Marine Geospatial Ecology Tools Open Source Geoprocessing for








- Slides: 40

Marine Geospatial Ecology Tools Open Source Geoprocessing for Marine Ecology Jason Roberts, Ben Best, Dan Dunn, Eric Treml, Pat Halpin Duke University Marine Geospatial Ecology Lab 4 -Mar-2009

Talk outline Overview of MGET Quick tour of the MGET tool collection Example application: habitat modeling

What is MGET? Also use fu for terre l stria problem l s! A collection of geoprocessing tools for marine ecology Oceanographic data management and analysis Habitat modeling, connectivity modeling, statistics Highly modular; designed to be used in many scenarios Emphasis on batch processing and interoperability Free, open source software Written in Python, R, MATLAB, C#, and C++ Minimum requirements: Win XP, Python 2. 4 Arc. GIS 9. 1 or later currently needed for many tools Arc. GIS and Windows are only non-free requirements

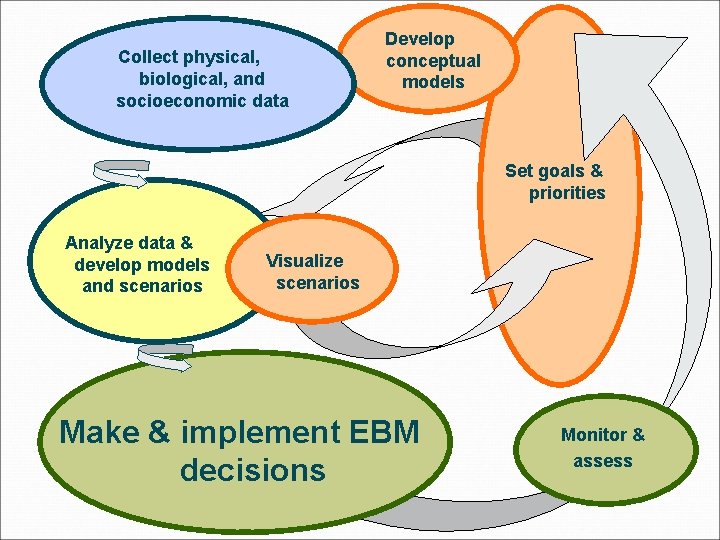
Collect physical, biological, and socioeconomic data Develop conceptual models Set goals & priorities Analyze data & develop models and scenarios Visualize scenarios Make & implement EBM decisions Monitor & assess

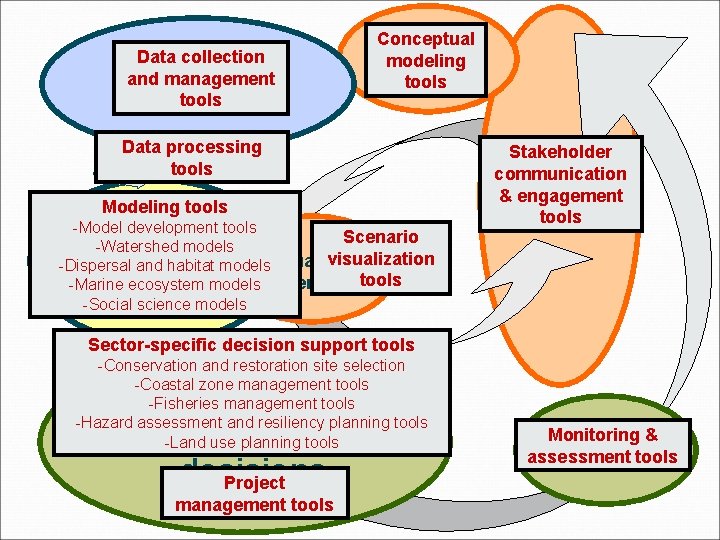
Data collection Collect physical, and management biological, and tools socioeconomic data Conceptual Develop modeling conceptual models tools Data processing tools Modeling tools -Model development tools Scenario Analyze data & -Watershed models visualization developand models -Dispersal habitat models. Visualize -Marine ecosystem models scenariostools and scenarios -Social science models Stakeholder Set goals & communication priorities & engagement tools Sector-specific decision support tools -Conservation and restoration site selection -Coastal zone management tools -Fisheries management tools -Hazard assessment and resiliency planning tools -Land use planning tools Make & implement EBM decisions Project management tools Monitoring Monitor && assessment assess tools

