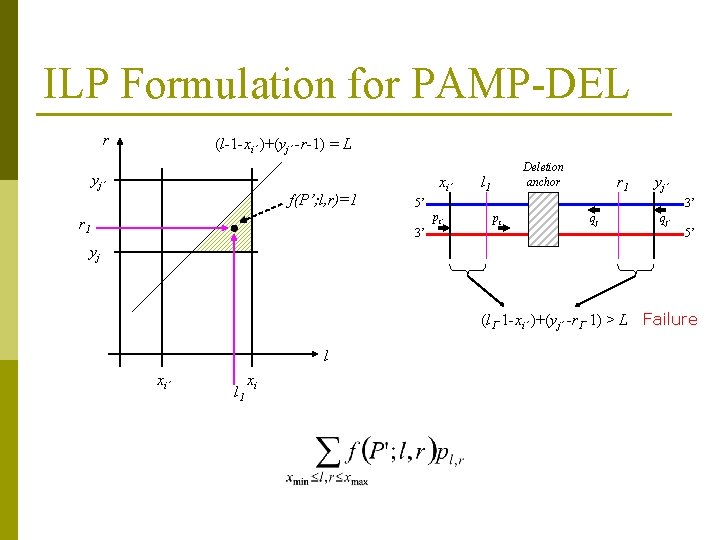
PAMP Primer Selection Problem for Anchored Deletion Detection (PAMP-DEL) p Given: n n n p Sets of forward and reverse candidate primers, {p 1, p 2, …, pm} and {q 1, q 2, …, qn} Set E of primer pairs that form dimers Maximum multiplexing degrees Nf and Nr, and amplification length upper-bound L Find: Subset P’ of at most Nf forward and at most Nr reverse primers such that 1. 2. P’ does not include any pair of primers in E P’ minimizes the failure probability § where f(P’; l, r) = 1 if P’ fails to yield a PCR product when the deletion with endpoints (l, r) is present in the sample, and f(P’; l, r) = 0 otherwise.

ILP Formulation for PAMP-DEL r (l-1 -xi’ )+(yj’ -r-1) = L yj’ f(P’; l, r)=1 xi’ l 1 r 1 yj’ 5’ 3’ pi’ r 1 Deletion anchor 3’ pi qj qj’ 5’ yj (l 1 -1 -xi’ )+(yj’ -r 1 -1) > L Failure l xi’ l 1 xi

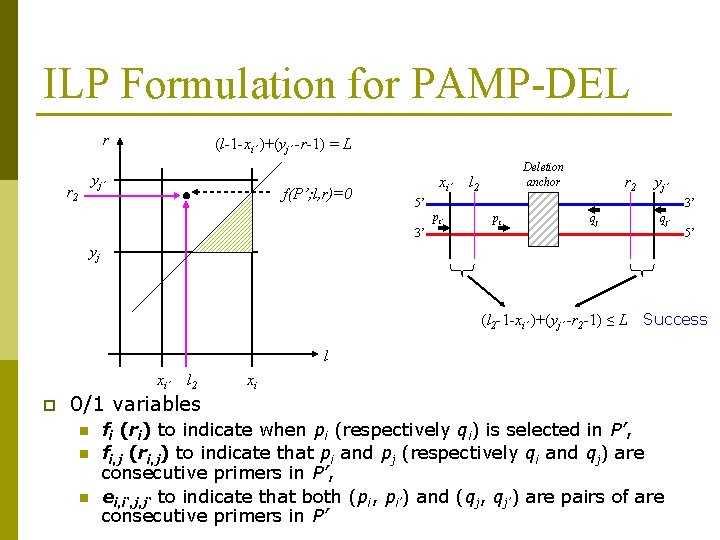
ILP Formulation for PAMP-DEL r (l-1 -xi’ )+(yj’ -r-1) = L yj’ r 2 f(P’; l, r)=0 xi’ Deletion anchor l 2 r 2 yj’ 5’ 3’ pi qj qj’ 3’ 5’ yj (l 2 -1 -xi’ )+(yj’ -r 2 -1) ≤ L Success l xi’ p l 2 xi 0/1 variables n n n fi (ri) to indicate when pi (respectively qi) is selected in P’, fi, j (ri, j) to indicate that pi and pj (respectively qi and qj) are consecutive primers in P’, ei, i‘, j, j‘ to indicate that both (pi, pi’) and (qj, qj’) are pairs of are consecutive primers in P’

