Pairwise Alignment How do we tell whether two






























- Slides: 50

Pairwise Alignment • How do we tell whether two sequences are similar? Assigned reading: Ch 4. 1 -4. 7, Ch 5. 1, get what you can out of 5. 2, 5. 4 BIO 520 Bioinformatics Jim Lund

Pairwise alignment • DNA: DNA • polypeptide: polypeptide The BASIC Sequence Analysis Operation

Alignments • Pairwise sequence alignments – One-to-One – One-to-Database • Multiple sequence alignments – Many-to-Many

Origins of Sequence Similarity • Homology – common evolutionary descent • Chance – Short similar segments are very common. • Similarity in function – Convergence (very rare)


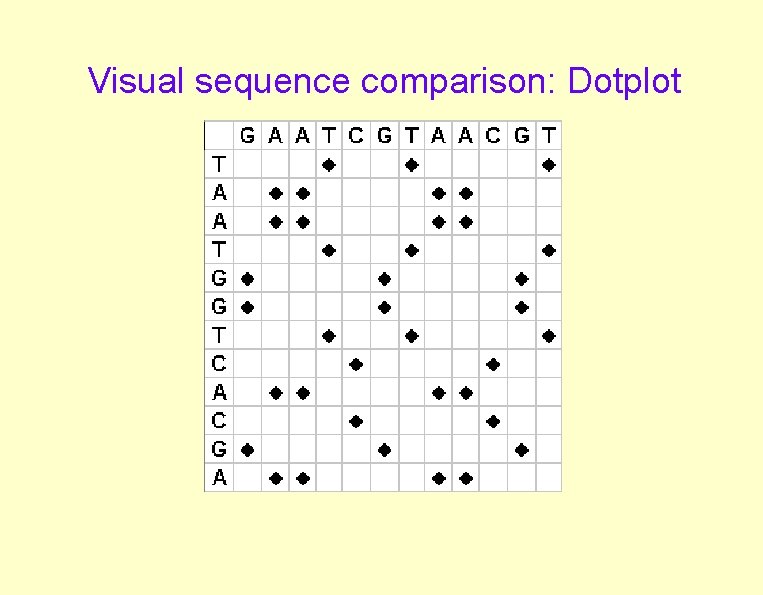
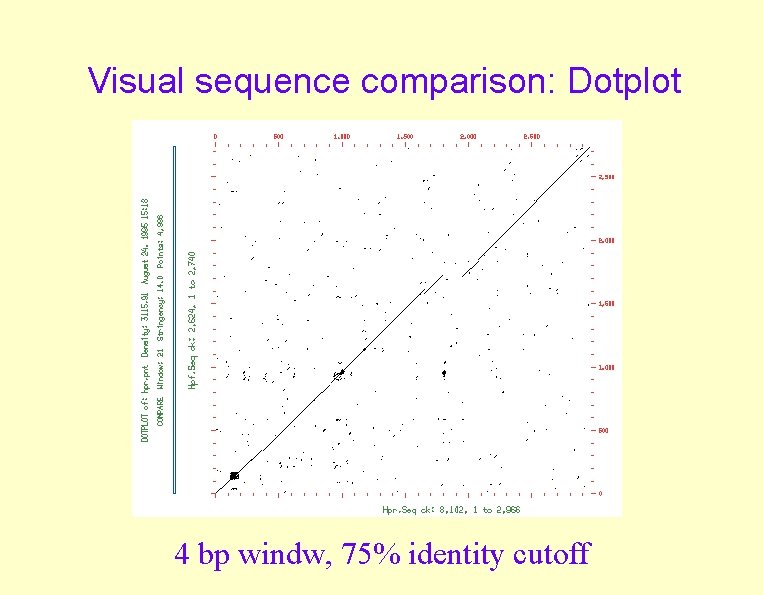
Visual sequence comparison: Dotplot

