PLAZA 2 5 a resource for plant comparative



- Slides: 32

PLAZA 2. 5 – a resource for plant comparative genomics Michiel Van Bel Bioinformatics & Evolutionary Genomics group SPICY workshop 08/03/0212 Wageningen, Netherlands Comparative & Integrative Genomics group plaza@psb. vib-ugent. be VIB – Ghent University, Belgium

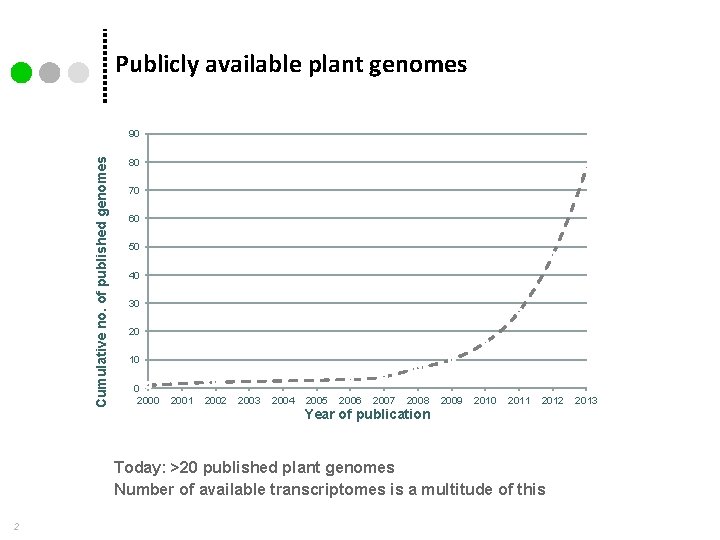
Publicly available plant genomes Cumulative no. of published genomes 90 80 70 60 50 40 30 20 10 0 2001 2002 2003 2004 2005 2006 2007 2008 2009 2010 2011 2012 Year of publication Today: >20 published plant genomes Number of available transcriptomes is a multitude of this 2 2013

Exploiting cross-species genome information v Centralized infrastructure v Detailed gene catalog per species Structural annotation (gene models, UTRs) Functional annotation (experimental, sequence-based) v Intuitive & advanced data mining tools for non-expert users • • • 3 Gene function Genome organization Gene families Pathway evolution Data manipulation

PLAZA, a resource for plant comparative genomics http: //bioinformatics. psb. ugent. be/plaza/ 5

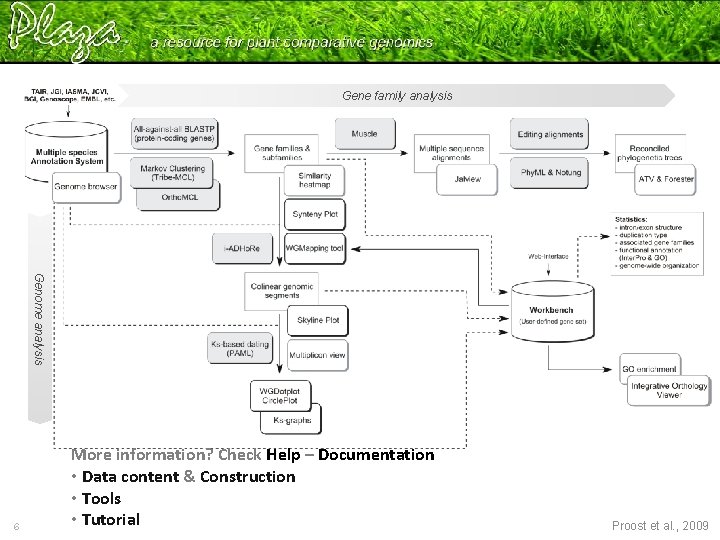
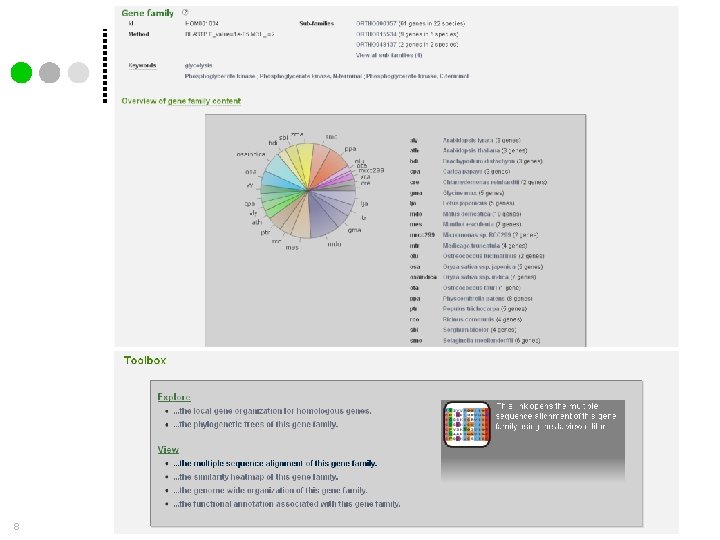
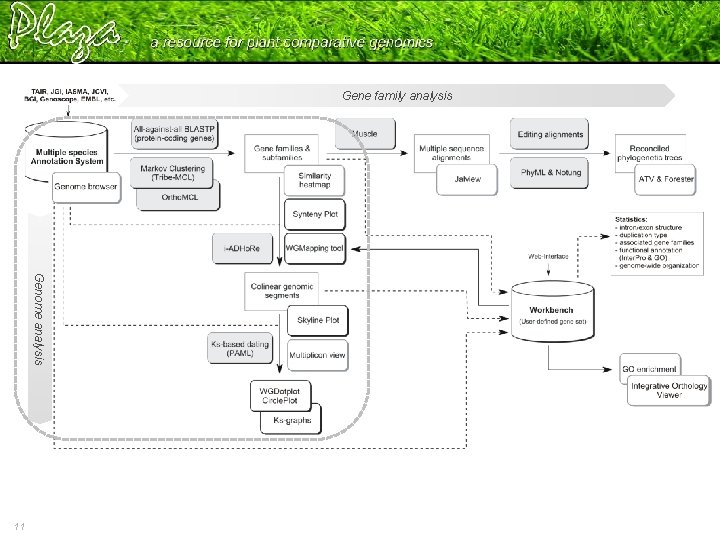
Gene family analysis Genome analysis 6 More information? Check Help – Documentation • Data content & Construction • Tools • Tutorial Proost et al. , 2009

