Naming shaming sequencing the Apicomplexans Outline of the







- Slides: 24

Naming & shaming & sequencing the ‘Apicomplexans’

Outline of the presentation • Eukaryotic pathogen sequencing in Sanger • ‘Apicomplexans’ – ‘the culprits’! • Sequencing Plasmodium in Sanger • Plasmodium biology: what makes P. falciparum unique • Comparitive genomics • Insights from other Apicomplexan genomes (theileria)

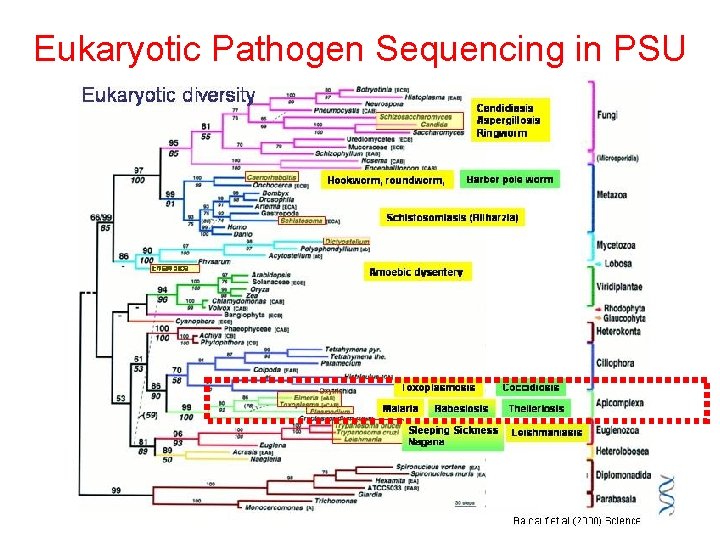
Eukaryotic Pathogen Sequencing in PSU

Why ‘Apicomplexans’?

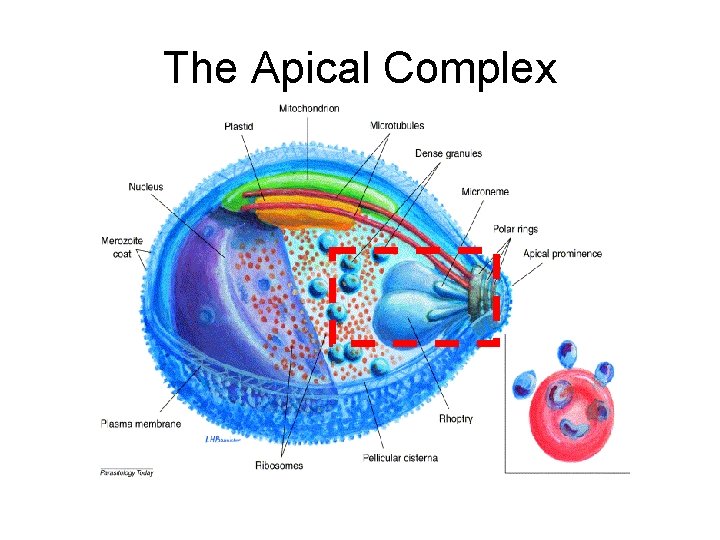
The Apical Complex

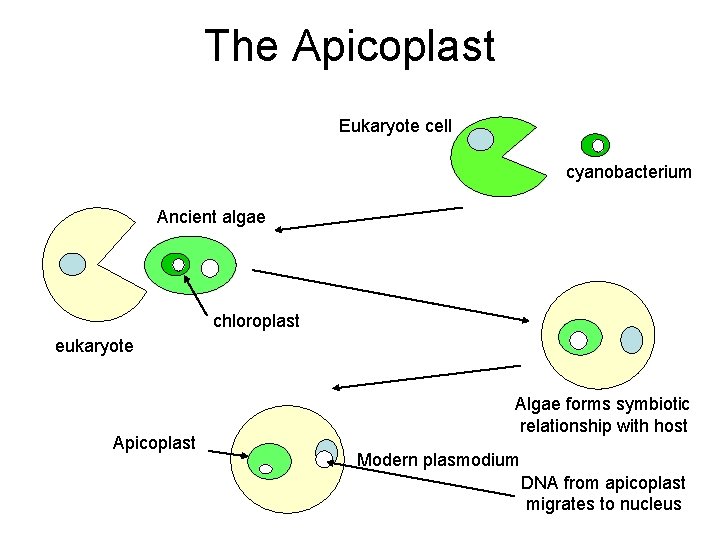
The Apicoplast Eukaryote cell cyanobacterium Ancient algae chloroplast eukaryote Apicoplast Algae forms symbiotic relationship with host Modern plasmodium DNA from apicoplast migrates to nucleus


