NonNegative Matrix Factorization for Statistical Analysis Stan Young




![Inference Strategy Non-negative matrix factorization is used to group genes. [Unsupervised training. ] The Inference Strategy Non-negative matrix factorization is used to group genes. [Unsupervised training. ] The](https://slidetodoc.com/presentation_image_h2/ea1429ed19016983dc1c6096a671b8cf/image-24.jpg)



- Slides: 32

Non-Negative Matrix Factorization for Statistical Analysis Stan Young Paul Fogel, Doug Hawkins NISS MPC Vienna 11 July 2007 1

Outline 1. Introduction 2. Robust singular value decomposition 3. Non-negative matrix factorization 4. Inference with NMF. 2

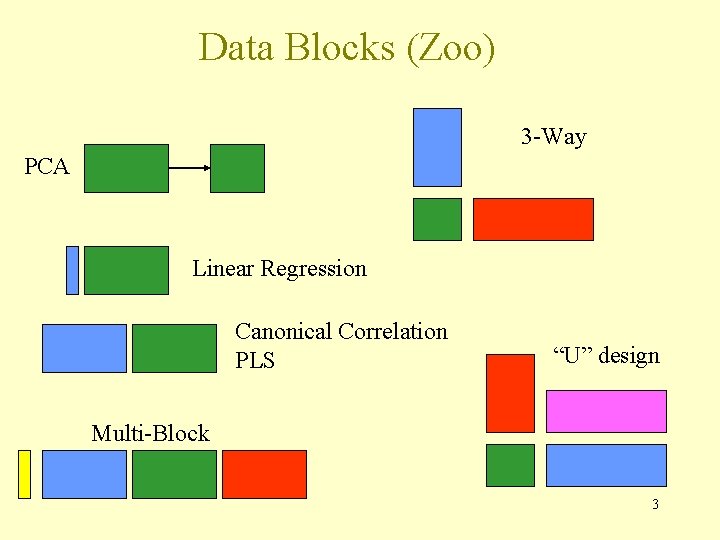
Data Blocks (Zoo) 3 -Way PCA Linear Regression Canonical Correlation PLS “U” design Multi-Block 3

Multiple Blocks 2 -way tables of data are ubiquitous, PCA. One response and a table of predictors is common, linear regression. Multiple 2 -way tables are becoming important: Gene expression, proteomics, metabololomics. 4

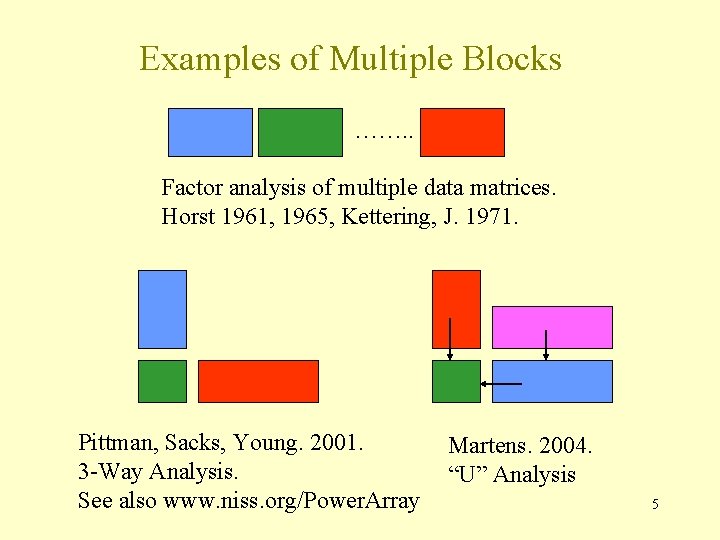
Examples of Multiple Blocks ……. . Factor analysis of multiple data matrices. Horst 1961, 1965, Kettering, J. 1971. Pittman, Sacks, Young. 2001. 3 -Way Analysis. See also www. niss. org/Power. Array Martens. 2004. “U” Analysis 5

Motivating Problem Permute the rows and columns to find patterns. Problems: 1. Large, 10 s to 100 s of rows and 1000 s of columns. 2. Missing data. 3. Outliers. 6

Matrix Factroization Methods 1. Principle component analysis. 2. Singular value decomposition. 3. Non-negative matrix factorization. 4. Independent component analysis. 5. Inference using NMF. 6. Area of active research. 7