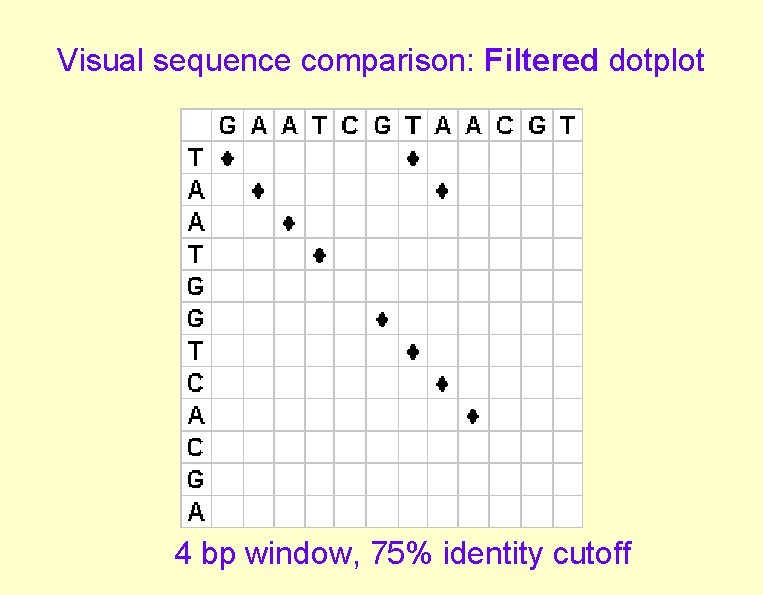
Visual sequence comparison: Filtered dotplot 4 bp window, 75% identity cutoff

Visual sequence comparison: Dotplot 4 bp windw, 75% identity cutoff

Dotplots of sequence rearrangements

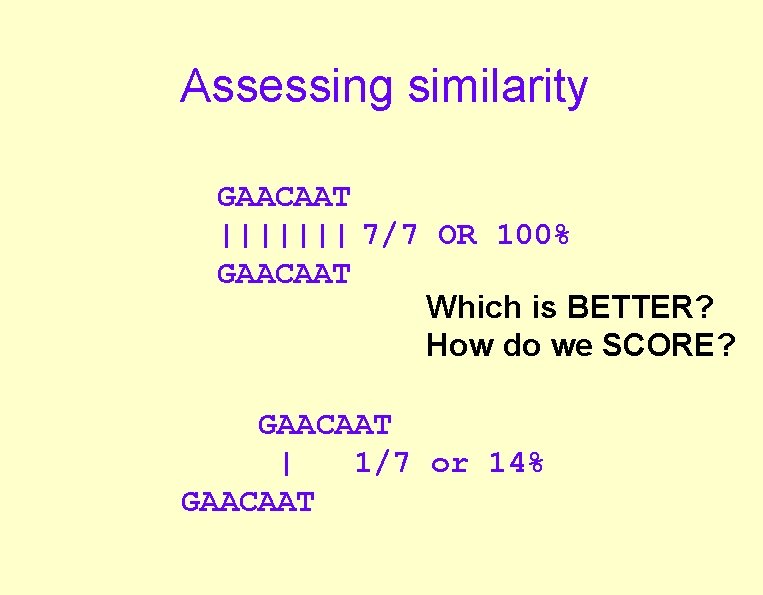
Assessing similarity GAACAAT ||||||| 7/7 OR 100% GAACAAT Which is BETTER? How do we SCORE? GAACAAT | 1/7 or 14% GAACAAT

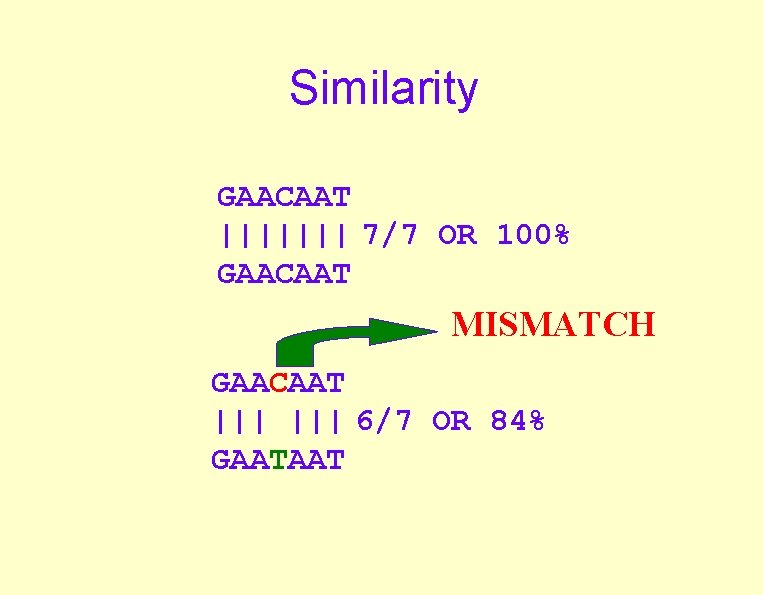
Similarity GAACAAT ||||||| 7/7 OR 100% GAACAAT MISMATCH GAACAAT ||| 6/7 OR 84% GAATAAT

