Preprocessing HCS data using Nonnegative Matrix Factorization S




![Optimization Criteria Minimize (xij – whij)2 [xij log (xij / whij) + (Xij– whij)] Optimization Criteria Minimize (xij – whij)2 [xij log (xij / whij) + (Xij– whij)]](https://slidetodoc.com/presentation_image_h/25a0400cb767ebf83068c233687ddf56/image-13.jpg)




- Slides: 35

Pre-processing HCS data using Non-negative Matrix Factorization S. Stanley Young National Institute of Statistical Sciences MBSW, Muncie 19 May 2009 1

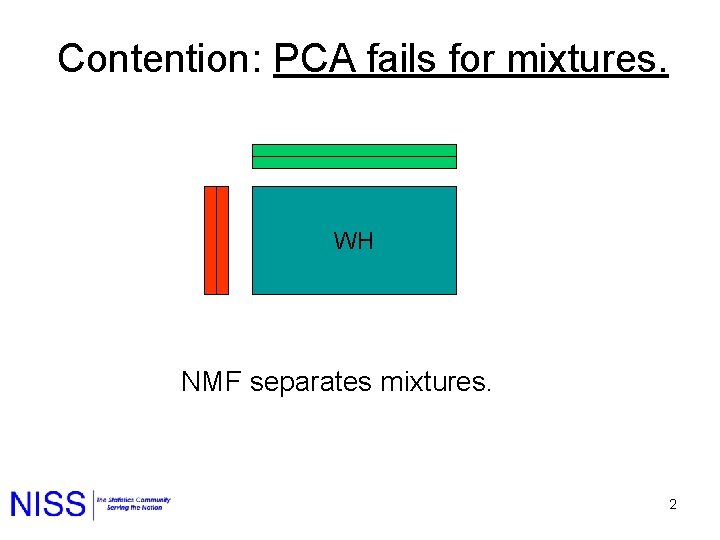
Contention: PCA fails for mixtures. WH NMF separates mixtures. 2

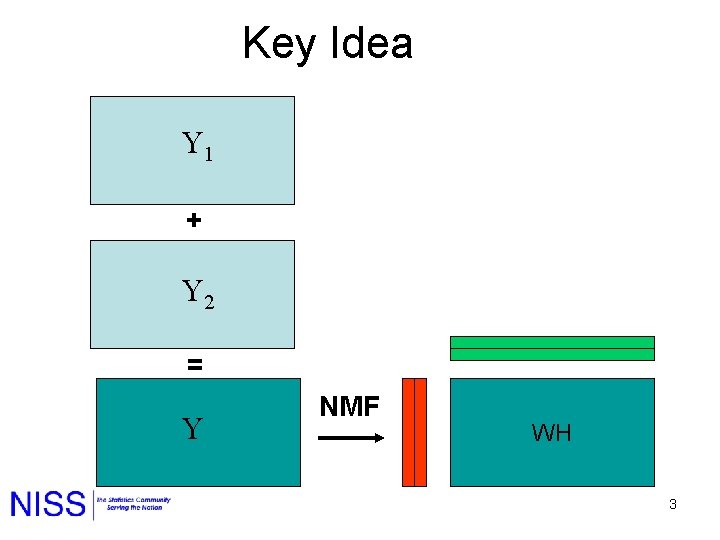
Key Idea Y 1 + Y 2 = Y NMF WH 3

Outline 1. 2. 3. 4. 5. 6. Basics of HCS Non-negative matrix factorization The experiment/simulation NMF versus PCA Analysis of experiment Literature 4

Basic Experimental Setup 1. Multiple cells within a well. 2. Treat the wells. 3. Image each well. 4. Image analysis yields a vector for each cell. 5. Summarize the well. 6. Analyze the well summaries. 5