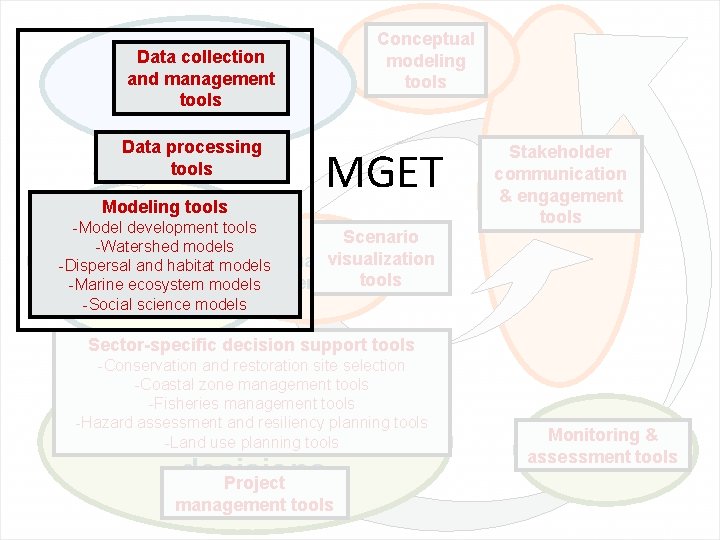
Conceptual Develop modeling conceptual models tools Data collection Collect physical, and management biological, and tools socioeconomic data Data processing tools Modeling tools MGET -Model development tools Scenario Analyze data & -Watershed models visualization developand models -Dispersal habitat models. Visualize -Marine ecosystem models scenariostools and scenarios -Social science models Stakeholder Set goals & communication priorities & engagement tools Sector-specific decision support tools -Conservation and restoration site selection -Coastal zone management tools -Fisheries management tools -Hazard assessment and resiliency planning tools -Land use planning tools Make & implement EBM decisions Project management tools Monitoring Monitor && assessment assess tools

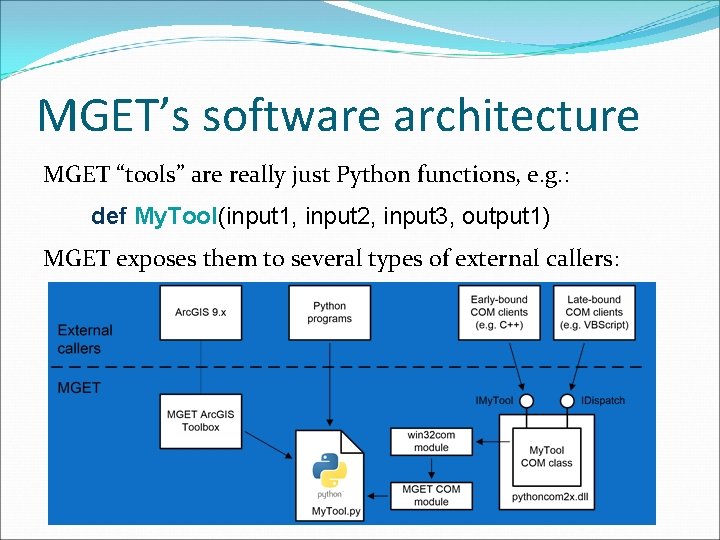
MGET’s software architecture MGET “tools” are really just Python functions, e. g. : def My. Tool(input 1, input 2, input 3, output 1) MGET exposes them to several types of external callers:

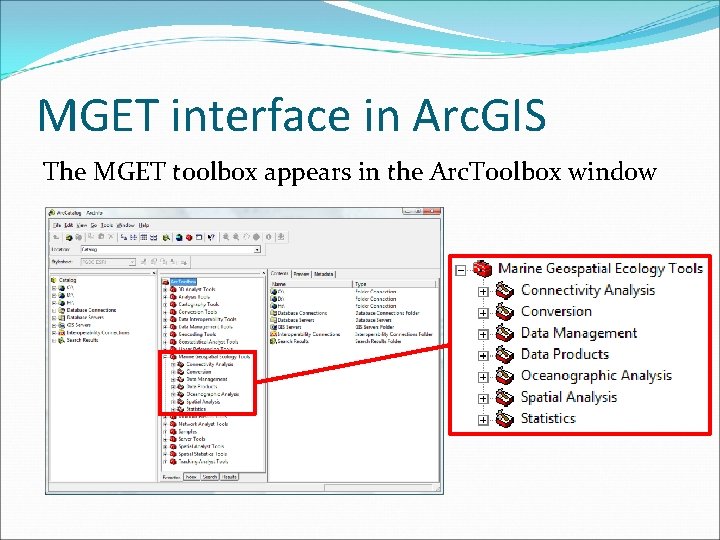
MGET interface in Arc. GIS The MGET toolbox appears in the Arc. Toolbox window

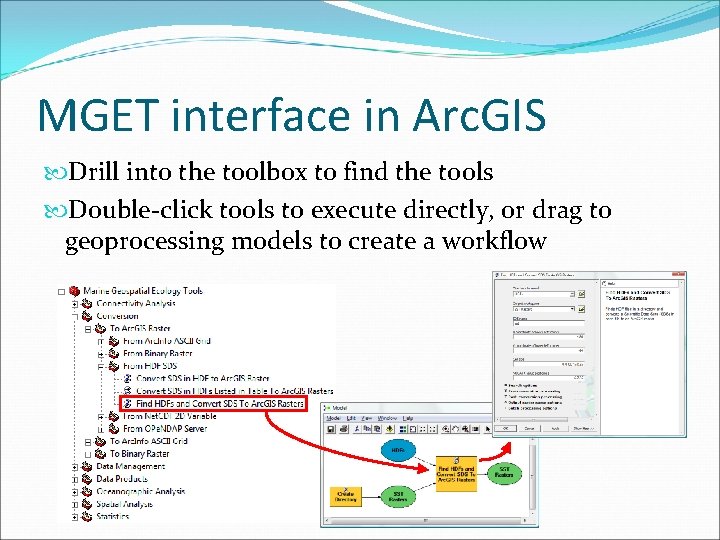
MGET interface in Arc. GIS Drill into the toolbox to find the tools Double-click tools to execute directly, or drag to geoprocessing models to create a workflow