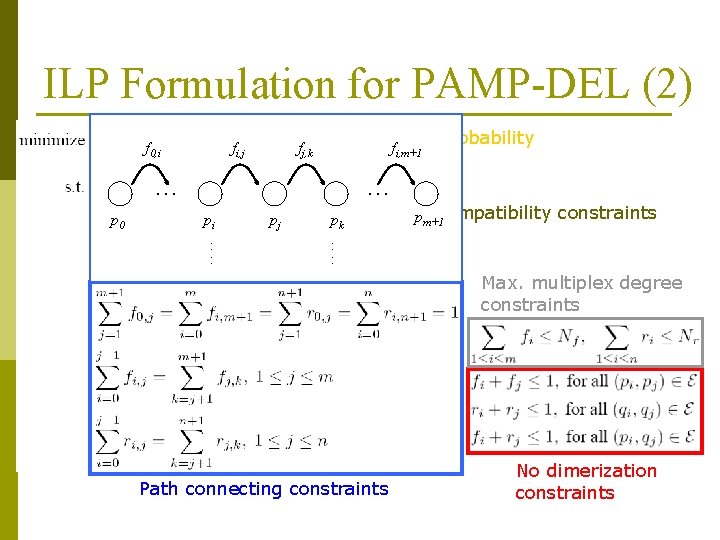
ILP Formulation for PAMP-DEL (2) f 0, i fi, j Failure probability f fj, k i, m+1 . . . p 0 . . . pi : : pj pk Compatibility constraints pm+1 : : Max. multiplex degree constraints Path connecting constraints No dimerization constraints

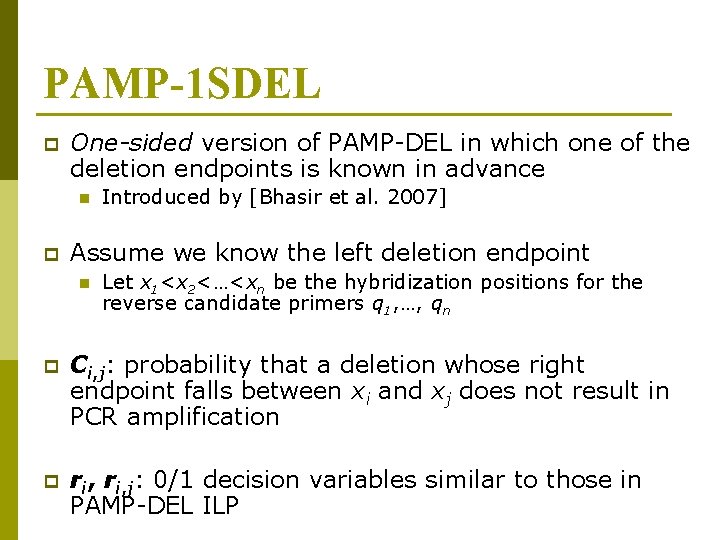
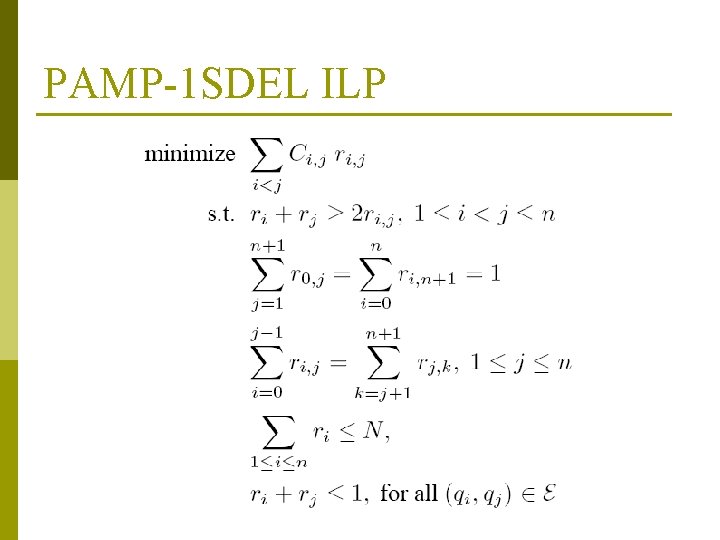
PAMP-1 SDEL p One-sided version of PAMP-DEL in which one of the deletion endpoints is known in advance n p Introduced by [Bhasir et al. 2007] Assume we know the left deletion endpoint n Let x 1<x 2<…<xn be the hybridization positions for the reverse candidate primers q 1, …, qn p Ci, j: probability that a deletion whose right endpoint falls between xi and xj does not result in PCR amplification p ri, j: 0/1 decision variables similar to those in PAMP-DEL ILP