7

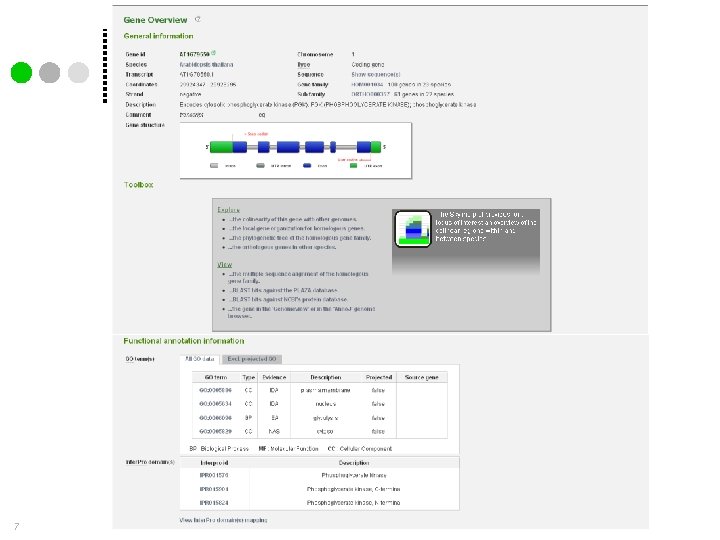
Gene family page 8

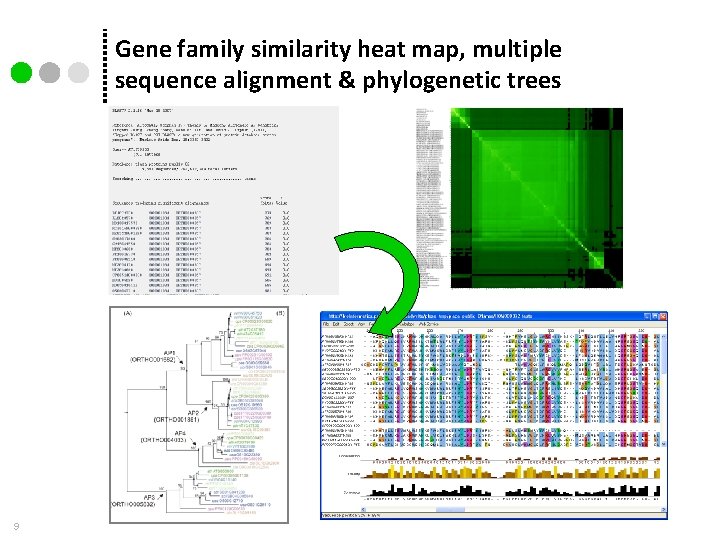
Gene family similarity heat map, multiple sequence alignment & phylogenetic trees 9

