Sequence Alignment I Dot Matrices Reading Mount Chapters






- Slides: 15

Sequence Alignment I Dot Matrices

Reading • Mount, Chapters 1, 2, and 3 (up to page 94) 2

Why compare sequences? • To find whether two (or more) genes or proteins are evolutionarily related to each other • To find structurally or functionally similar regions within proteins 3

Similar genes arise by gene duplication • Copy of a gene inserted next to the original • Two copies mutate independently • Each can take on separate functions • All or part can be transferred from one part of genome to another 4

Sequence Comparison Methods • Dot matrix analysis • Dynamic Programming • Word or k-tuple methods (FASTA and BLAST) 5

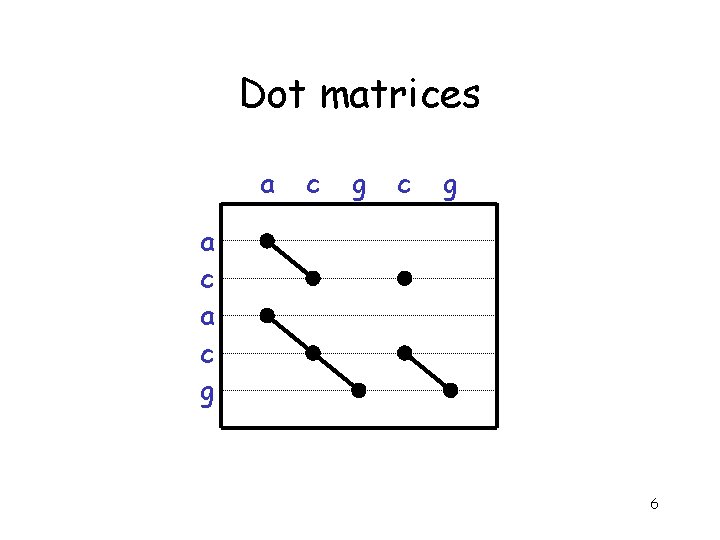
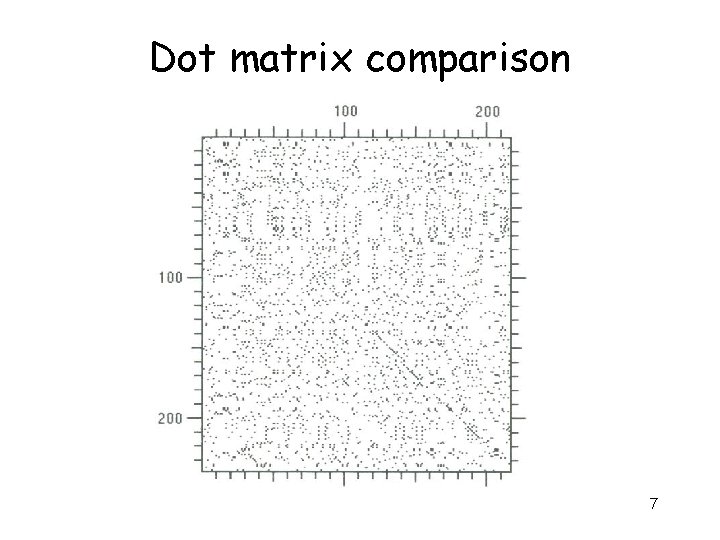
Dot matrices a c g 6

Dot matrix comparison 7

Interpretation • Regions of similarity appear as diagonal runs of dots • Reverse diagonals (perpendicular to diagonal) indicate inversions • Reverse diagonals crossing diagonals (Xs) indicate palindromes 8

Interpretation • Can link separate diagonals to form alignment with gaps – Each a. a. or base can only be used once • Can't double back – A gap is introduced by each vertical or horizontal skip 9