PAMP-1 SDEL ILP

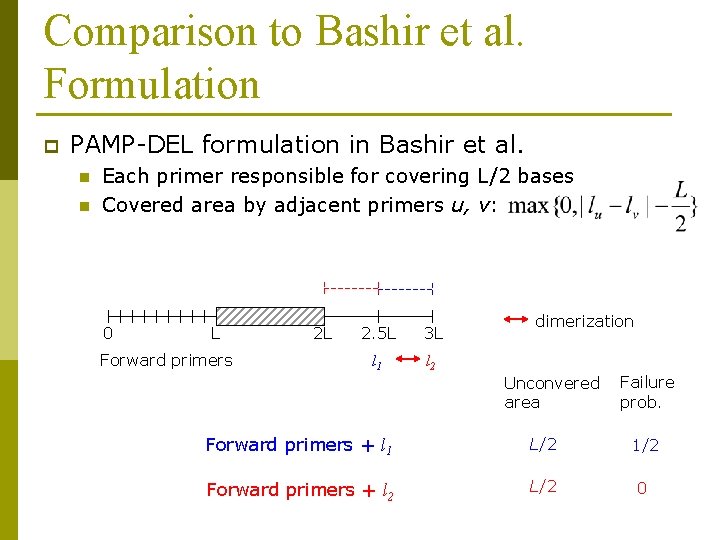
Comparison to Bashir et al. Formulation p PAMP-DEL formulation in Bashir et al. n n Each primer responsible for covering L/2 bases Covered area by adjacent primers u, v: 0 L Forward primers 2 L 2. 5 L 3 L l 1 l 2 dimerization Unconvered area Failure prob. Forward primers + l 1 L/2 1/2 Forward primers + l 2 L/2 0

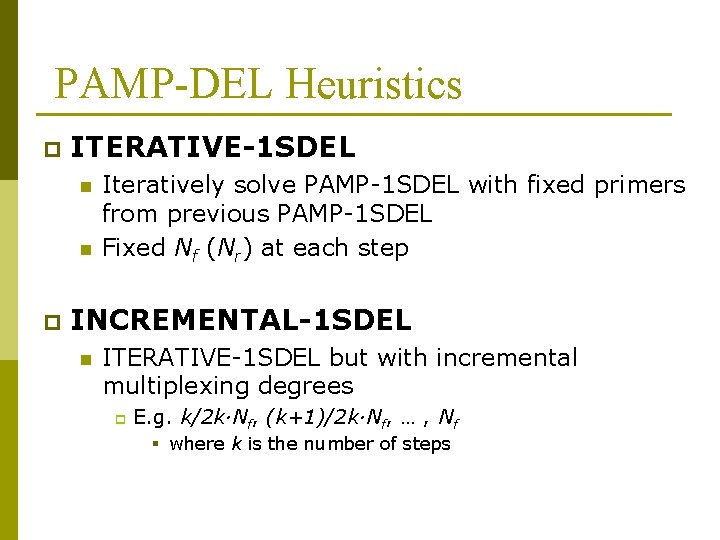
PAMP-DEL Heuristics p ITERATIVE-1 SDEL n n p Iteratively solve PAMP-1 SDEL with fixed primers from previous PAMP-1 SDEL Fixed Nf (Nr) at each step INCREMENTAL-1 SDEL n ITERATIVE-1 SDEL but with incremental multiplexing degrees p E. g. k/2 k·Nf, (k+1)/2 k·Nf, … , Nf § where k is the number of steps