Naming & shaming the ‘Apicomplexans’

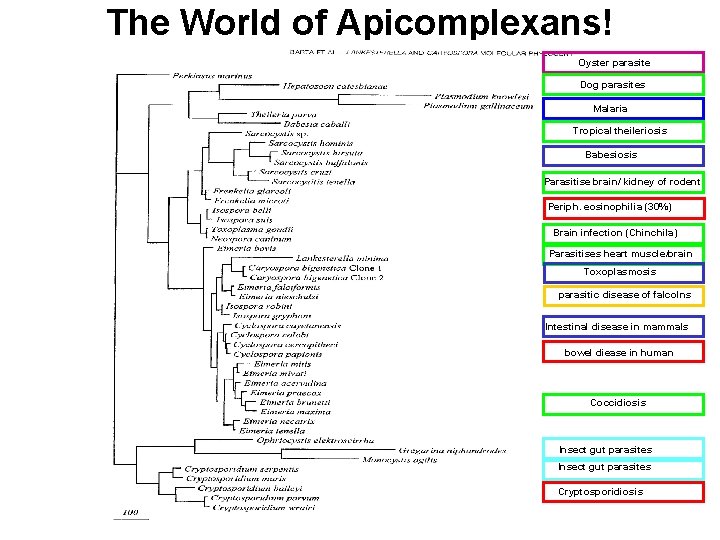
The World of Apicomplexans! Oyster parasite Dog parasites Malaria Tropical theileriosis Babesiosis Parasitise brain/ kidney of rodent Periph. eosinophilia (30%) Brain infection (Chinchila) Parasitises heart muscle/brain Toxoplasmosis parasitic disease of falcolns Intestinal disease in mammals bowel diease in human Coccidiosis Insect gut parasites Cryptosporidiosis

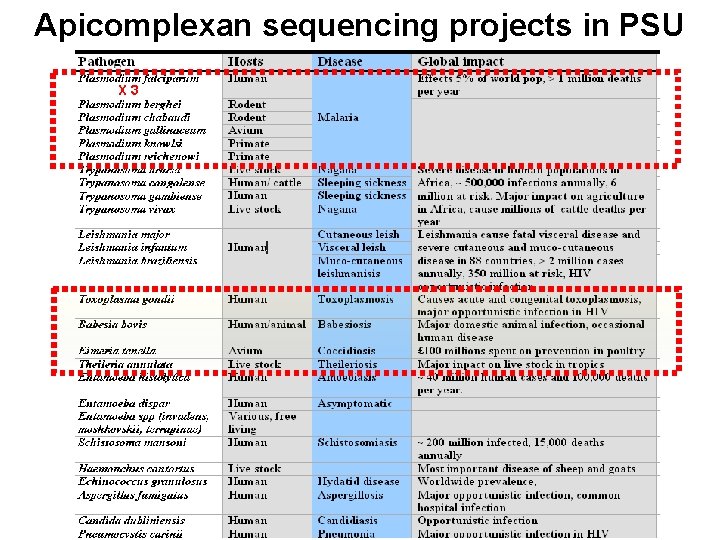
Apicomplexan Euk. pathogen sequencing projects in PSU X 3