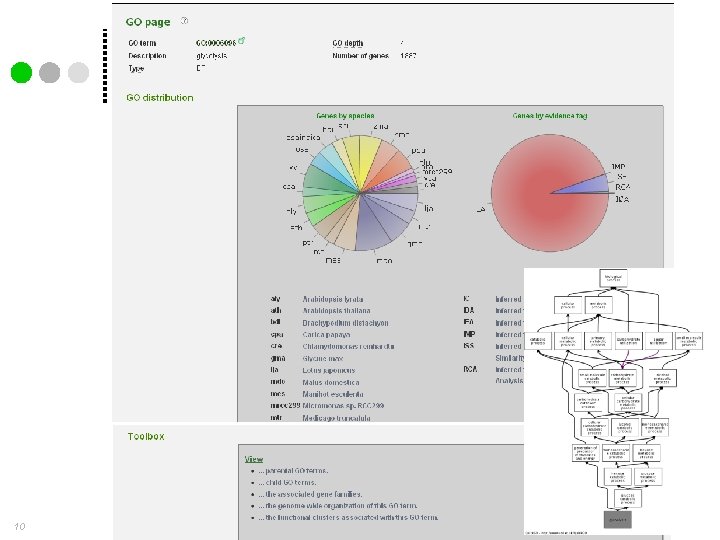
Gene Ontology annotation 10

Gene family analysis Genome analysis 11

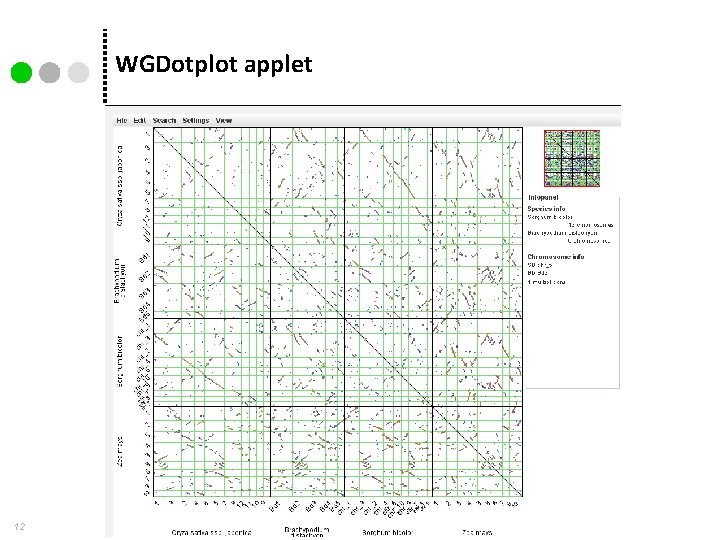
WGDotplot applet 12

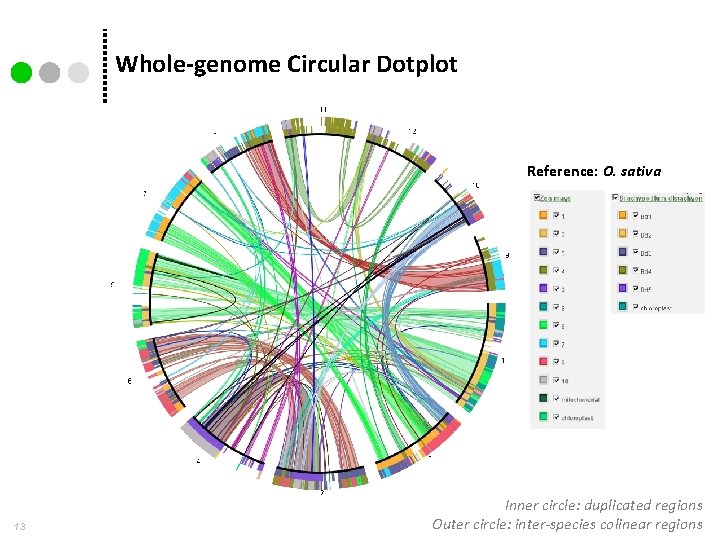
Whole-genome Circular Dotplot Reference: O. sativa 13 Inner circle: duplicated regions Outer circle: inter-species colinear regions