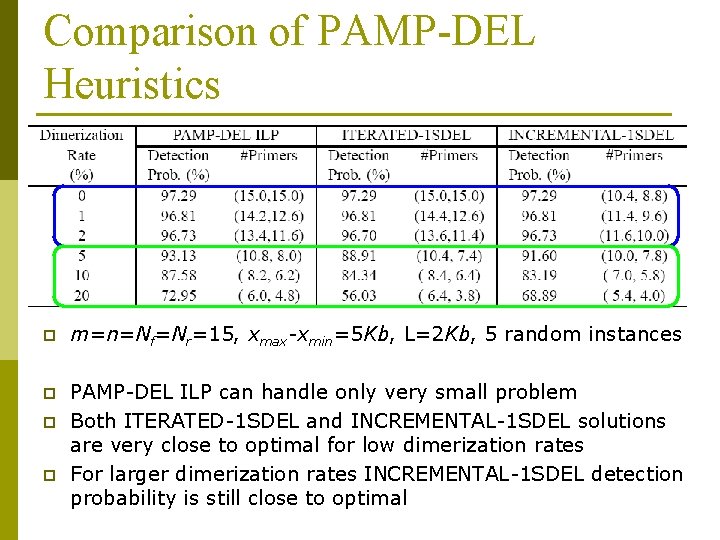
Comparison of PAMP-DEL Heuristics p m=n=Nf=Nr=15, xmax-xmin=5 Kb, L=2 Kb, 5 random instances p PAMP-DEL ILP can handle only very small problem Both ITERATED-1 SDEL and INCREMENTAL-1 SDEL solutions are very close to optimal for low dimerization rates For larger dimerization rates INCREMENTAL-1 SDEL detection probability is still close to optimal p p

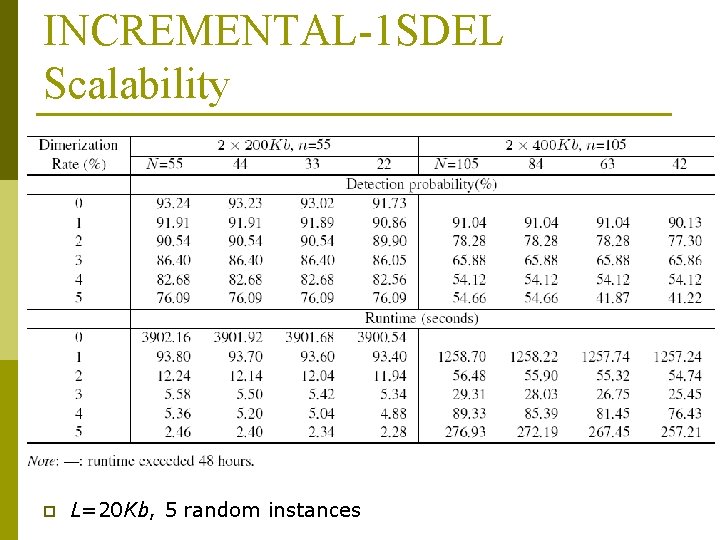
INCREMENTAL-1 SDEL Scalability p L=20 Kb, 5 random instances

Outline p Introduction p Anchored Deletion Detection p Inversion Detection p Conclusions