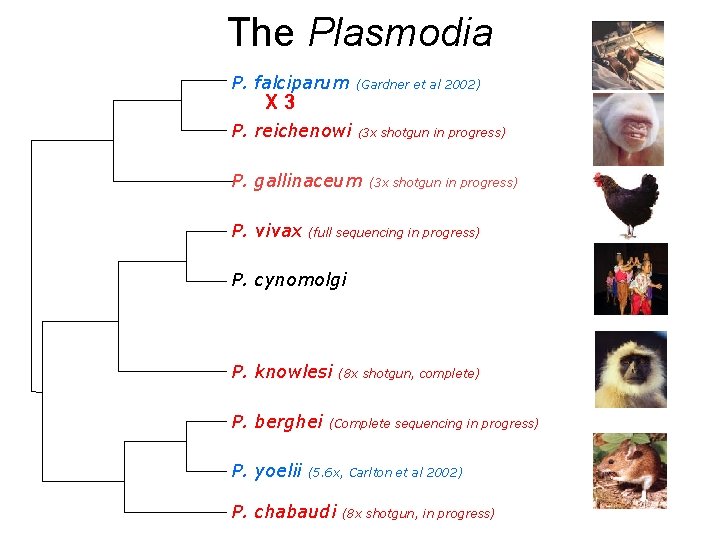
The Plasmodia P. falciparum (Gardner et al 2002) P. reichenowi (3 x X 3 P. gallinaceum P. vivax shotgun in progress) (3 x shotgun in progress) (full sequencing in progress) P. cynomolgi P. knowlesi P. berghei P. yoelii (8 x shotgun, complete) (Complete sequencing in progress) (5. 6 x, Carlton et al 2002) P. chabaudi (8 x shotgun, in progress)

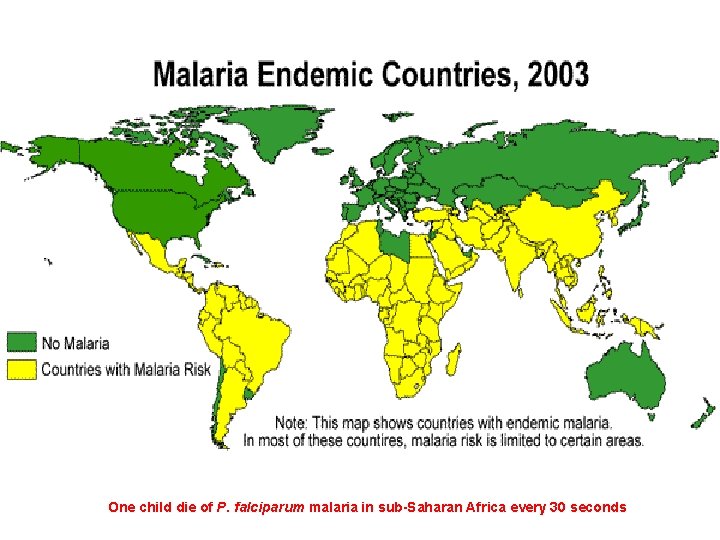
One child die of P. falciparum malaria in sub-Saharan Africa every 30 seconds

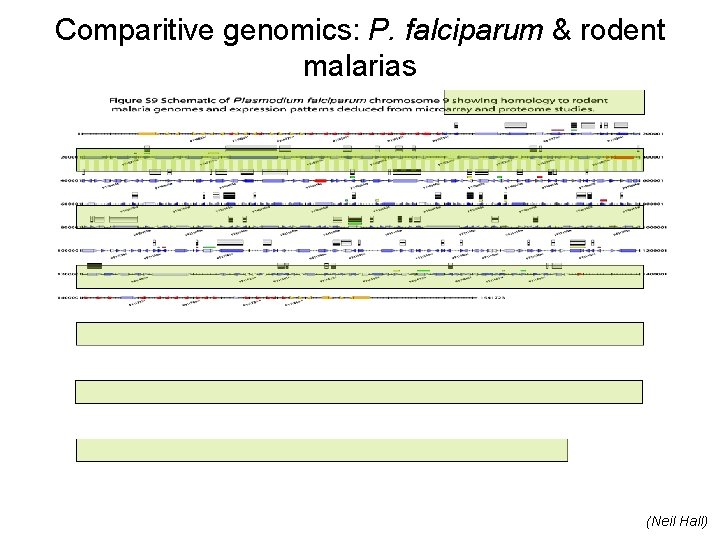
Comparitive genomics: P. falciparum & rodent malarias (Neil Hall)