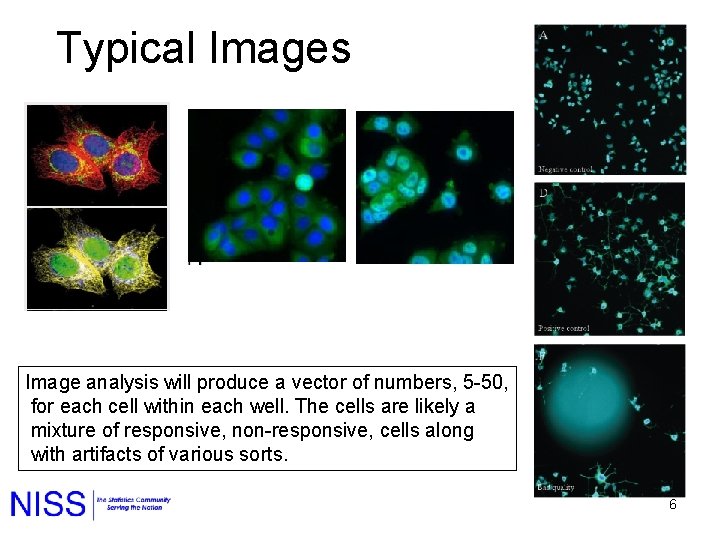
Typical Images Image analysis will produce a vector of numbers, 5 -50, for each cell within each well. The cells are likely a mixture of responsive, non-responsive, cells along with artifacts of various sorts. 6

Equipment 7

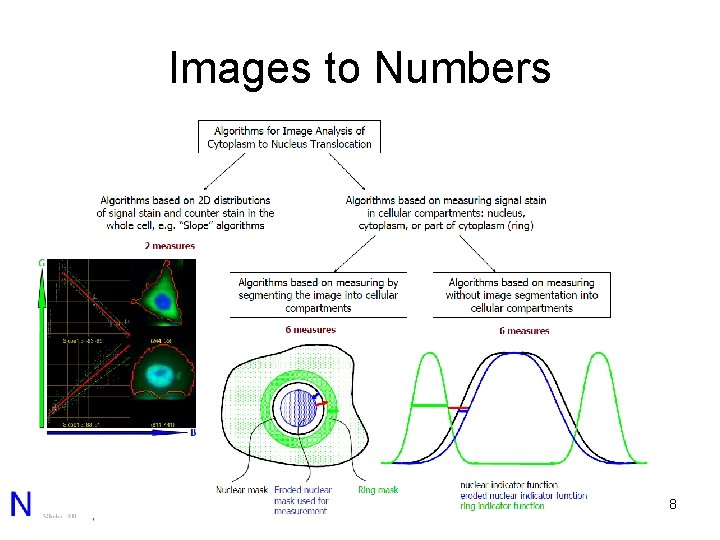
Images to Numbers 8

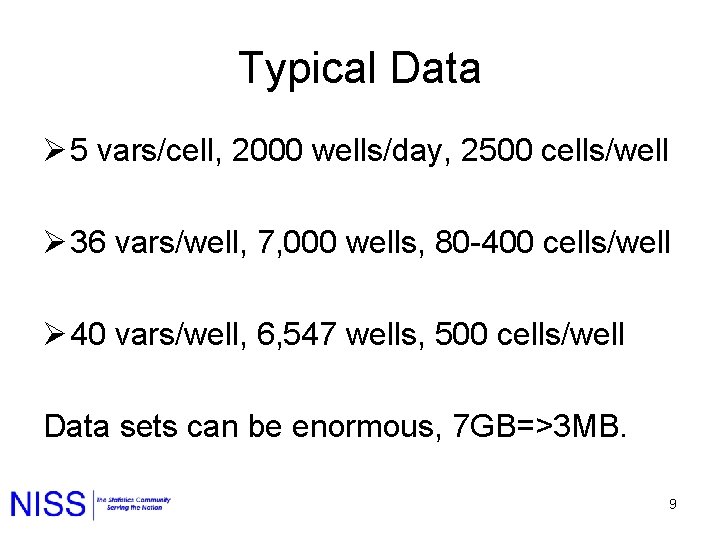
Typical Data Ø 5 vars/cell, 2000 wells/day, 2500 cells/well Ø 36 vars/well, 7, 000 wells, 80 -400 cells/well Ø 40 vars/well, 6, 547 wells, 500 cells/well Data sets can be enormous, 7 GB=>3 MB. 9