Mismatches GAACAAT ||| 6/7 OR 84% GAATAAT GAACAAT ||| 6/7 OR 84% GAAGAAT

Terminal Mismatch GAACAATttttt ||| aaacc. GAATAAT 6/7 OR 84%

INDELS GAAg. CAAT |||| GAA*CAAT 7/7 OR 100%

Indels, cont’d GAAg. CAAT |||| GAA*CAAT GAAgggg. CAAT |||| GAA****CAAT

Similarity Scoring Common Method: • • • Terminal mismatches (0) Match score (1) Mismatch penalty (-3) Gap penalty (-1) Gap extension penalty (-1) DNA Defaults

DNA Scoring GGGGGGAGAA |||||*|*|| GGGGGAAAAAGGGGGGAGAA--GGG |||||*|*|| ||| GGGGGAAAAAGGGGG 8(1)+2(-3)=2 11(1)+2(-3)+1(-1)=3

Absurdity of Low Gap Penalty GATCGCTACGCTCAGC A. C. C. . T Perfect similarity, Every time!

Sequence alignment algorithms • Local alignment – Smith-Waterman • Global alignment – Needleman-Wunsch

Alignment Programs • Local alignment (Smith-Waterman) – BLAST (simplified Smith-Waterman) – FASTA (simplified Smith-Waterman) – BESTFIT (GCG program) • Global alignment (Needleman-Wunsch) – GAP

Local vs. global alignment 10 gaggc 15 ||||| 3 gaggc 7 Local alignment: alignment of regions of substantial similarity 1 gggggaaaaagtggccccc 19 || || 1 gggggttttgtggtttcc 22 Global alignment: alignment of the full length of the sequences

Local vs. global alignment

BLAST Algorithm Look for local alignment, a High Scoring Pair (HSP) • Finding word (W) in query and subject. Score > T. • Extend local alignment until score reaches maximum-X. • Keep High Scoring Segment Pairs (HSPs) with scores > S. • Find multiple HSPs per query if present • Expectation value (E value) using Karlin-Altschul stats

BLAST statistical significance: assessing the likelihood a match occurs by chance Karlin-Altschul statistic: E = k m N exp(-Lambda S) m = Size of query seqeunce N = Size of database k = Search space scaling parameter Lambda = scoring scaling parameter S = BLAST HSP score Low E -> good match

BLAST statistical significance: Rule of thumb for a good match: • Nucleotide match • E < 1 e-6 • Identity > 70% • Protein match • E < 1 e-3 • Identity > 25%

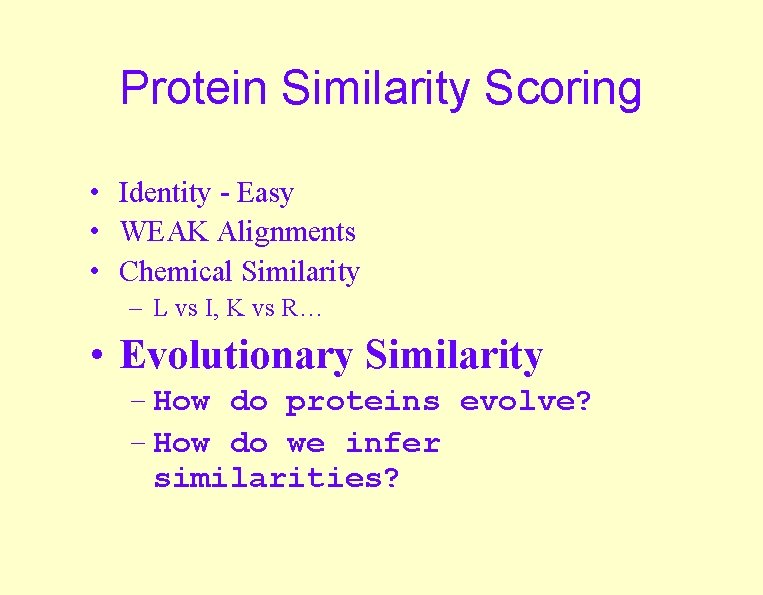
Protein Similarity Scoring • Identity - Easy • WEAK Alignments • Chemical Similarity – L vs I, K vs R… • Evolutionary Similarity – How do proteins evolve? – How do we infer similarities?