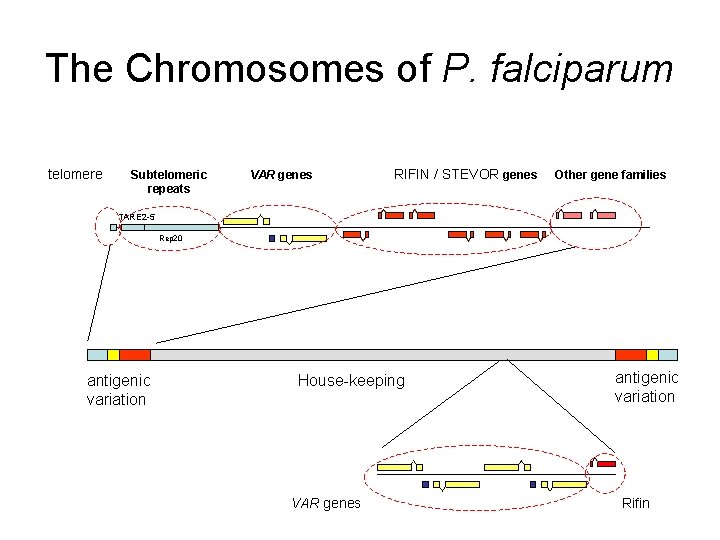
The Chromosomes of P. falciparum telomere Subtelomeric repeats VAR genes RIFIN / STEVOR genes Other gene families TARE 2 -5 Rep 20 antigenic variation House-keeping VAR genes antigenic variation Rifin

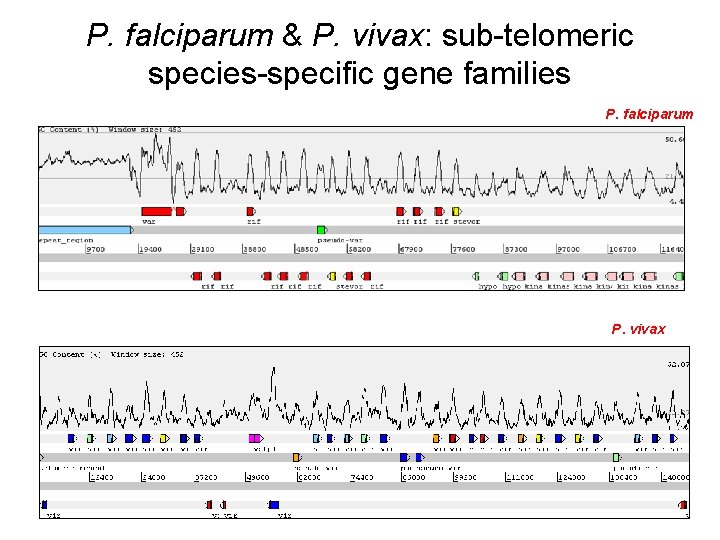
P. falciparum & P. vivax: sub-telomeric species-specific gene families P. falciparum P. vivax

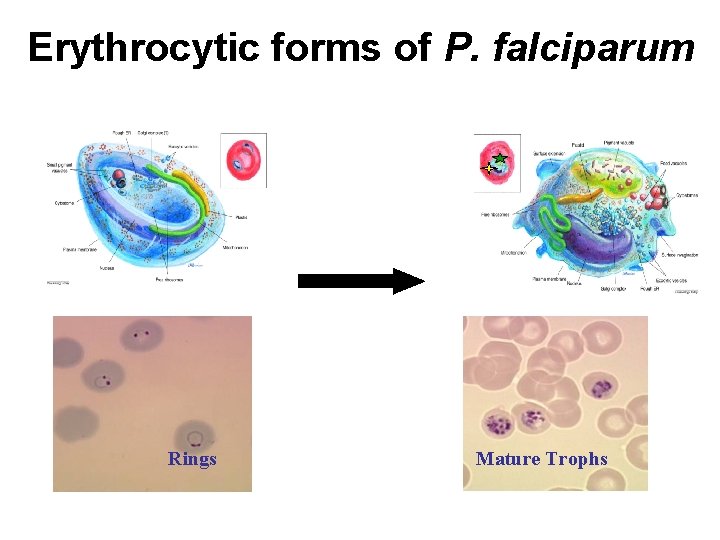
Erythrocytic forms of P. falciparum Rings Mature Trophs