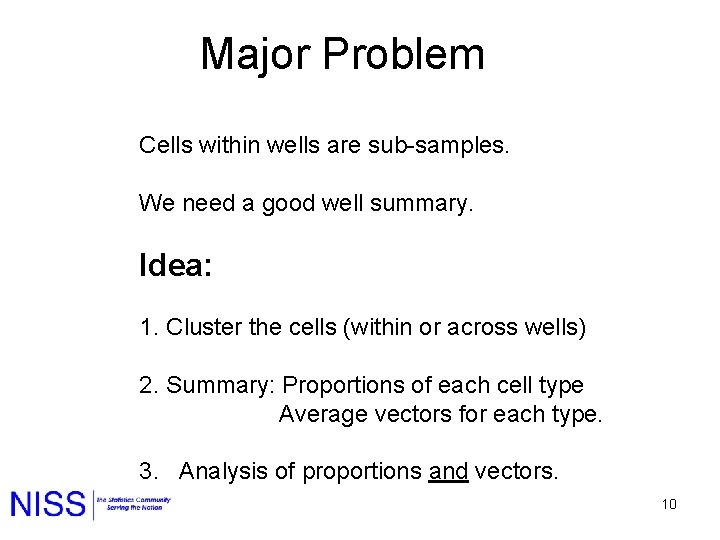
Major Problem Cells within wells are sub-samples. We need a good well summary. Idea: 1. Cluster the cells (within or across wells) 2. Summary: Proportions of each cell type Average vectors for each type. 3. Analysis of proportions and vectors. 10

Matrix Factorization Methods 1. Principle component analysis. 2. Singular value decomposition. 3. Non-negative matrix factorization. 4. Independent component analysis. NMF is an area of active research. 11

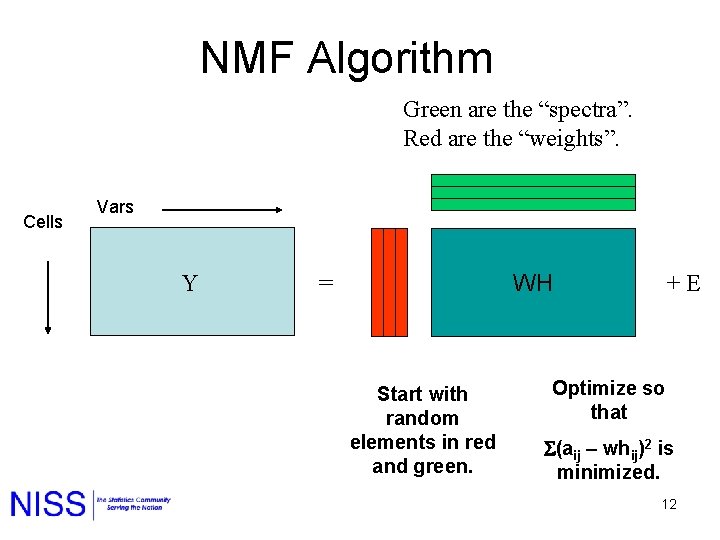
NMF Algorithm Green are the “spectra”. Red are the “weights”. Cells Vars Y WH = Start with random elements in red and green. + E Optimize so that (aij – whij)2 is minimized. 12
![Optimization Criteria Minimize xij whij2 xij log xij whij Xij whij Optimization Criteria Minimize (xij – whij)2 [xij log (xij / whij) + (Xij– whij)]](https://slidetodoc.com/presentation_image_h/25a0400cb767ebf83068c233687ddf56/image-13.jpg)
Optimization Criteria Minimize (xij – whij)2 [xij log (xij / whij) + (Xij– whij)] 13