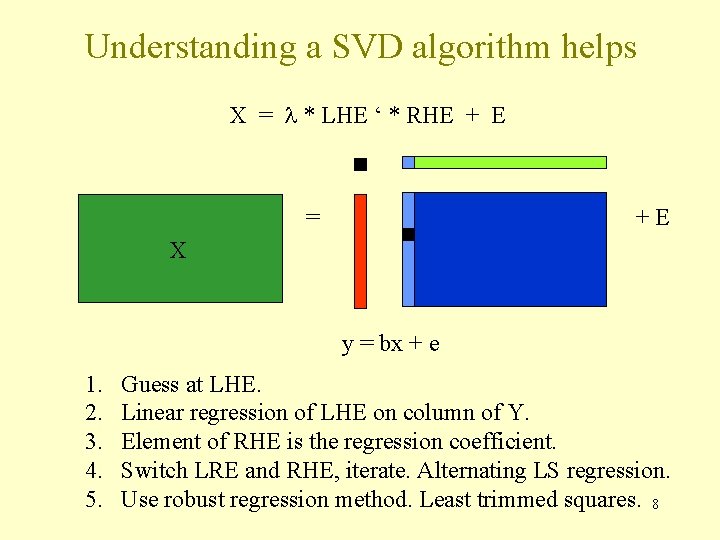
Understanding a SVD algorithm helps X = l * LHE ‘ * RHE + E = +E X y = bx + e 1. 2. 3. 4. 5. Guess at LHE. Linear regression of LHE on column of Y. Element of RHE is the regression coefficient. Switch LRE and RHE, iterate. Alternating LS regression. Use robust regression method. Least trimmed squares. 8

California Versus All Challengers, The 1999 Cabernet Challenge 1. 47 wines judged by 32 wine experts 2. No data for 1 wine/expert 3. One missing data point 4. Results are ranks of wine by each judge 9

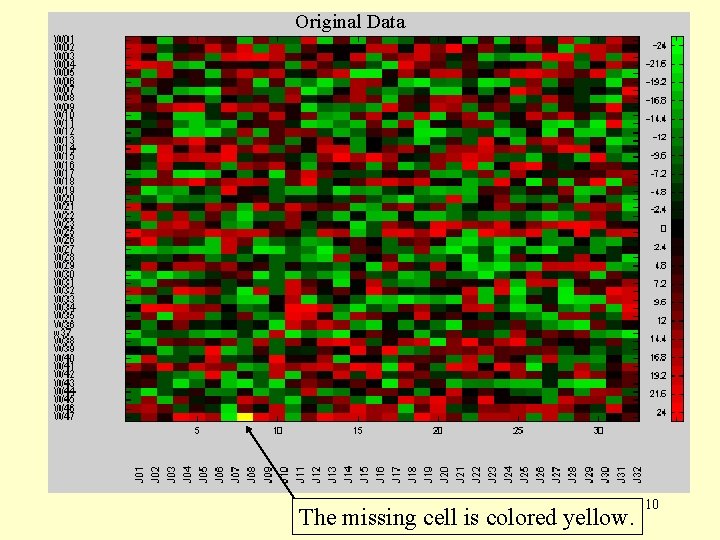
Original Data The missing cell is colored yellow. 10

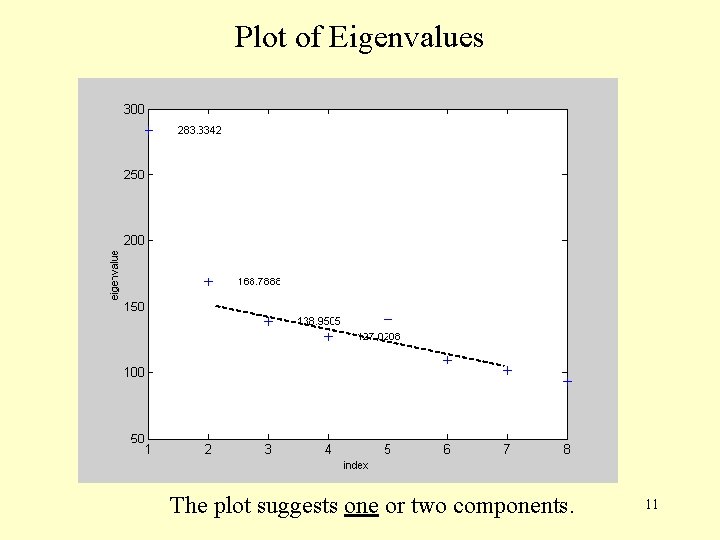
Plot of Eigenvalues The plot suggests one or two components. 11