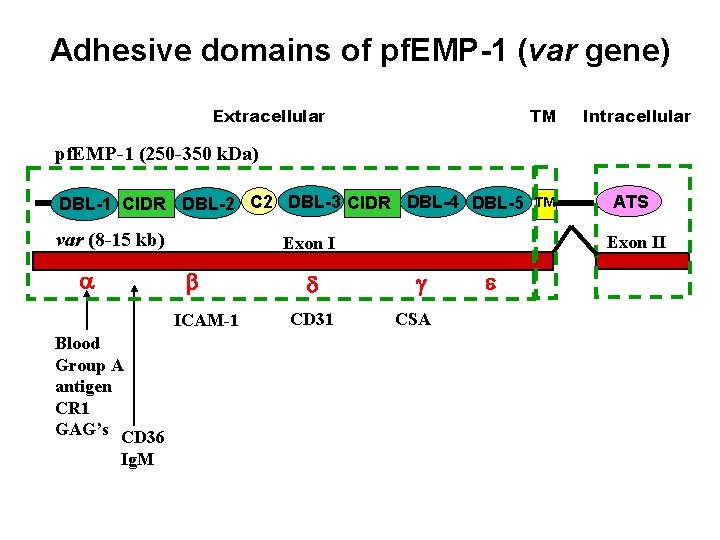
Adhesive domains of pf. EMP-1 (var gene) Extracellular TM Intracellular pf. EMP-1 (250 -350 k. Da) DBL-1 CIDR DBL-2 C 2 DBL-3 CIDR DBL-4 DBL-5 var (8 -15 kb) a ICAM-1 Blood Group A antigen CR 1 GAG’s CD 36 Ig. M d CD 31 ATS Exon II Exon I b TM g CSA e

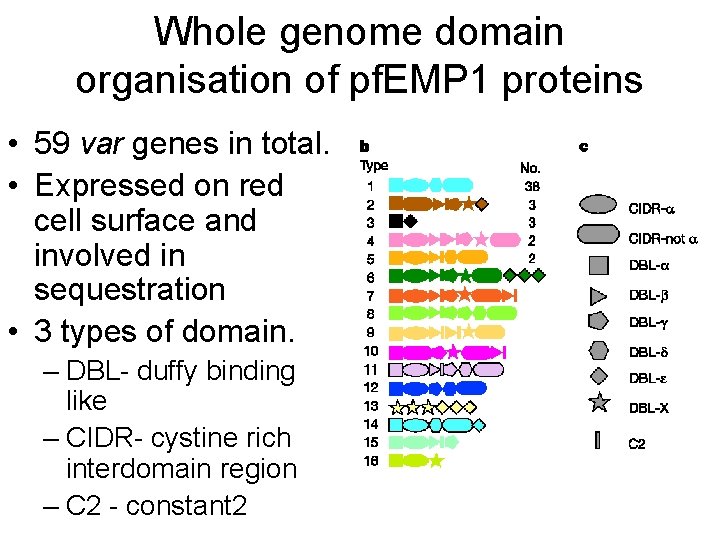
Whole genome domain organisation of pf. EMP 1 proteins • 59 var genes in total. • Expressed on red cell surface and involved in sequestration • 3 types of domain. – DBL- duffy binding like – CIDR- cystine rich interdomain region – C 2 - constant 2

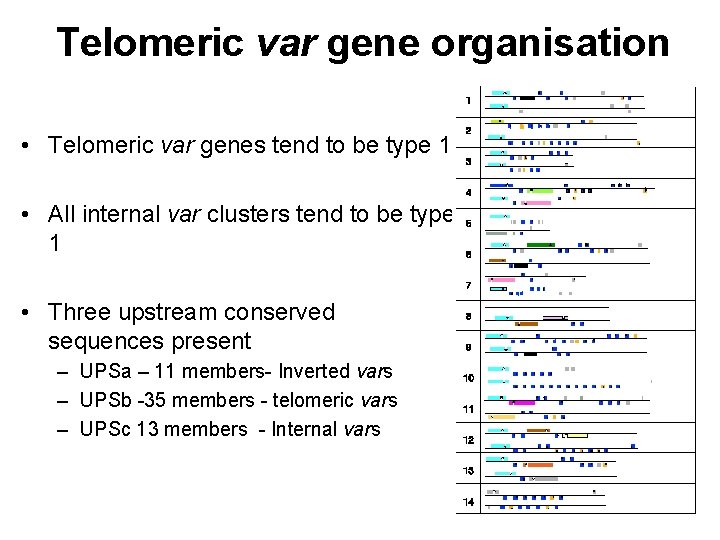
Telomeric var gene organisation • Telomeric var genes tend to be type 1 • All internal var clusters tend to be type 1 • Three upstream conserved sequences present – UPSa – 11 members- Inverted vars – UPSb -35 members - telomeric vars – UPSc 13 members - Internal vars