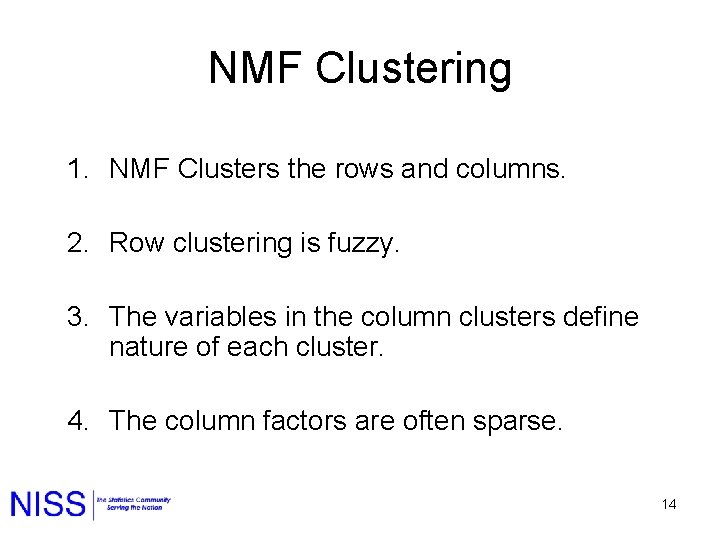
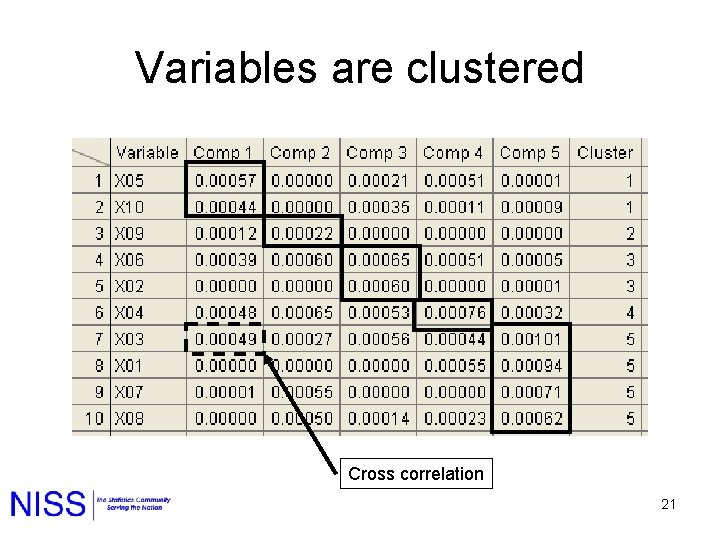
NMF Clustering 1. NMF Clusters the rows and columns. 2. Row clustering is fuzzy. 3. The variables in the column clusters define nature of each cluster. 4. The column factors are often sparse. 14

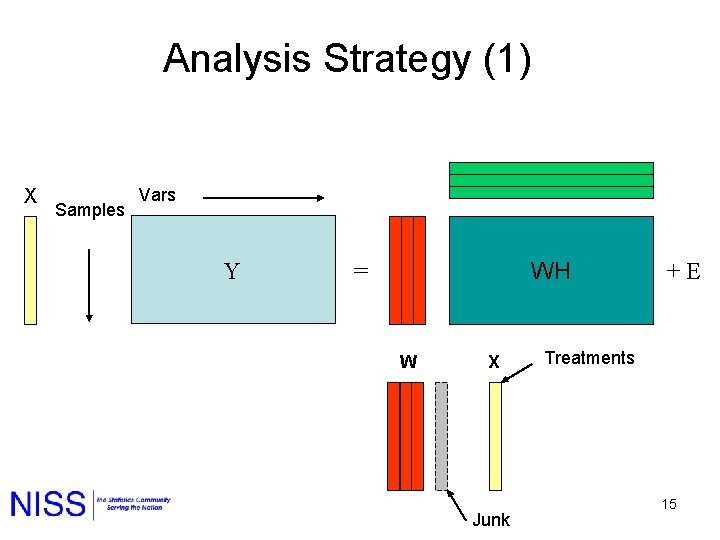
Analysis Strategy (1) X Samples Vars Y WH = W X Junk + E Treatments 15

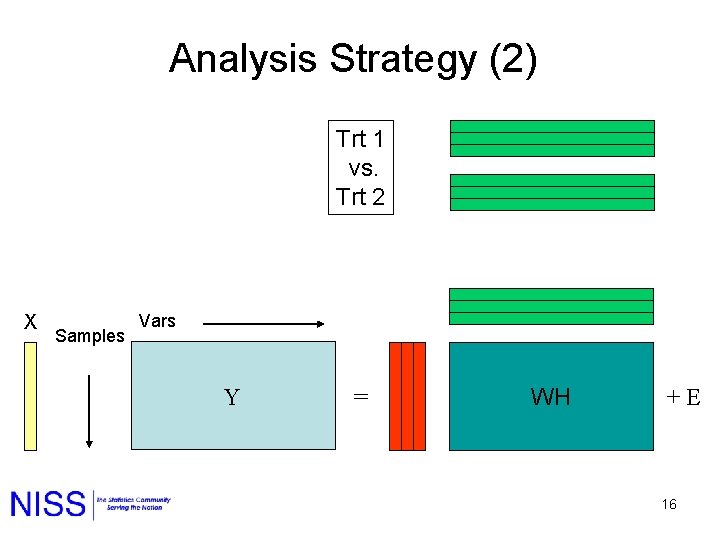
Analysis Strategy (2) Trt 1 vs. Trt 2 X Samples Vars Y = WH + E 16