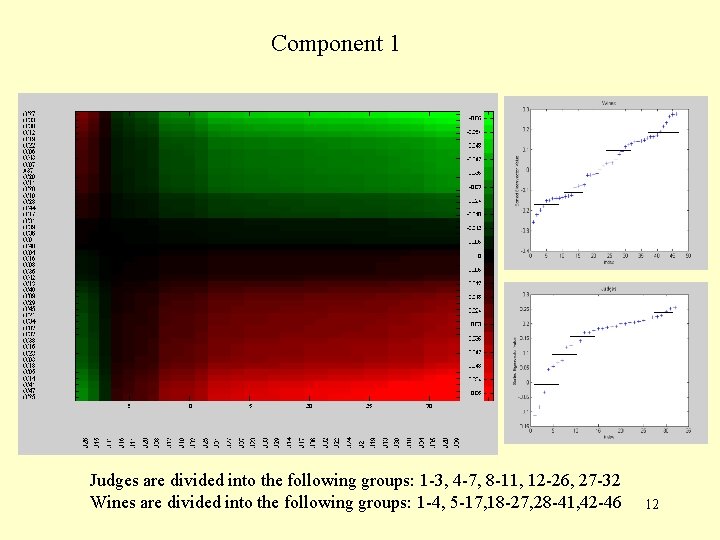
Component 1 Judges are divided into the following groups: 1 -3, 4 -7, 8 -11, 12 -26, 27 -32 Wines are divided into the following groups: 1 -4, 5 -17, 18 -27, 28 -41, 42 -46 12

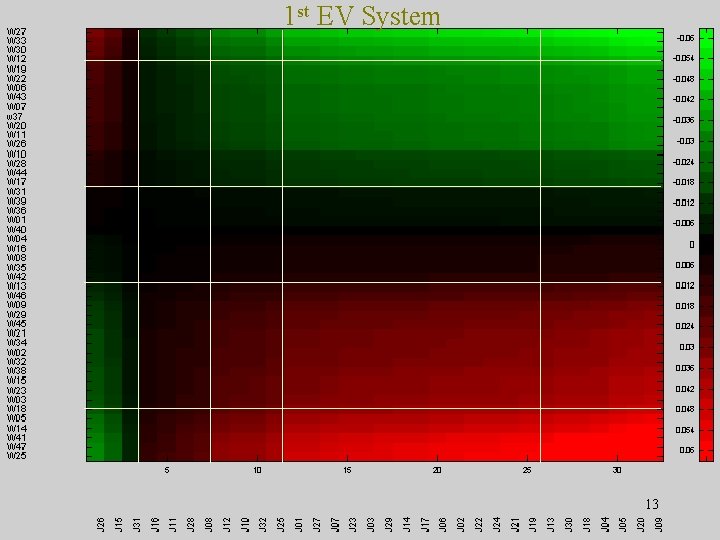
1 st EV System 13

Comments Wine Dataset Most judges were consistent. Three judges are at odds with the rest. The wines divided into 6 classes; six wines group very well. There is an apparent interaction of wines and judges. One eigen system captures most of the variance. 14

Key Matrix Factorization Papers 1. Good (1969) Technometrics – SVD. 2. Liu et al. (2003) PNAS – r. SVD. 3. Lee and Seung (1999) Nature – NMF. 4. Kim and Tidor (2003) Genome Research. 5. Brunet et al. (2004) PNAS – Micro array. 6. Fogel et al. (2007) Bioinformatics. 15