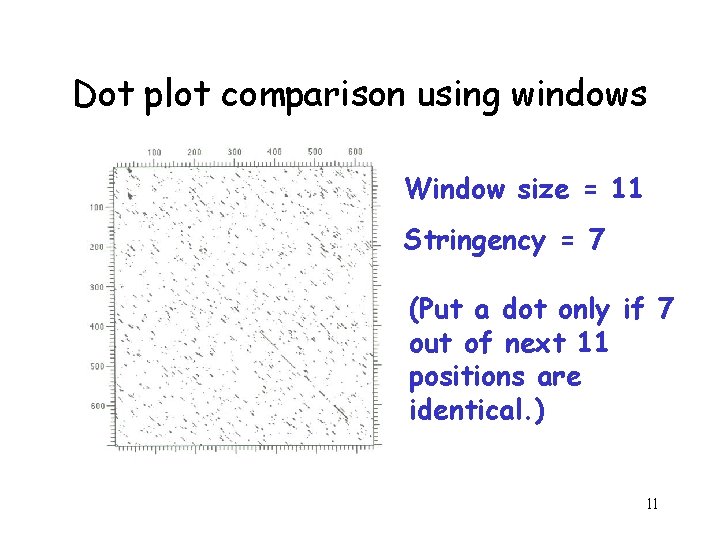
Filtering • Dot matrices for long sequences can be noisy due to insignificant matches • Solution: use a window and a threshold – compare character by character within a window (have to choose window size) – require certain fraction of matches within window in order to display it with a dot 10

Dot plot comparison using windows Window size = 11 Stringency = 7 (Put a dot only if 7 out of next 11 positions are identical. ) 11

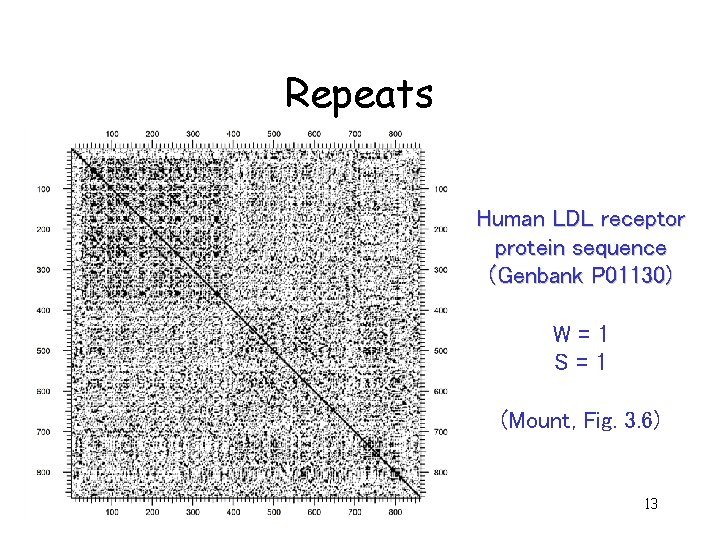
Uses for dot matrices • Aligning two proteins or two nucleic acid sequences • Finding amino acid repeats within a protein by comparing a protein sequence to itself – Repeats appear as a set of diagonal runs stacked vertically and/or horizontally 12

Repeats Human LDL receptor protein sequence (Genbank P 01130) W=1 S=1 (Mount, Fig. 3. 6) 13

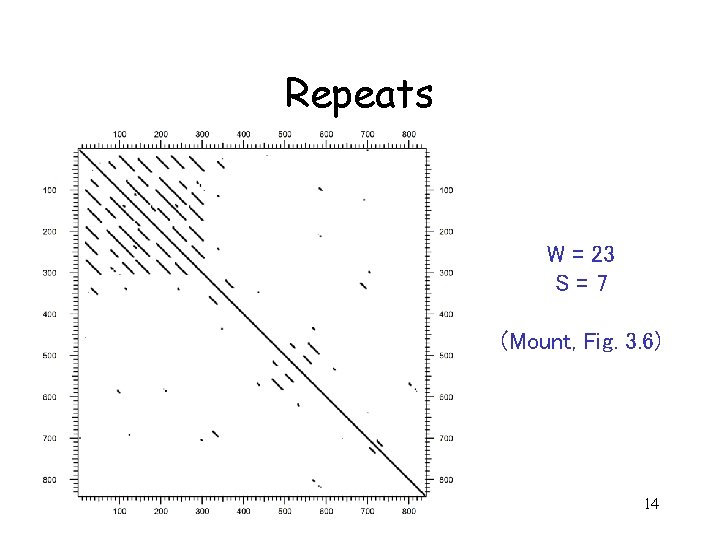
Repeats W = 23 S=7 (Mount, Fig. 3. 6) 14

Using substitution matrices • Dots can have weights • Some matches are rewarded more than others, depending on likelihood – Use PAM or BLOSUM matrix (more on these later) • Put a dot only if a minimum total or average weight is achieved – See Mount, Fig. 3. 5 15