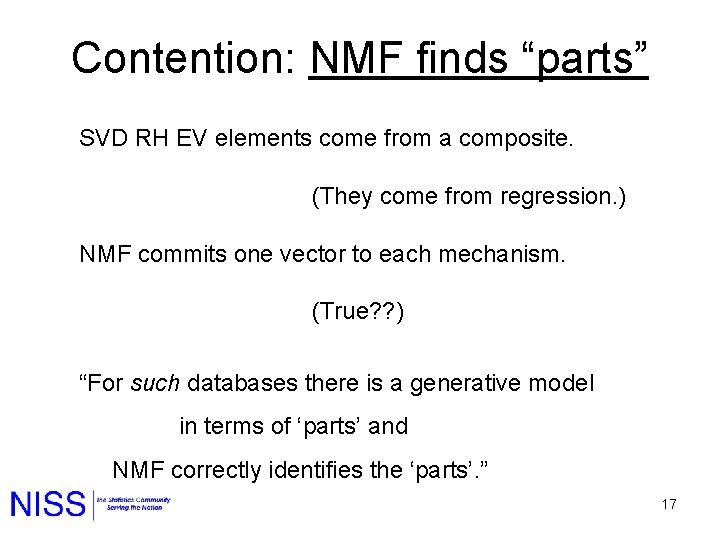
Contention: NMF finds “parts” SVD RH EV elements come from a composite. (They come from regression. ) NMF commits one vector to each mechanism. (True? ? ) “For such databases there is a generative model in terms of ‘parts’ and NMF correctly identifies the ‘parts’. ” 17

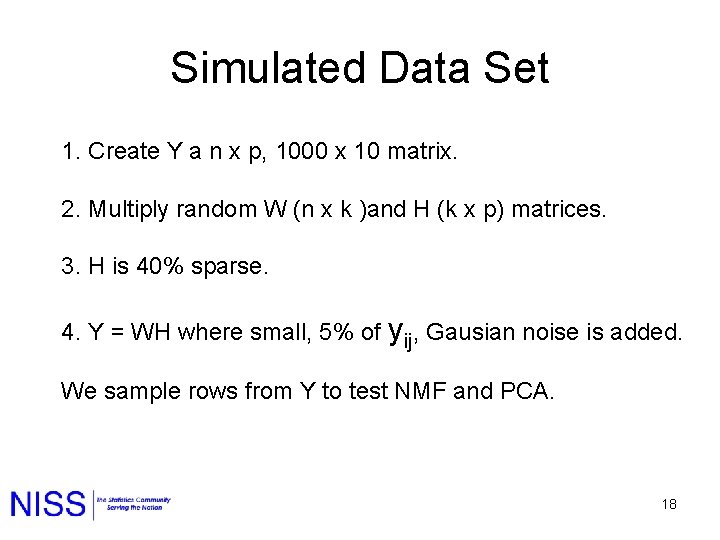
Simulated Data Set 1. Create Y a n x p, 1000 x 10 matrix. 2. Multiply random W (n x k )and H (k x p) matrices. 3. H is 40% sparse. 4. Y = WH where small, 5% of yij, Gausian noise is added. We sample rows from Y to test NMF and PCA. 18