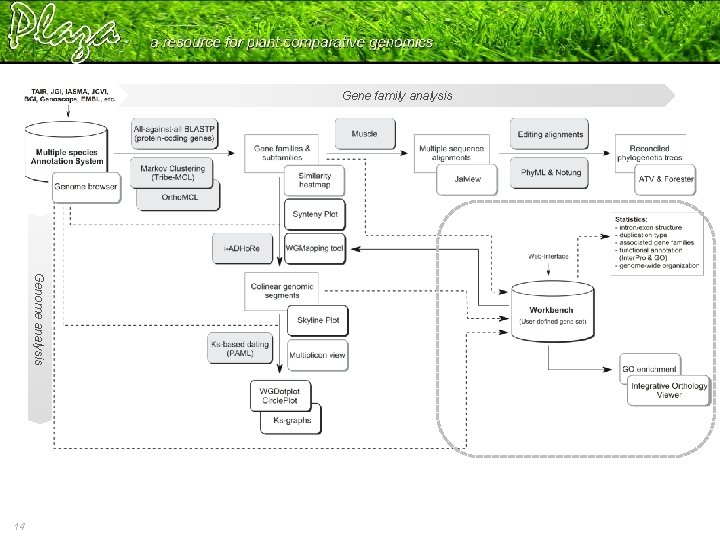
Gene family analysis Genome analysis 14

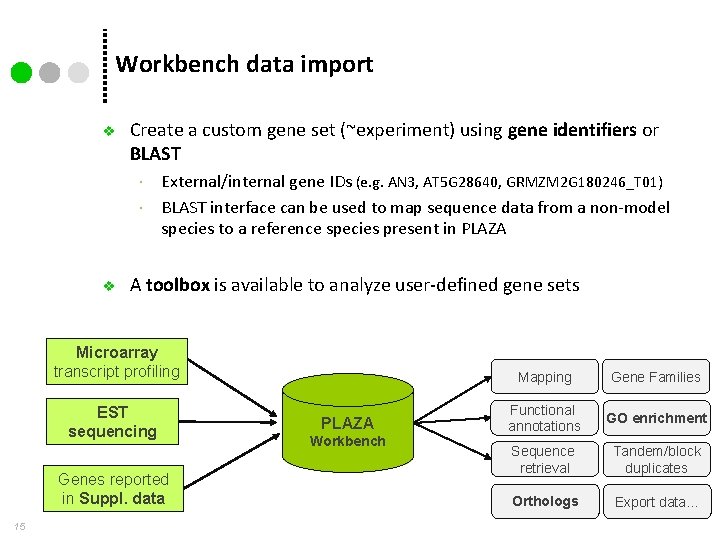
Workbench data import v Create a custom gene set (~experiment) using gene identifiers or BLAST v External/internal gene IDs (e. g. AN 3, AT 5 G 28640, GRMZM 2 G 180246_T 01) BLAST interface can be used to map sequence data from a non-model species to a reference species present in PLAZA A toolbox is available to analyze user-defined gene sets Microarray transcript profiling EST sequencing Genes reported in Suppl. data 15 PLAZA Workbench Mapping Gene Families Functional annotations GO enrichment Sequence retrieval Tandem/block duplicates Orthologs Export data…

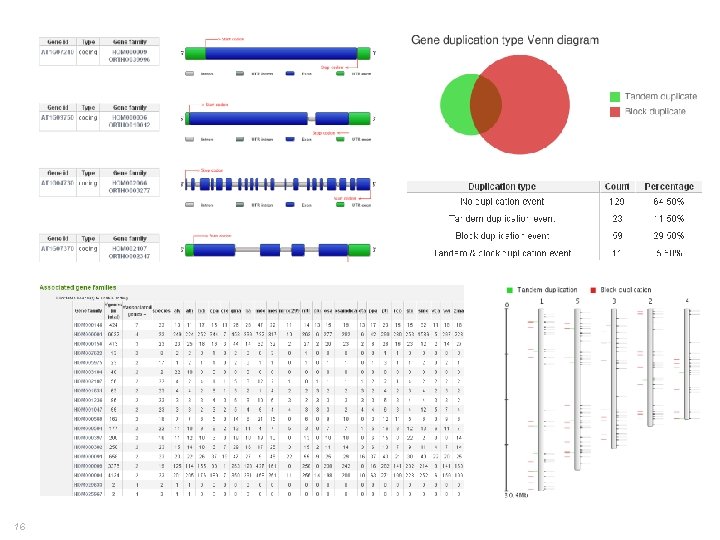
16

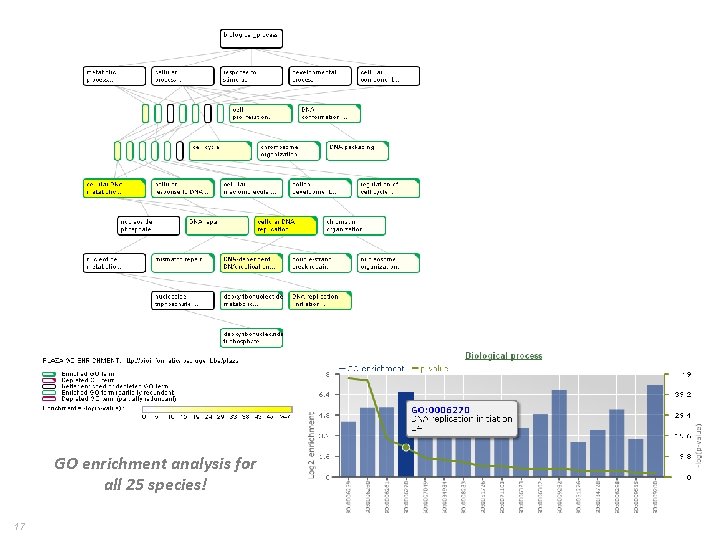
GO enrichment analysis for all 25 species! 17