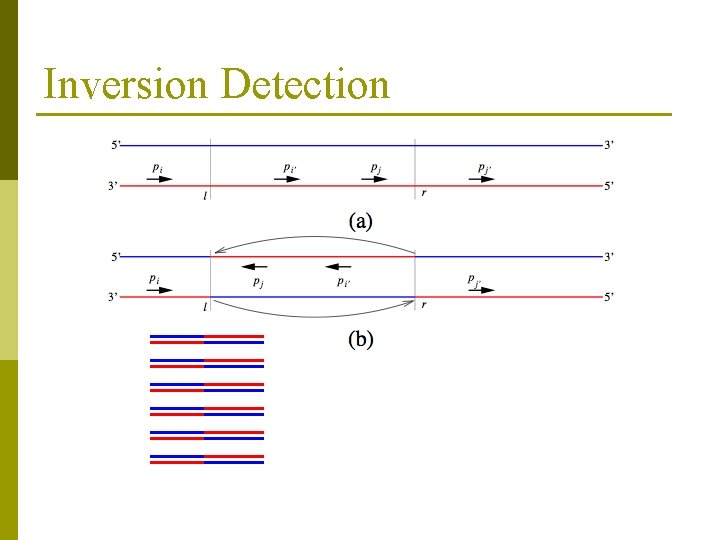
Inversion Detection

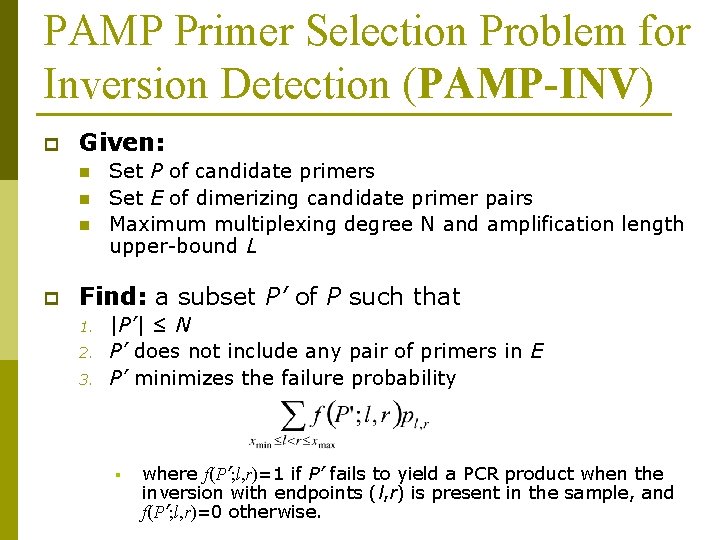
PAMP Primer Selection Problem for Inversion Detection (PAMP-INV) p Given: n n n p Set P of candidate primers Set E of dimerizing candidate primer pairs Maximum multiplexing degree N and amplification length upper-bound L Find: a subset P’ of P such that 1. 2. 3. |P’| ≤ N P’ does not include any pair of primers in E P’ minimizes the failure probability § where f(P’; l, r)=1 if P’ fails to yield a PCR product when the inversion with endpoints (l, r) is present in the sample, and f(P’; l, r)=0 otherwise.

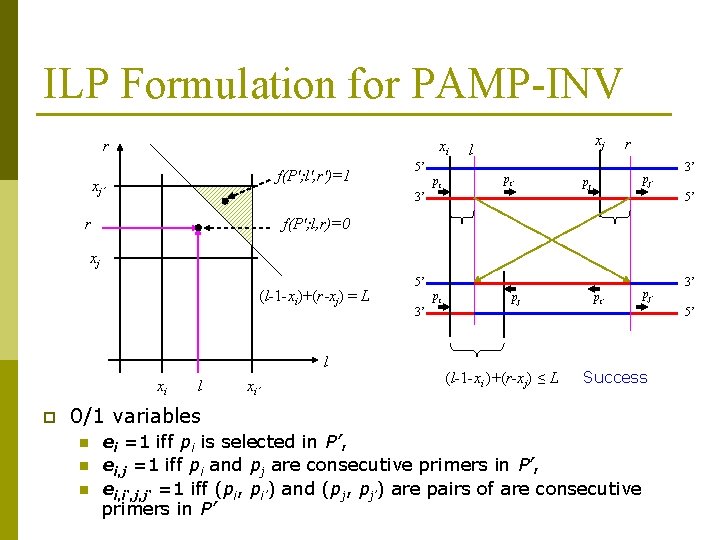
ILP Formulation for PAMP-INV r xi f(P'; l', r')=1 xj’ 5’ pi xj l pi’ r pj’ pj 3’ r 3’ 5’ f(P'; l, r)=0 xj (l-1 -xi)+(r-xj) = L l xi p l xi’ 5’ pi pj pi’ 3’ n n 3’ 5’ (l-1 -xi )+(r-xj) ≤ L Success 0/1 variables n pj’ ei =1 iff pi is selected in P’, ei, j =1 iff pi and pj are consecutive primers in P’, ei, i‘, j, j‘ =1 iff (pi, pi’) and (pj, pj’) are pairs of are consecutive primers in P’

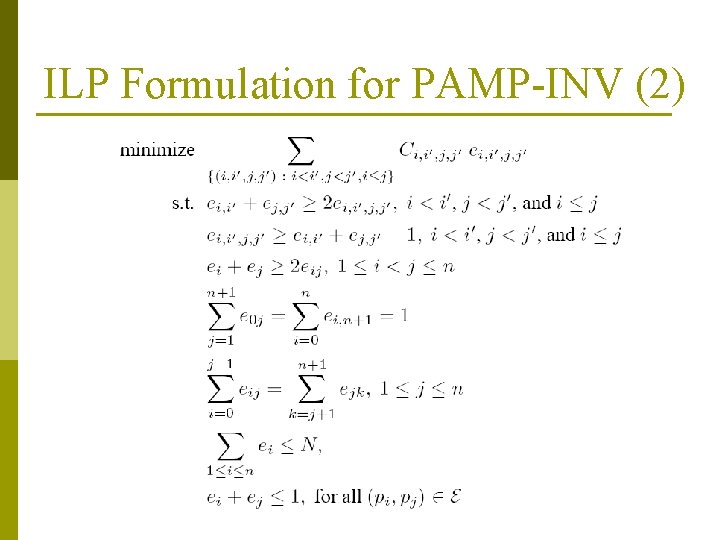
ILP Formulation for PAMP-INV (2)