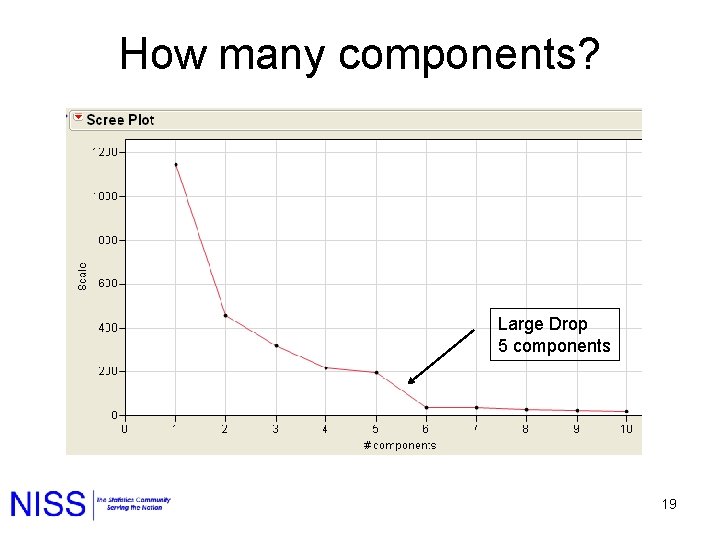
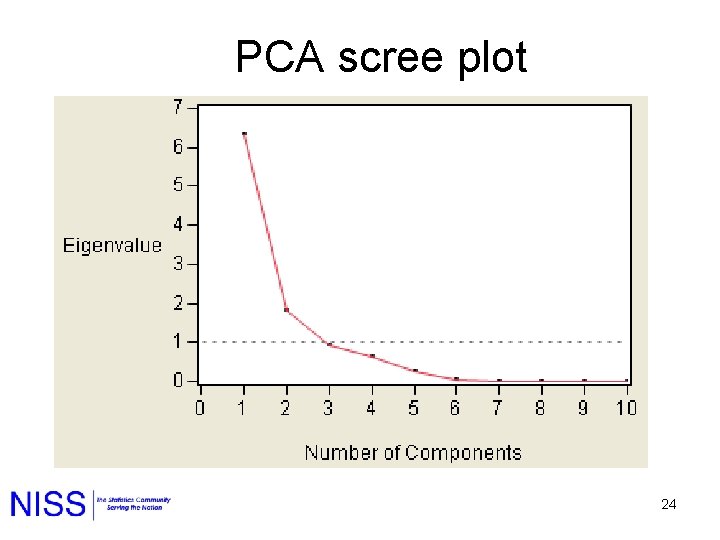
How many components? Large Drop 5 components 19

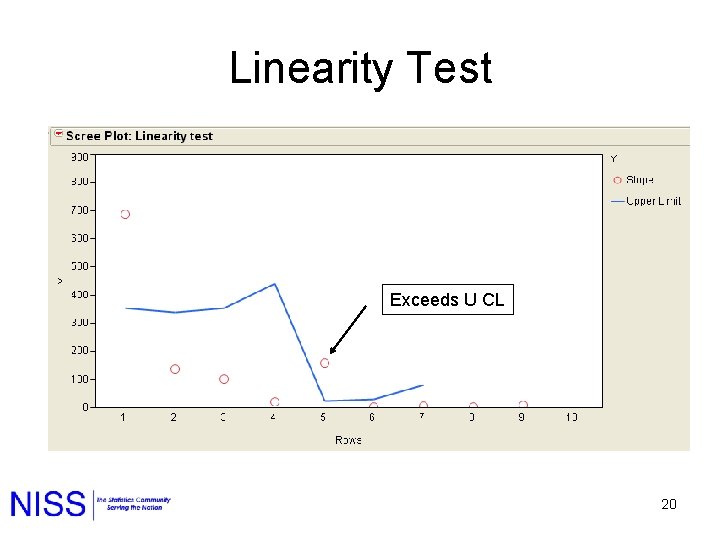
Linearity Test Exceeds U CL 20

Variables are clustered Cross correlation 21

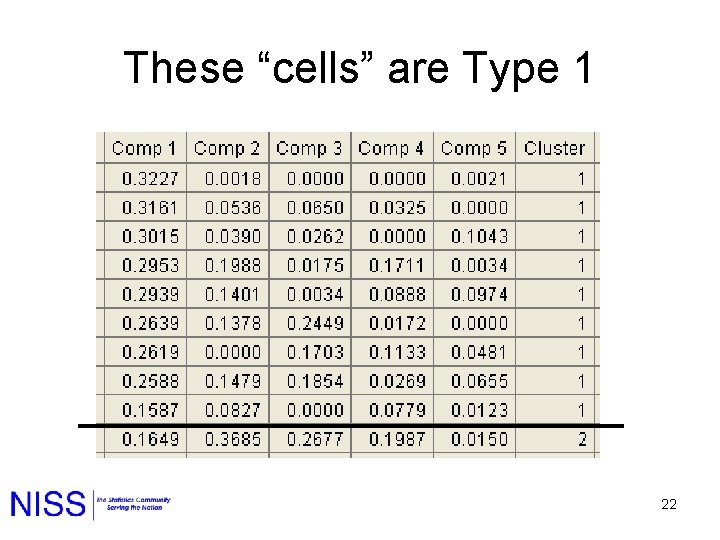
These “cells” are Type 1 22

NMF Summary 1. NMF honors the non-negative nature of the data. 2. Variables are grouped. 3. Samples are clustered. 4. The clustering is “fuzzy”. 5. Sparseness makes interpretation easier. 23