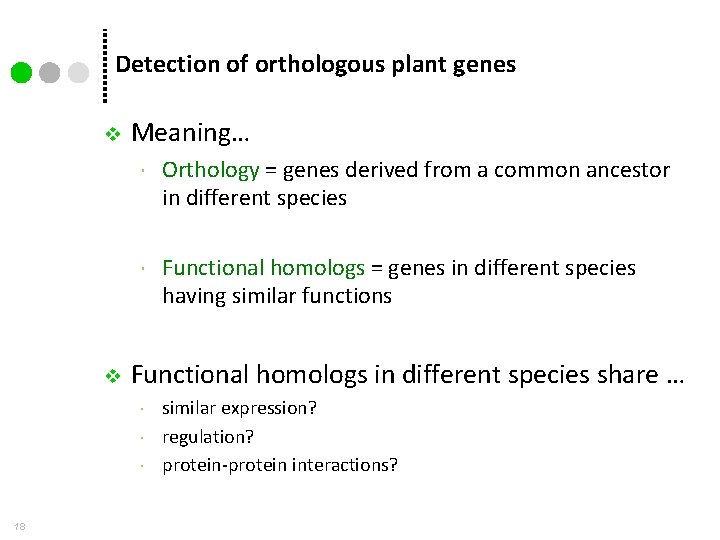
Detection of orthologous plant genes v v Meaning… Orthology = genes derived from a common ancestor in different species Functional homologs = genes in different species having similar functions Functional homologs in different species share … 18 similar expression? regulation? protein-protein interactions?

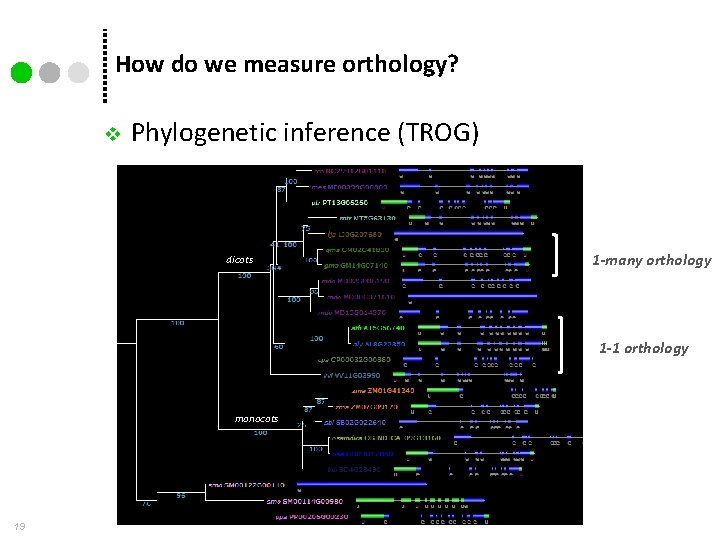
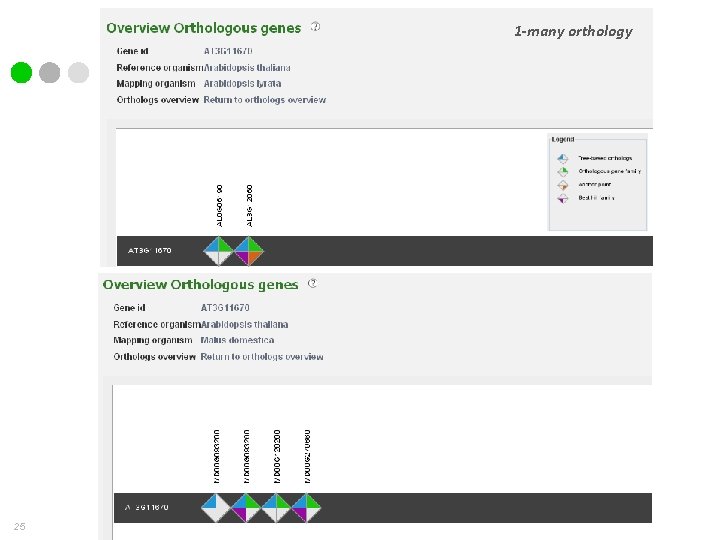
How do we measure orthology? v Phylogenetic inference (TROG) dicots 1 -many orthology 1 -1 orthology monocots 19

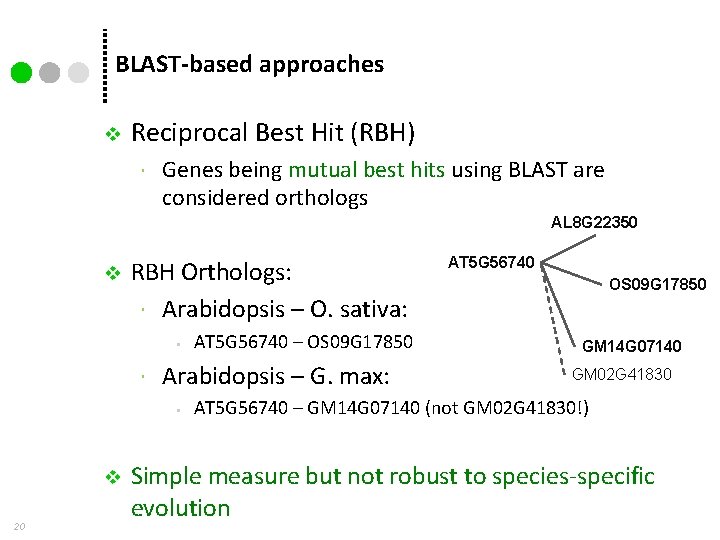
BLAST-based approaches v Reciprocal Best Hit (RBH) Genes being mutual best hits using BLAST are considered orthologs AL 8 G 22350 v RBH Orthologs: Arabidopsis – O. sativa: • Arabidopsis – G. max: • v 20 AT 5 G 56740 – OS 09 G 17850 AT 5 G 56740 OS 09 G 17850 GM 14 G 07140 GM 02 G 41830 AT 5 G 56740 – GM 14 G 07140 (not GM 02 G 41830!) Simple measure but not robust to species-specific evolution