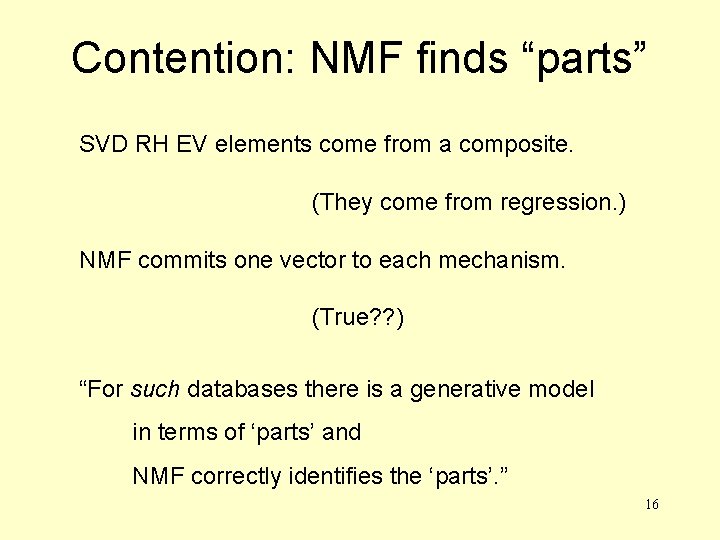
Contention: NMF finds “parts” SVD RH EV elements come from a composite. (They come from regression. ) NMF commits one vector to each mechanism. (True? ? ) “For such databases there is a generative model in terms of ‘parts’ and NMF correctly identifies the ‘parts’. ” 16

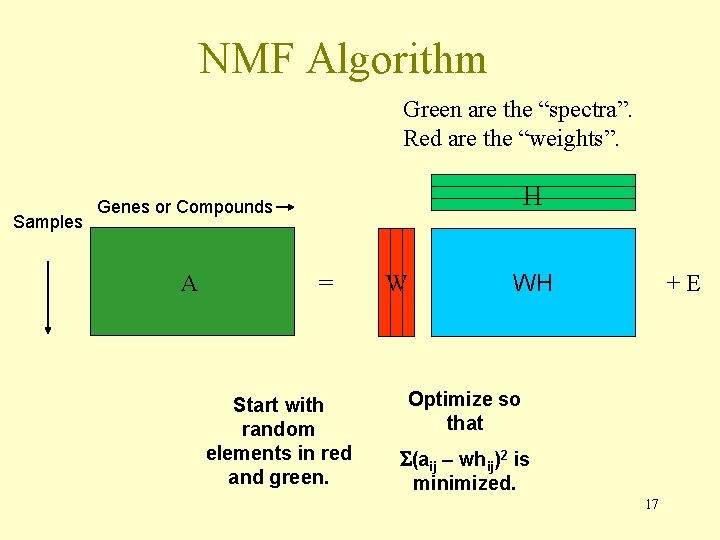
NMF Algorithm Green are the “spectra”. Red are the “weights”. Samples H Genes or Compounds A = Start with random elements in red and green. W WH +E Optimize so that (aij – whij)2 is minimized. 17

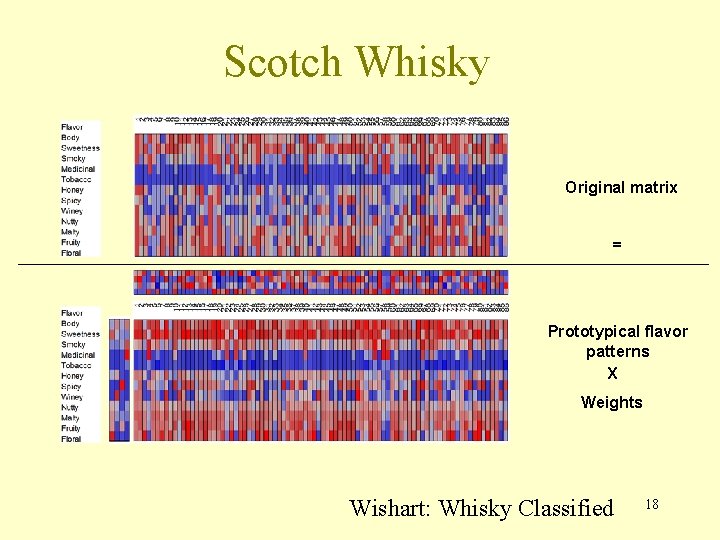
Scotch Whisky Original matrix = Prototypical flavor patterns X Weights Wishart: Whisky Classified 18