BLOSUM 62

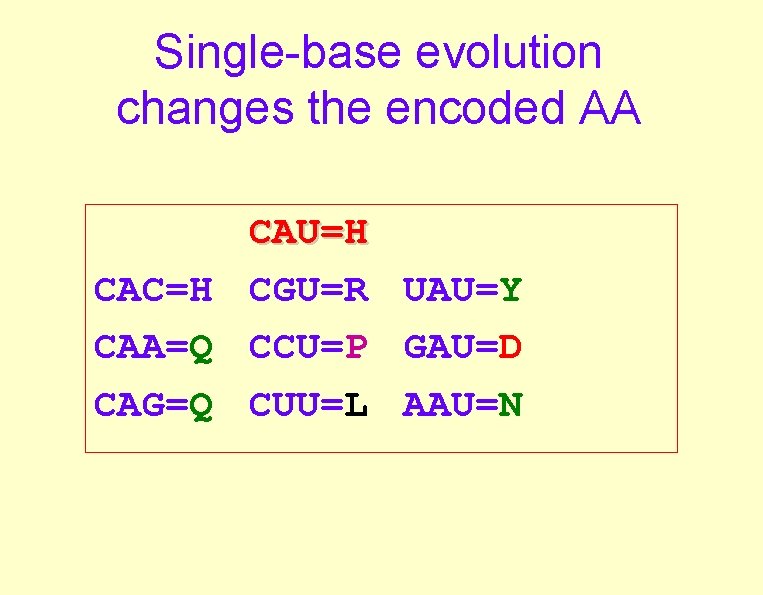
Single-base evolution changes the encoded AA CAU=H CAC=H CGU=R UAU=Y CAA=Q CCU=P GAU=D CAG=Q CUU=L AAU=N

Substitution Matrices Two main classes: • PAM-Dayhoff • BLOSUM-Henikoff

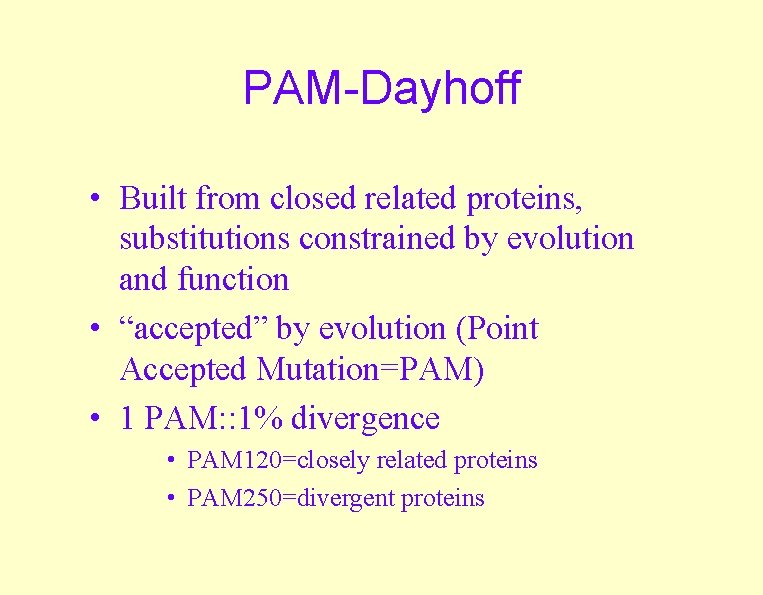
PAM-Dayhoff • Built from closed related proteins, substitutions constrained by evolution and function • “accepted” by evolution (Point Accepted Mutation=PAM) • 1 PAM: : 1% divergence • PAM 120=closely related proteins • PAM 250=divergent proteins

BLOSUMHenikoff&Henikoff • Built from ungapped alignments in proteins: “BLOCKS” • Merge blocks at given % similar to one sequence • Calculate “target” frequencies • BLOSUM 62=62% similar blocks – good general purpose • BLOSUM 30 – Detects weak similarities, used for distantly related proteins

BLOSUM 62

Gapped alignments • No general theory for significance of matches!! • G+L(n) – indel mutations rare – variation in gap length “easy”, G > L