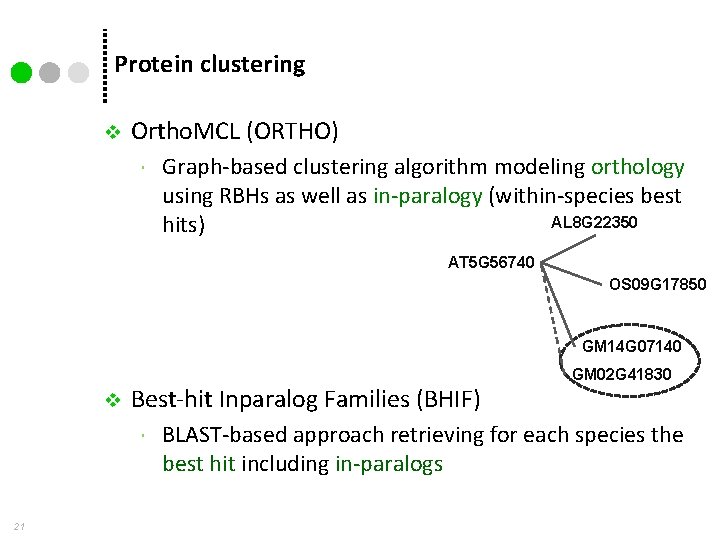
Protein clustering v Ortho. MCL (ORTHO) Graph-based clustering algorithm modeling orthology using RBHs as well as in-paralogy (within-species best AL 8 G 22350 hits) AT 5 G 56740 OS 09 G 17850 GM 14 G 07140 v Best-hit Inparalog Families (BHIF) 21 GM 02 G 41830 BLAST-based approach retrieving for each species the best hit including in-paralogs

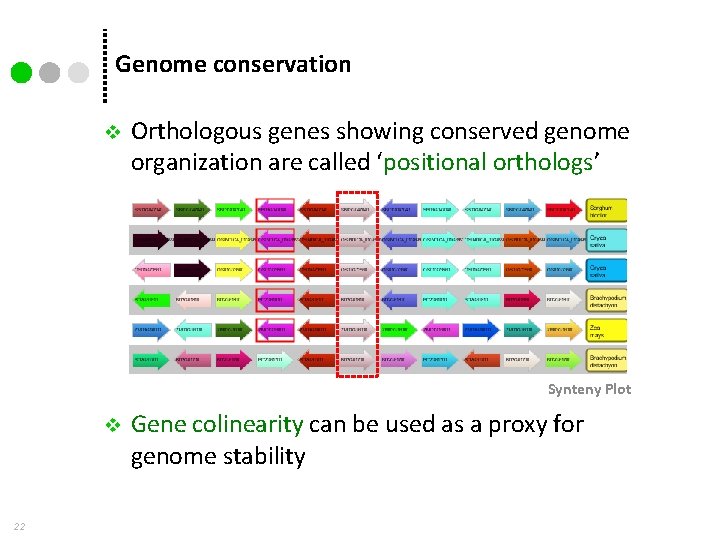
Genome conservation v Orthologous genes showing conserved genome organization are called ‘positional orthologs’ Synteny Plot v 22 Gene colinearity can be used as a proxy for genome stability

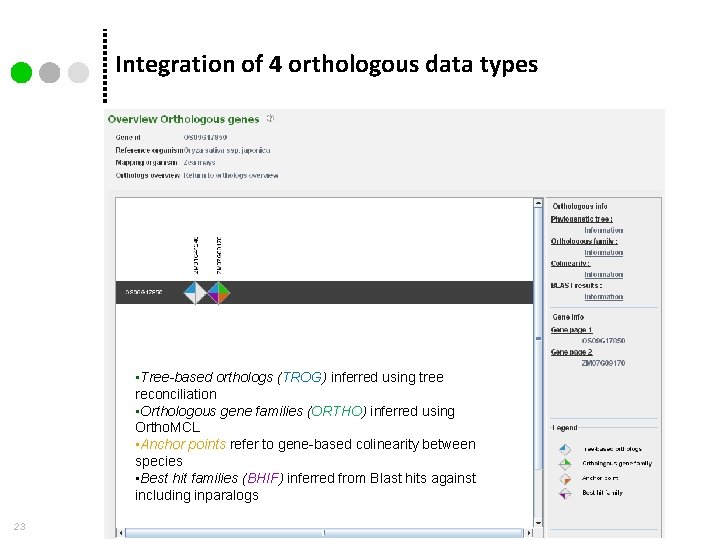
Integration of 4 orthologous data types • Tree-based orthologs (TROG) inferred using tree reconciliation • Orthologous gene families (ORTHO) inferred using Ortho. MCL • Anchor points refer to gene-based colinearity between species • Best hit families (BHIF) inferred from Blast hits against including inparalogs 23