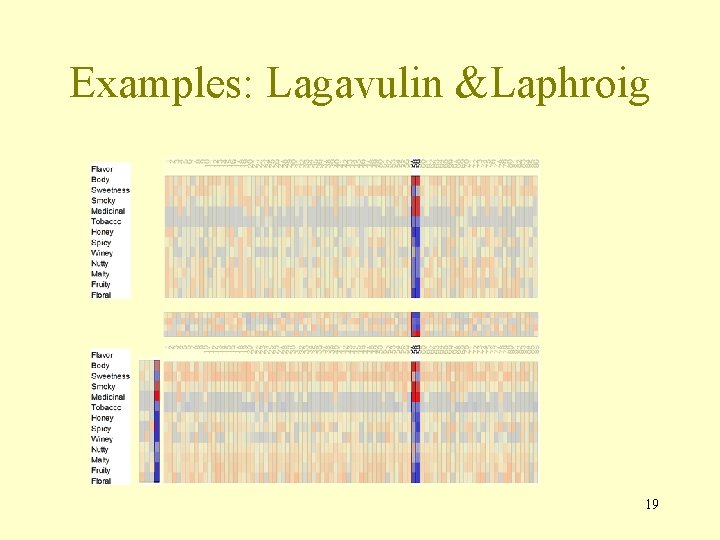
Examples: Lagavulin &Laphroig 19

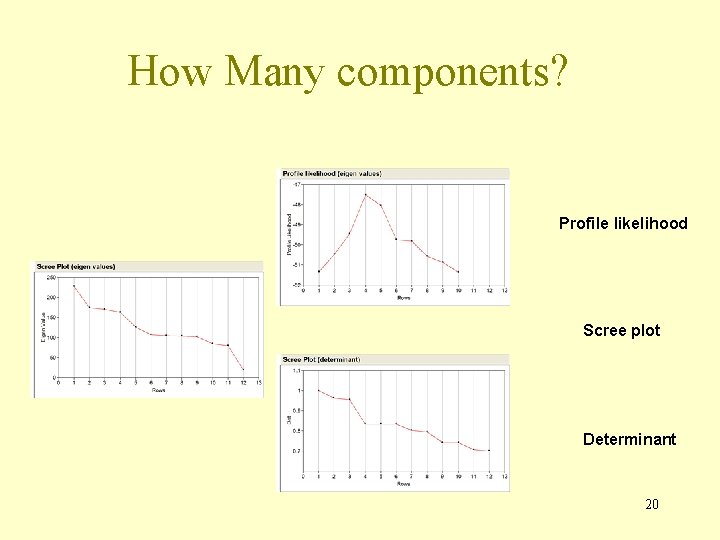
How Many components? Profile likelihood Scree plot Determinant 20

Golub, T. R. et al. (1999) Group AML: acute myeloid leukemia Group ALL: acute lymphoblastic leukemia Subgroup ALL-T: T cell subtypes Subgroup ALL-B: B cell subtypes 21

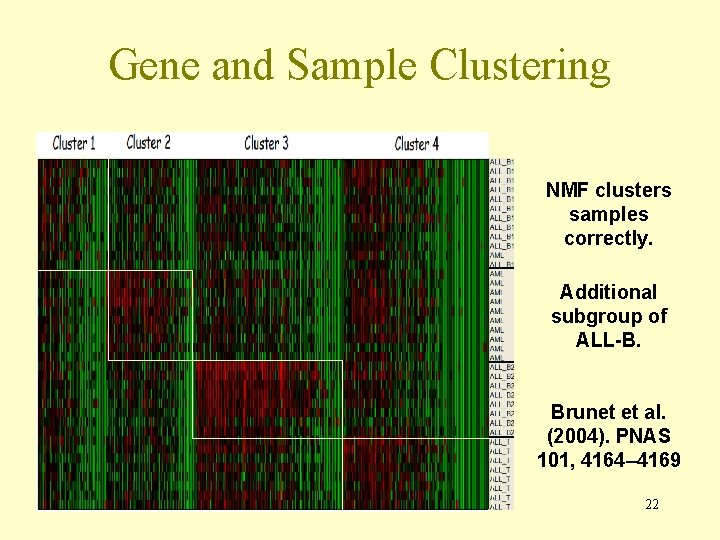
Gene and Sample Clustering NMF clusters samples correctly. Additional subgroup of ALL-B. Brunet et al. (2004). PNAS 101, 4164– 4169 22