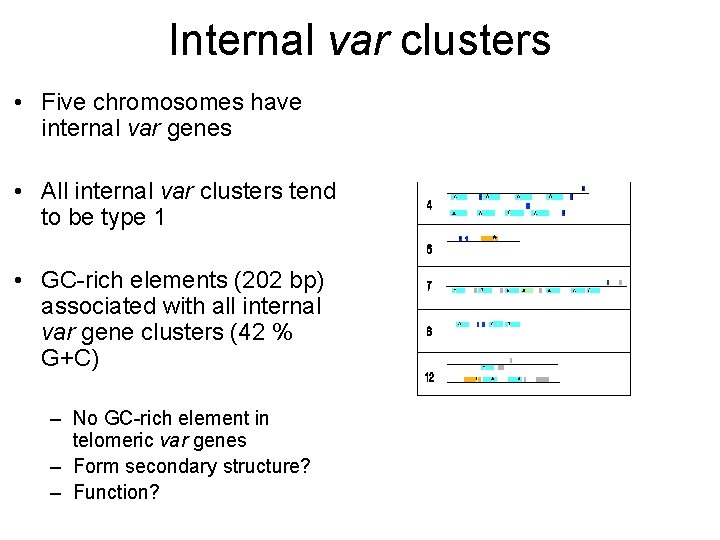
Internal var clusters • Five chromosomes have internal var genes • All internal var clusters tend to be type 1 • GC-rich elements (202 bp) associated with all internal var gene clusters (42 % G+C) – No GC-rich element in telomeric var genes – Form secondary structure? – Function?

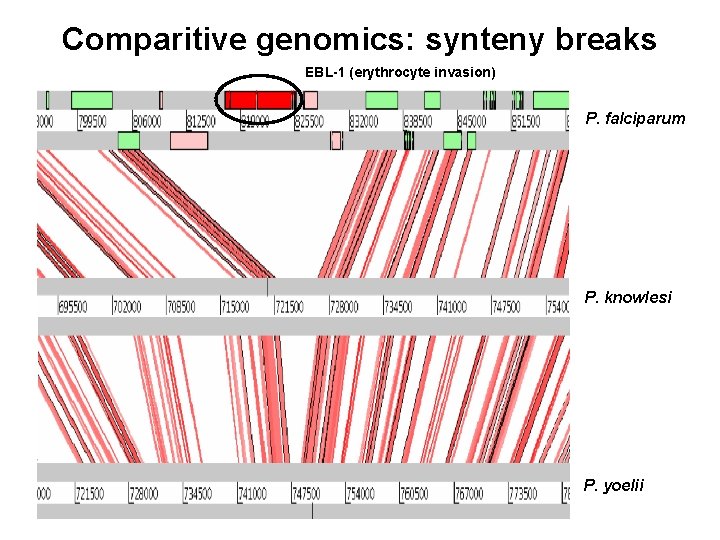
Comparitive genomics: synteny breaks EBL-1 (erythrocyte invasion) P. falciparum P. knowlesi P. yoelii