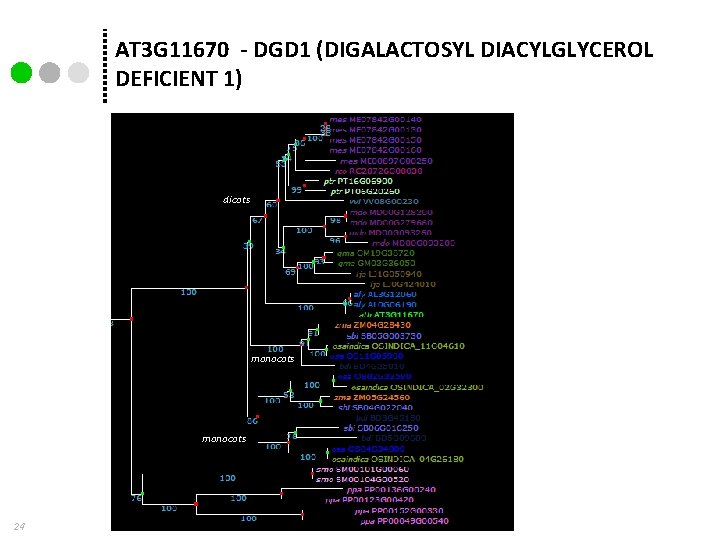
AT 3 G 11670 - DGD 1 (DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1) dicots monocots 24

1 -many orthology 25

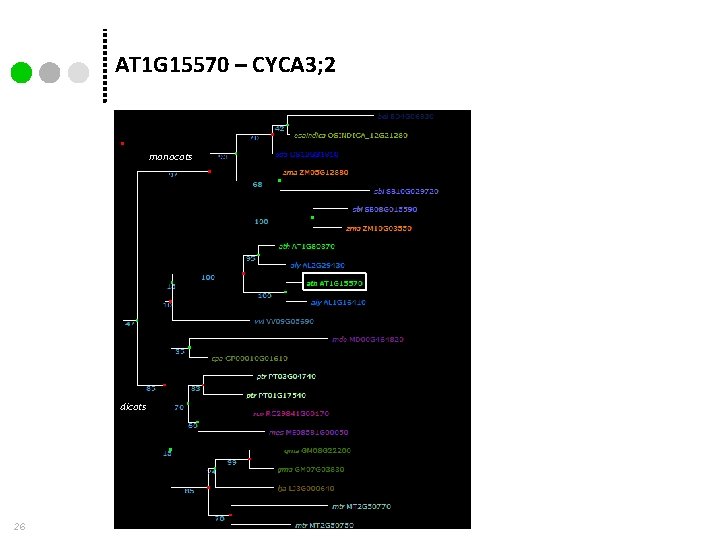
AT 1 G 15570 – CYCA 3; 2 monocots dicots 26