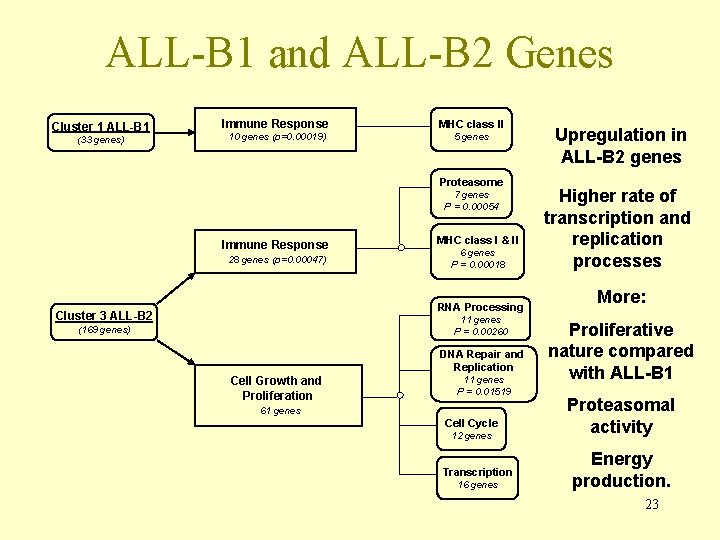
ALL-B 1 and ALL-B 2 Genes Cluster 1 ALL-B 1 (33 genes) Immune Response MHC class II 10 genes (p=0. 00019) 5 genes Proteasome 7 genes P = 0. 00054 Immune Response 28 genes (p=0. 00047) MHC class I & II 6 genes P = 0. 00018 RNA Processing Cluster 3 ALL-B 2 11 genes P = 0. 00260 (169 genes) DNA Repair and Replication Cell Growth and Proliferation 11 genes P = 0. 01519 61 genes Cell Cycle 12 genes Transcription 16 genes Upregulation in ALL-B 2 genes Higher rate of transcription and replication processes More: Proliferative nature compared with ALL-B 1 Proteasomal activity Energy production. 23
![Inference Strategy Nonnegative matrix factorization is used to group genes Unsupervised training The Inference Strategy Non-negative matrix factorization is used to group genes. [Unsupervised training. ] The](https://slidetodoc.com/presentation_image_h2/ea1429ed19016983dc1c6096a671b8cf/image-24.jpg)
Inference Strategy Non-negative matrix factorization is used to group genes. [Unsupervised training. ] The testing alpha is allocated over these groups/vectors. Within each group, genes are tested sequentially; there is no multiple testing adjustment!!!. 24

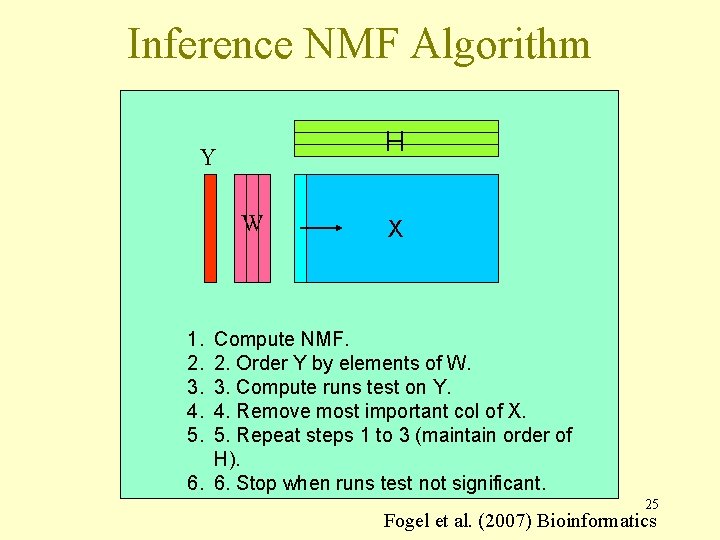
Inference NMF Algorithm H Y W X 1. 2. 3. 4. 5. Compute NMF. 2. Order Y by elements of W. 3. Compute runs test on Y. 4. Remove most important col of X. 5. Repeat steps 1 to 3 (maintain order of H). 6. 6. Stop when runs test not significant. 25 Fogel et al. (2007) Bioinformatics