Real Alignments

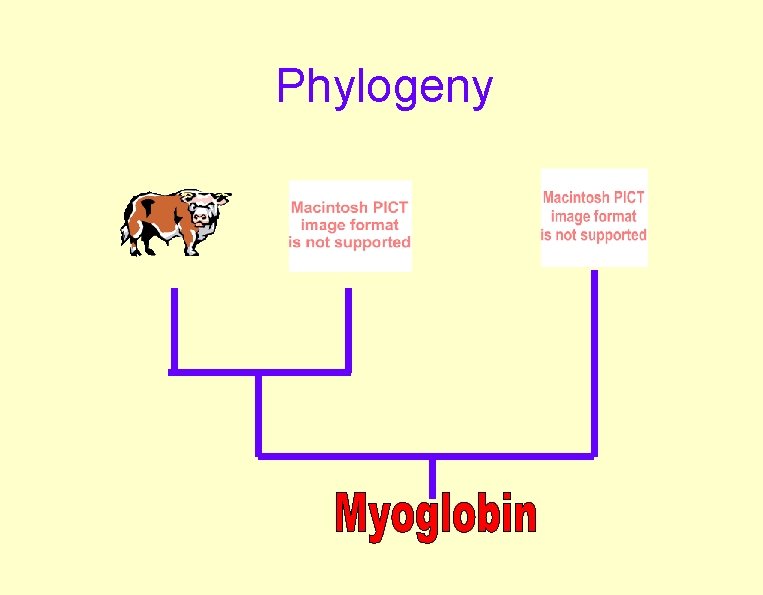
Phylogeny

Cow-to-Pig Protein

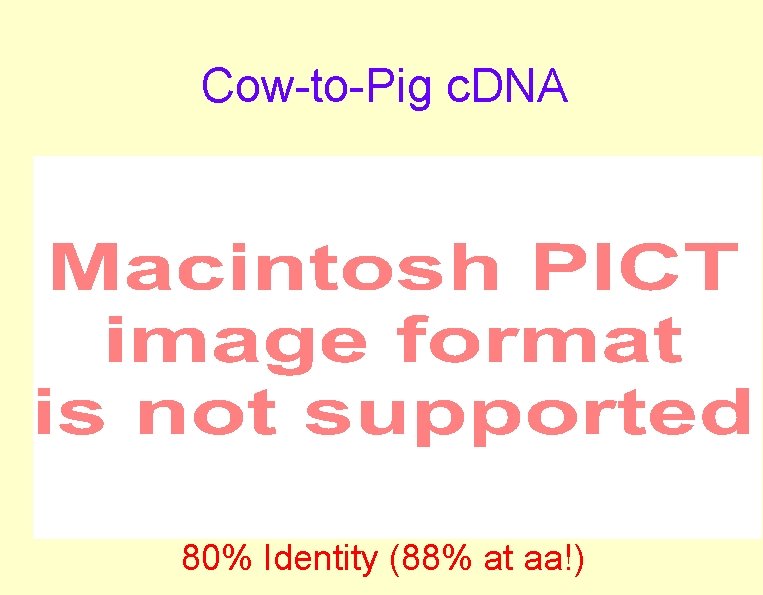
Cow-to-Pig c. DNA 80% Identity (88% at aa!)

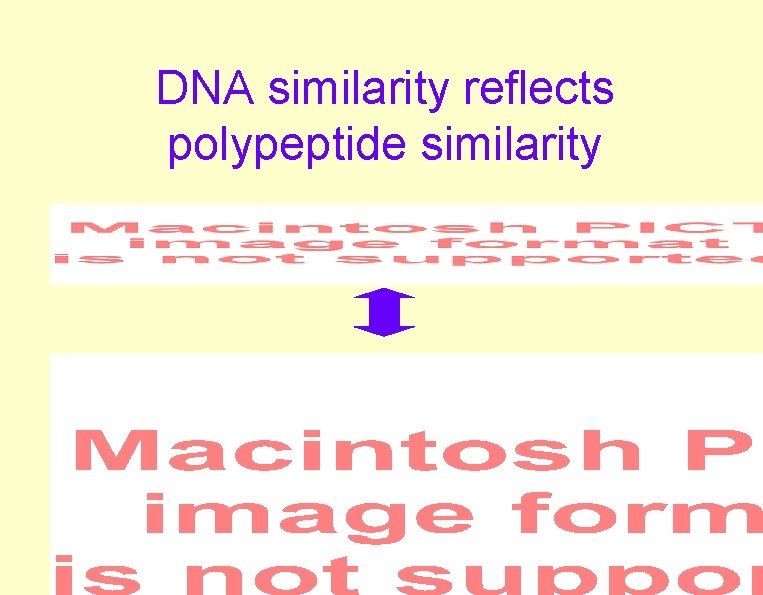
DNA similarity reflects polypeptide similarity