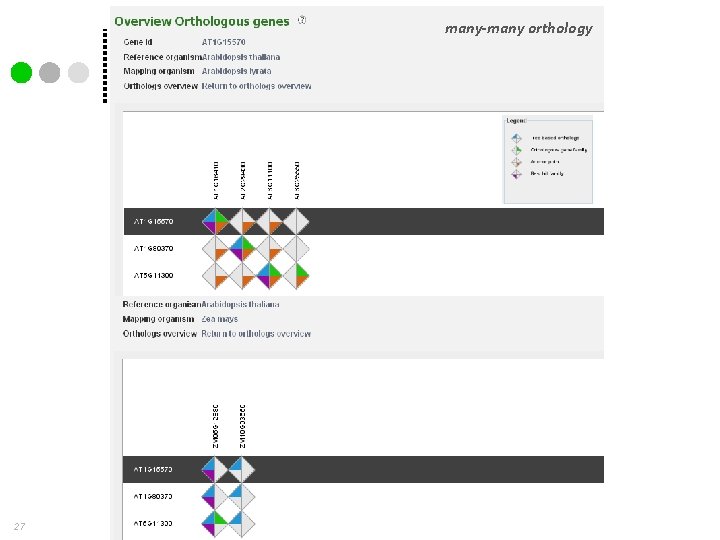
many-many orthology 27

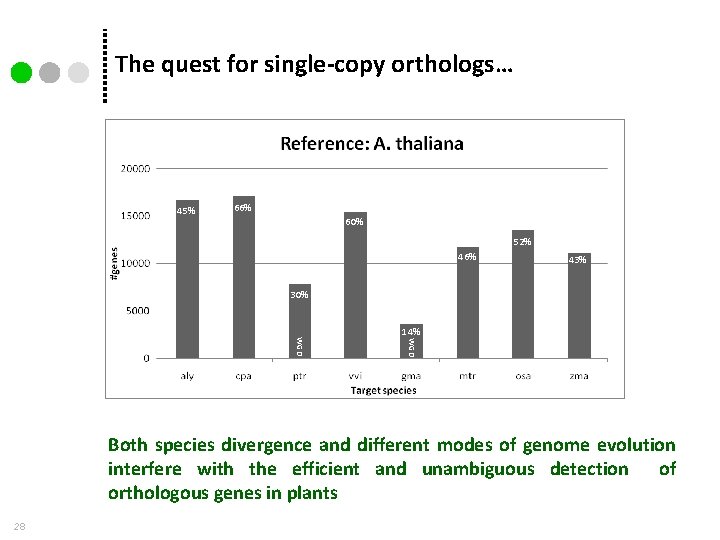
The quest for single-copy orthologs… 45% 66% 60% 52% 46% 43% 30% WGD 14% Both species divergence and different modes of genome evolution interfere with the efficient and unambiguous detection of orthologous genes in plants 28

Conclusions v 29 PLAZA provides an integrated and intuitive framework that can function as a data warehouse for plant genomes a comparative research environment for genomic data mining an easy access point for non-expert users to explore orthologous genes

Acknowledgments 30 v Prof. Dr. Klaas Vandepoele v Sebastian Proost v Prof. Dr. Yves Van de Peer http: //bioinformatics. psb. ugent. be/plaza/

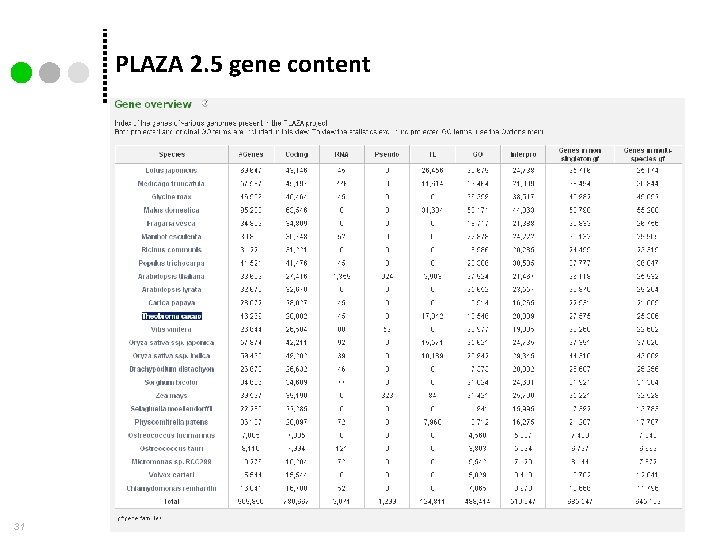
PLAZA 2. 5 gene content 31

Sequencing in progress 32 Eucalyptus grandis JGI Arabidopsis arenosa JGI Gossypium (cotton) genome Phase II JGI Gossypium raimondii JGI Brassica rapa B 3 JGI Zea mays ssp. mays Mo 17 JGI Salix purpurea L JGI Arabidposis halleri JGI Capsella rubella JGI Boechera holboellii Panther JGI Miscanthus giganteus Sequencing Pilot Project JGI Manihot esculenta CV AM 560 -2 JGI Setaria italica Yugu 1 JGI Aquilegia coerulea Goldsmith JGI Brassica ? MGBP Lycopersicum esculentum ITGSP Solanum tuberosum PGSC Musa acuminata GMGC Mimulus guttatus JGI Triphysaria versicolor JGI

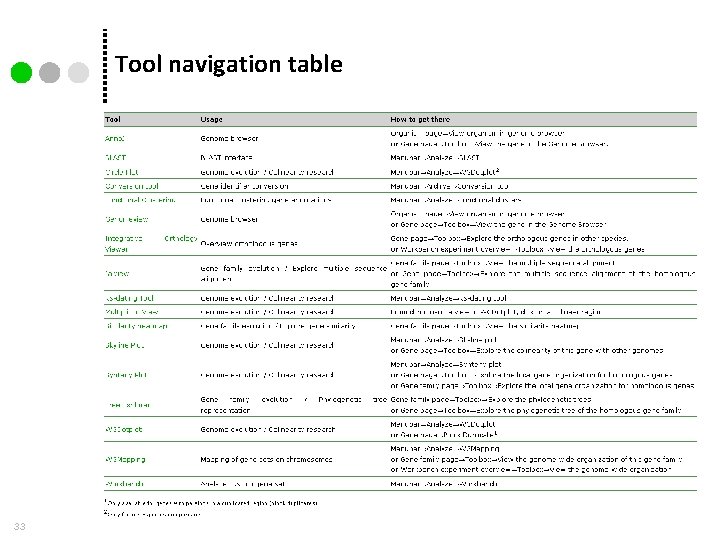
Tool navigation table 33