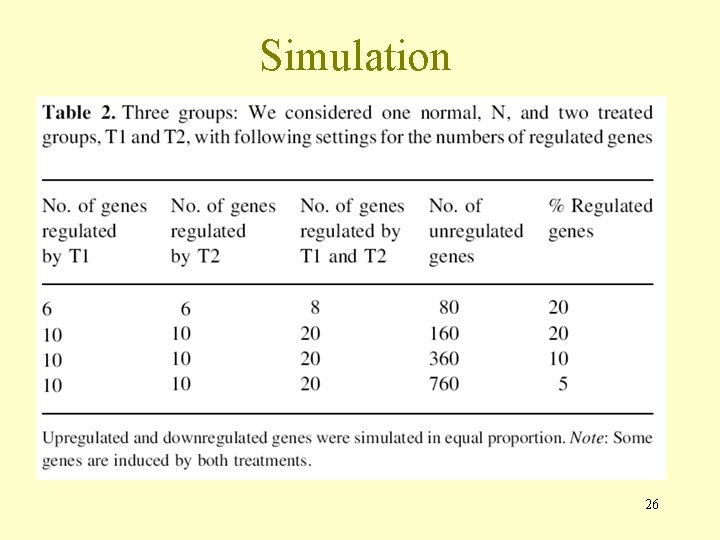
Simulation 26

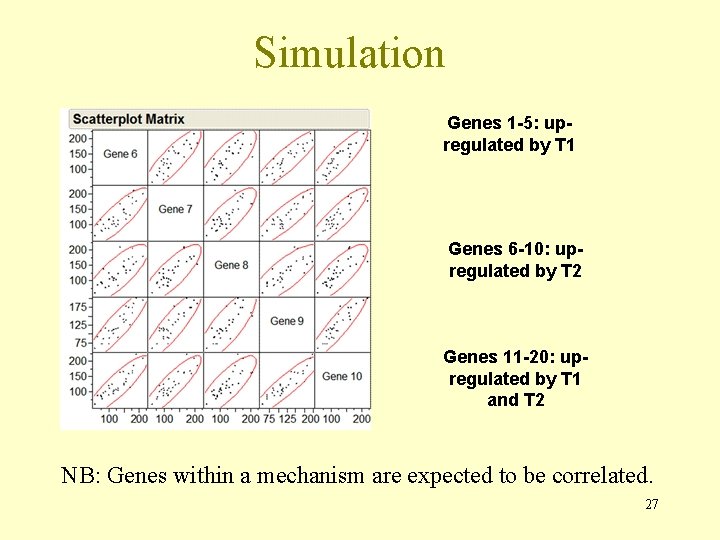
Simulation Genes 1 -5: upregulated by T 1 Genes 6 -10: upregulated by T 2 Genes 11 -20: upregulated by T 1 and T 2 NB: Genes within a mechanism are expected to be correlated. 27

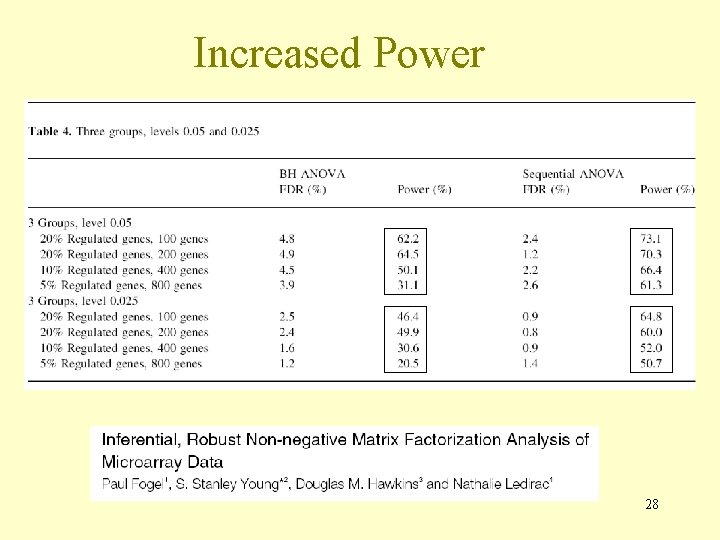
Increased Power 28

General Comments SVD is the basis for most linear statistical systems. Non-negative matrix factorization will become increasingly important. Data sets are getting much bigger. We are seeing complex, multi-block data sets. We need good software to expand data analysis. 29

ir. MF Summary 1. NMF is an attractive alternative to SVD. 2. Mechanisms appear to be captured in separate vectors. 3. Genes can be tested sequentially within a right vectors. 4. Many statistical problems are open for research. 30

More Information NMF program and papers at www. niss. org/ir. MF Stan Young : young@niss. org Paul Fogel : paul. fogel@wanadoo. fr 31

More Information NMF Code and papers at www. niss. org/ir. MF Analysis of “L” design: www. niss. org/Power. Array NMF roundtable luncheon at JSM 2007. See also: www. niss. org/Power. MV http: //eccr. stat. ncsu. edu/ young@niss. org 32