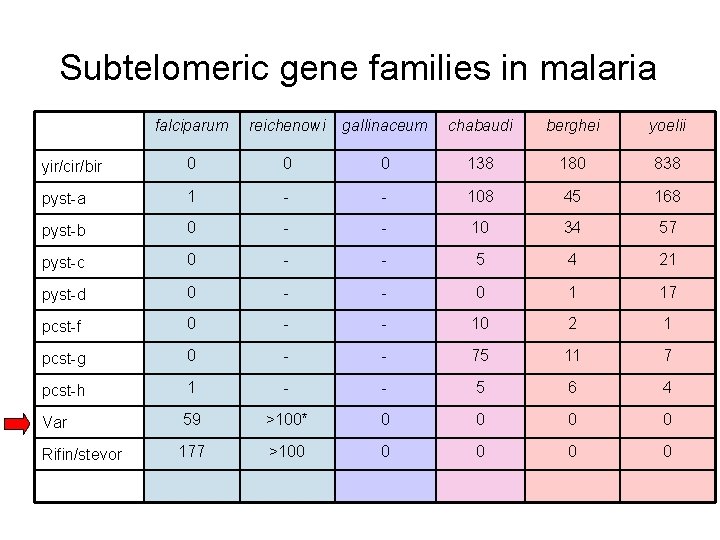
Subtelomeric gene families in malaria falciparum reichenowi gallinaceum chabaudi berghei yoelii yir/cir/bir 0 0 0 138 180 838 pyst-a 1 - - 108 45 168 pyst-b 0 - - 10 34 57 pyst-c 0 - - 5 4 21 pyst-d 0 - - 0 1 17 pcst-f 0 - - 10 2 1 pcst-g 0 - - 75 11 7 pcst-h 1 - - 5 6 4 Var 59 >100* 0 0 Rifin/stevor 177 >100 0 0

Summary

Sequencing Project Management Barrell Julian Parkhill Neil Hall Stephen Bentley Matthew Berriman Al Ivens Marie-Adèle Rajandream Nick Thomson Programming David Harper Rob Davies Arnaud Kerhornou Paul Mooney Kim Rutherford Ellen Scofield Adrian Tivey Ed Zuiderwijk Administration Analysis & Databases Yvonne Shaw Sarah L. ocke Microarrays Martin Aslett Celine Carret Andrew Berry Theresa Feltwell Ulrike Böhme Maria Fookes Ana Cerdeño Nefeli Nikolaidou. Lisa Crossman Katsaridou Christiane Fowler Matloob Qureshi Matthew Holden Jason Skelton Arnab Pain Chris Peacock Hubert Renauld Mohammed Sebaihia Valerie Wood Carol Churcher Becky Atkin Karen Brooks Inna Cherevach Tracey Chillingworth Paul Davis Nancy Hamlin Kay Jagels Sharon Moule Brian White Sally Whitehead David Harris Nigel Fosker Arlette Goble Lee Murphy Susan O’Neil Simon Rutter David Saunders Kathy Seeger Robert Squares Subcloning Mike Quail Angela Lord Halina Norbertczak Claire Price Ester Rabbinowitsch John Woodward Karen Mungall Ian Goodhead Zahra Hance Heidi Hauser Mandy Sanders Mark Simmonds Danielle Walker Barbara Harris Andrew Barron Louise Clark Craig Corton Jonathan Doggett Nicola Lennard Alexandra Line Doug Ormond Comparative Genomics Alison Dennis Emily Kay Helena Seth-Smith
 Stigmatic shaming
Stigmatic shaming Labeling and reintegrative shaming theory
Labeling and reintegrative shaming theory What is the
What is the Fig 28
Fig 28 Quote sandwich example
Quote sandwich example Johnson rule for 3 machines
Johnson rule for 3 machines Difference between ngs and sanger sequencing
Difference between ngs and sanger sequencing Sequencing analysis viewer
Sequencing analysis viewer Dna
Dna Sequence / process writing
Sequence / process writing Ngs sequencing data analysis
Ngs sequencing data analysis Sequencing activities asl
Sequencing activities asl The very lonely firefly sequencing
The very lonely firefly sequencing Days of the week sequencing
Days of the week sequencing Address sequencing in computer architecture
Address sequencing in computer architecture Rna sequencing steps
Rna sequencing steps Greedy algorithm for job sequencing with deadlines
Greedy algorithm for job sequencing with deadlines History of sequencing
History of sequencing Sequencing batch reactor advantages and disadvantages
Sequencing batch reactor advantages and disadvantages Illumina
Illumina Sanger vs maxam gilbert sequencing
Sanger vs maxam gilbert sequencing 3rd generation dna sequencing
3rd generation dna sequencing Cloning and sequencing explorer series
Cloning and sequencing explorer series Address sequencing in computer architecture
Address sequencing in computer architecture Explain priority sequencing
Explain priority sequencing