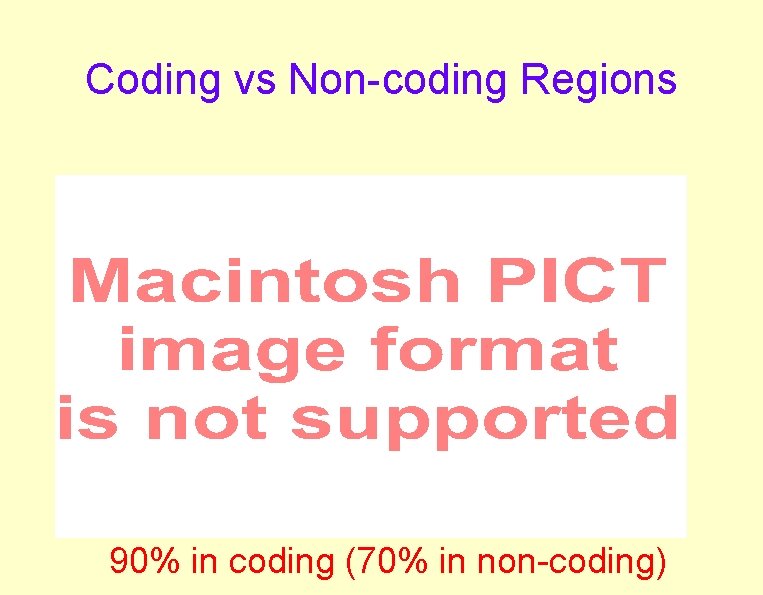
Coding vs Non-coding Regions 90% in coding (70% in non-coding)

Third Base of Codon is Hypervariable

Cow-to-Fish Protein 42% identity, 51% similarity

Cow-to-Fish DNA 48% similarity

Protein vs. DNA Alignments • Polypeptide similarity > DNA • Coding DNA > Non-coding • 3 rd base of codon hypervariable • Moderate Distance poor DNA similarity

Rules of Thumb • DNA-DNA similarities – 50% significant if “long” – E < 1 e-6, 70% identity • Protein-protein similarities – 80% end-end: same structure, same function – 30% over domain, similar function, structure overall similar – 15 -30% “twilight zone” – Short, strong match…could be a “motif”

Basic BLAST Family • BLASTN – DNA to DNA database • BLASTP – protein to protein database • TBLASTN – DNA (translated) to protein database • BLASTX – protein to DNA database (translated) • TBLASTX – DNA (translated) to DNA database (translated)

DNA Databases • nr (non-redundantish merge of Genbank, EMBL, etc…) – EXCLUDES HTGS 0, 1, 2, EST, GSS, STS, PAT, WGS • • • est (expressed sequence tags) htgs (high throughput genome seq. ) gss (genome survey sequence) vector, yeast, ecoli, mito chromosome (complete genomes) And more http: //www. ncbi. nlm. nih. gov/BLAST/blastcgihelp. shtml#nucleotide_databases

Protein Databases • nr (non-redundant Swiss-prot, PIR, PDF, PDB, Genbank CDS) • swissprot • ecoli, yeast, fly • month • And more

BLAST Input • • Program Database Options - see more Sequence – FASTA – gi or accession#

BLAST Options • Algorithm and output options – # descriptions, # alignments returned – Probability cutoff – Strand • Alignment parameters – Scoring Matrix • PAM 30, PAM 70, BLOSUM 45, BLOSUM 62, BLOSUM 80 – Filter (low complexity) PPPPP->XXXXX

Extended BLAST Family • Gapped Blast (default) • PSI-Blast (Position-specific iterated blast) – “self” generated scoring matrix • PHI BLAST (motif plus BLAST) • BLAST 2 client (align two seqs) • megablast (genomic sequence) • rpsblast (search for domains)