PCA scree plot 24

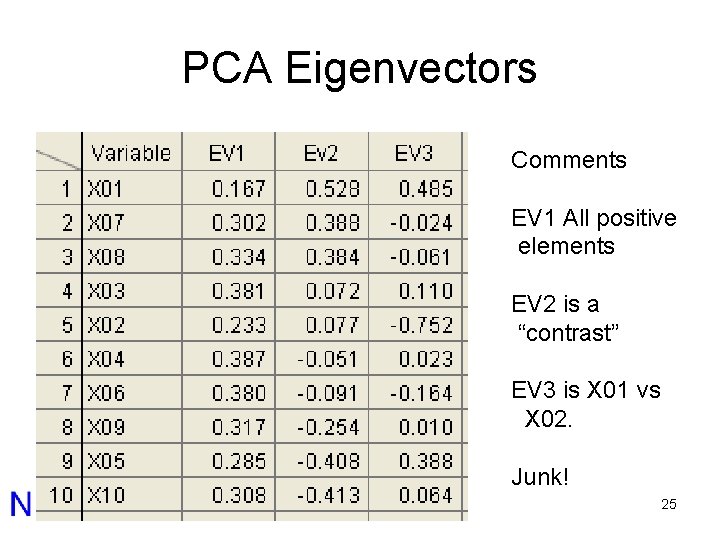
PCA Eigenvectors Comments EV 1 All positive elements EV 2 is a “contrast” EV 3 is X 01 vs X 02. Junk! 25

PCA Summary 1. 2 or 3 components. 2. 1 st component is general sum. 3. 2 nd component is a contrast. 4. Variables do not group cleanly. 26

General Comments SVD is the basis for most linear statistical methods. PCA is terrible for mixtures. Where NMF can replace SVD, it will become increasingly important. NMF can be extended to complex, multi-block data sets. We need good software to make NMF accessible. 27

Matrix Factorization References 1. Good (1969) Technometrics – SVD. 2. Liu et al. (2003) PNAS – r. SVD. 3. Lee and Seung (1999) Nature – NMF. 4. Brunet et al. (2004) PNAS – Micro array. 5. Fogel et al. (2007) Bioinformatics – Micro array. 28

HCS References Kümmel A, Gabriel D, Parker CN, Bender A. (2008) Computational methods to support high-content screening: from compound selection and data analysis to postulating target hypotheses. Expert Opin. Drug Discovery 4, 1 -9. Low J, Huang S, et al. (2008) High-content imaging characterization of cell cycle therapeutics through in vitro and in vivo subpopulation analysis. Mol Cancer Ther 7, 2455 -2463. Young DW, Bender A, et al. (2008) Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nature Chemical Biology 4, 59 -68. Dürr O, Duval D, et al. (2007) Robust hit identification by quality assurance and multivariate data analysis of a high-content, cell-based assay. Journal of Biomolecular Screening 12, 1042 -1049. 29

NMF Software 1. ir. MF: inferential, robust Matrix Factorization (JMP script) http: //www. niss. org/ir. MF/ 2. Array Studio: Software package which provides state of the art statistics and visualization for the analysis of high dimensional quantification data (e. g. Microarray or Taqman data). Omic. Soft Corporation www. omicsoft. com 3. Bio. NMF – free 30