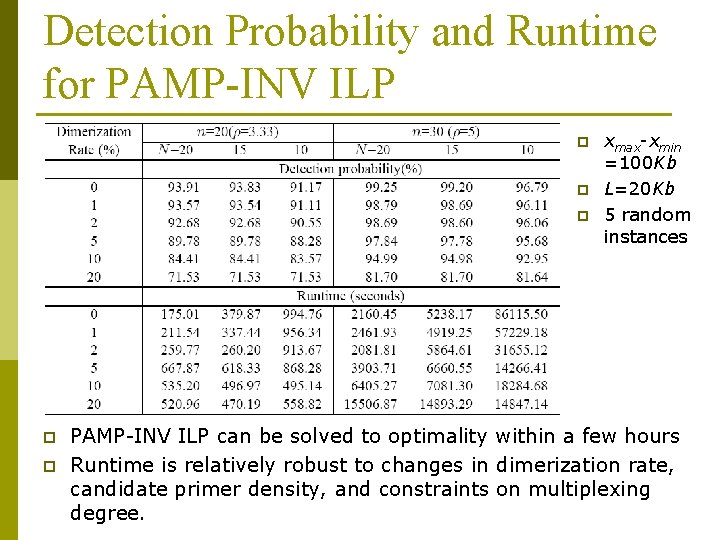
Detection Probability and Runtime for PAMP-INV ILP p p p xmax-xmin =100 Kb L=20 Kb 5 random instances PAMP-INV ILP can be solved to optimality within a few hours Runtime is relatively robust to changes in dimerization rate, candidate primer density, and constraints on multiplexing degree.

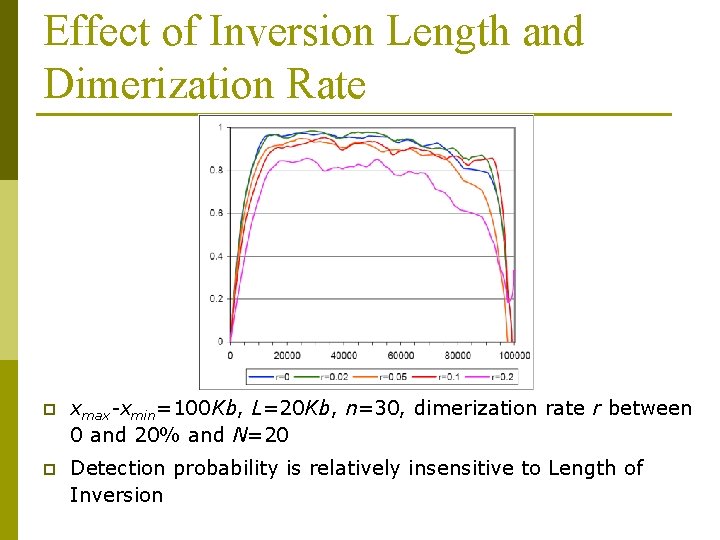
Effect of Inversion Length and Dimerization Rate p xmax-xmin=100 Kb, L=20 Kb, n=30, dimerization rate r between 0 and 20% and N=20 p Detection probability is relatively insensitive to Length of Inversion

Outline p Introduction p Anchored Deletion Detection p Inversion Detection p Conclusions

Summary p ILP formulations for PAMP primer selection n n p Anchored deletion detection (PAMP-DEL) 1 -sided anchored deletion detection (PAMP-1 SDEL) Inversion detection (PAMP-INV) Practical runtime for mid-sized PAMP-INV ILP, highly scalable PAMP-1 SDEL ILP Heuristics for PAMP-DEL based on PAMP 1 SDEL ILP n Near optimal solutions with highly scalable runtime

Questions?