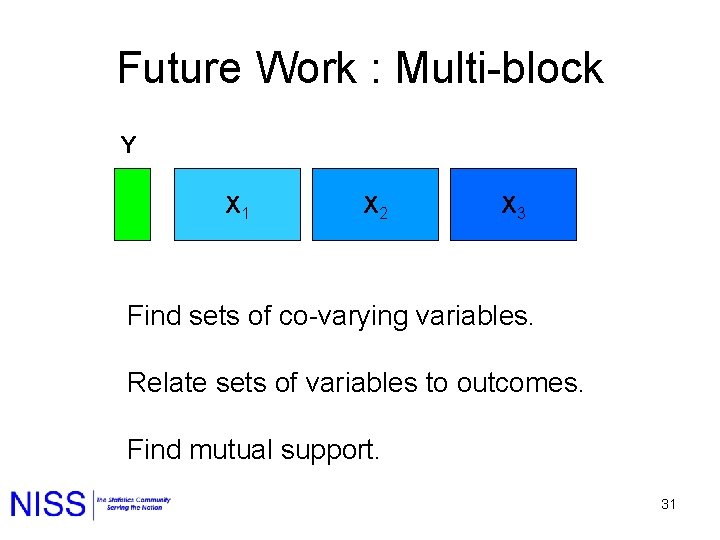
Future Work : Multi-block Y X 1 X 2 X 3 Find sets of co-varying variables. Relate sets of variables to outcomes. Find mutual support. 31

Co-Workers Stan Young, young@niss. org stan. young@omicsoft. com Paul Fogel, paul_fogel@hotmail. com George Luta, gl 77@georgetown. edu Joe Maisog, bravas 02@gmail. com 32

Useful Information Array Studio, www. omicsoft. com ir. MF, www. niss. org/ir. MF Google (Bio. NMF) 33

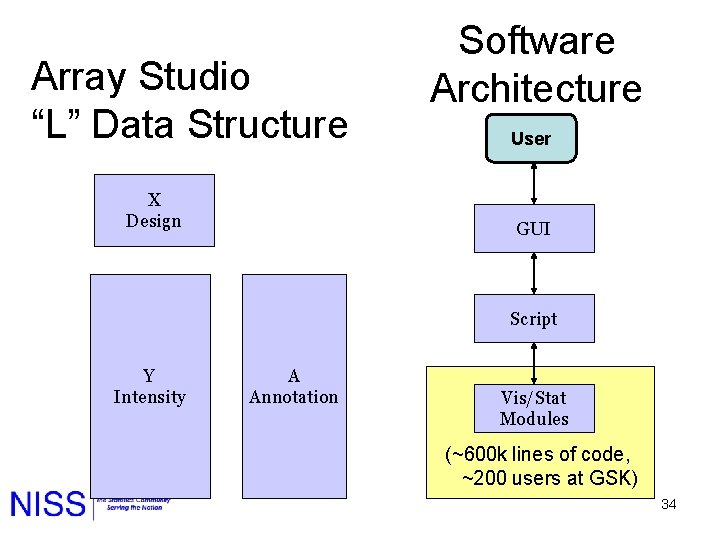
Array Studio “L” Data Structure X Design Software Architecture User GUI Script Y Intensity A Annotation Vis/Stat Modules (~600 k lines of code, ~200 users at GSK) 34

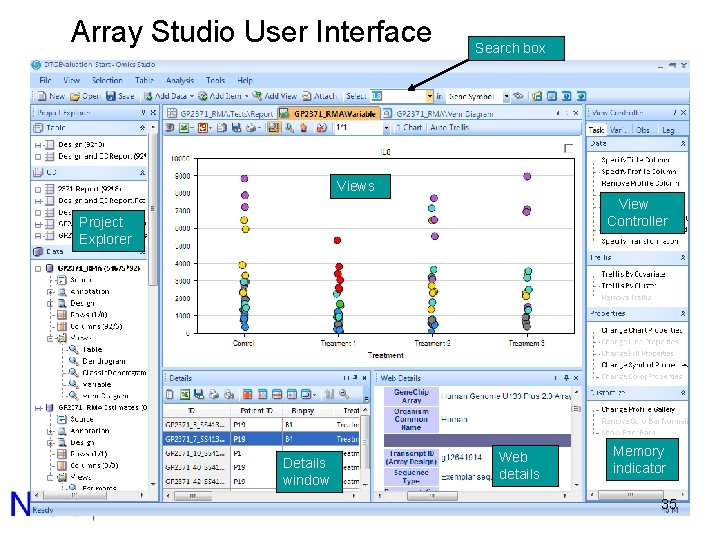
Array Studio User Interface Search box Views View Controller Project Explorer Details window Web details Memory indicator 35