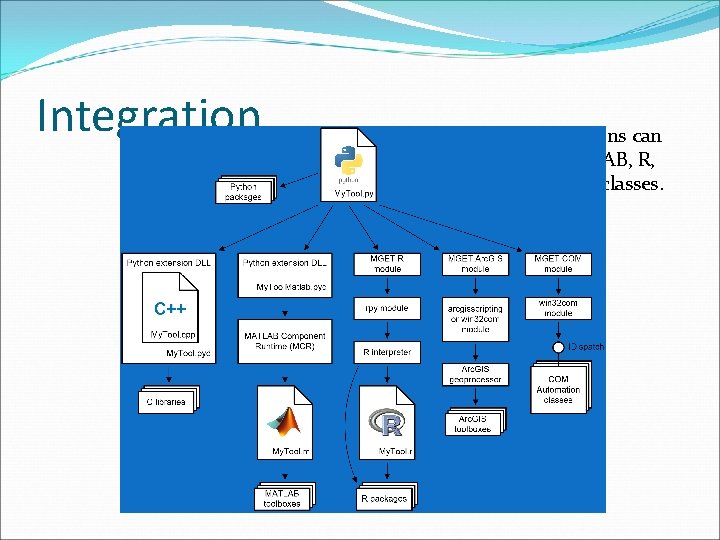
Integration The Python functions can invoke C++, MATLAB, R, Arc. GIS, and COM classes.

MGET utilizes a lot of other software Interpreters / Runtimes Python Packages C Libraries Python MATLAB Component Runtime R docutils httplib 2 lxml netcdf 4 numpy osgeo pydap pyparsing pyproj pywin 32 rpy setuptools GDAL/OGR gzip hdf libxml libxslt netcdf proj 4 zlib Applications Arc. GIS NOAA Coast. Watch Utilities All but one of these (pywin 32) are installed automatically R Packages gam MASS mgcv rgdal ROCR

Quick tour of the tools

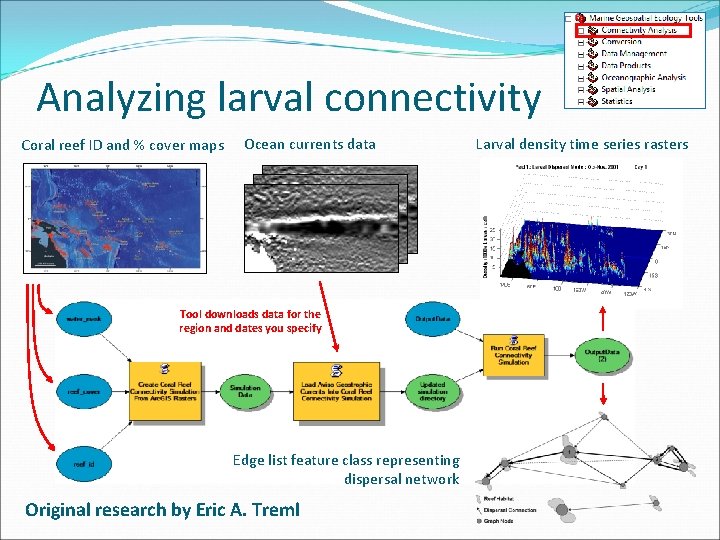
Analyzing larval connectivity Coral reef ID and % cover maps Ocean currents data Tool downloads data for the region and dates you specify Edge list feature class representing dispersal network Original research by Eric A. Treml Larval density time series rasters

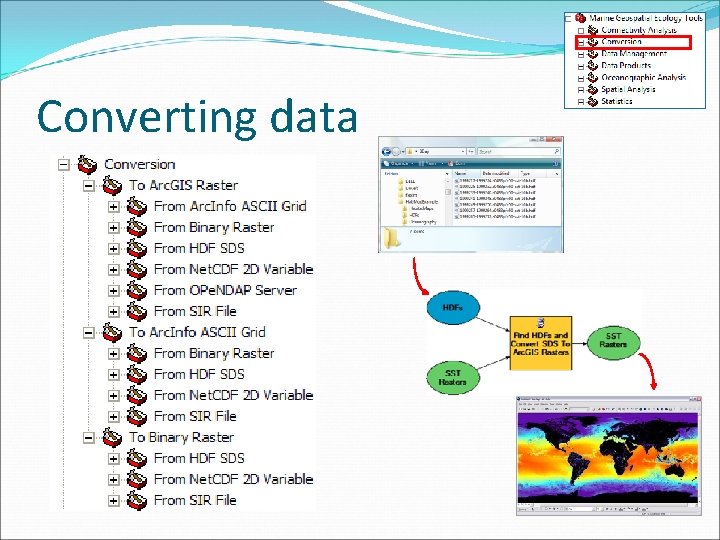
Converting data

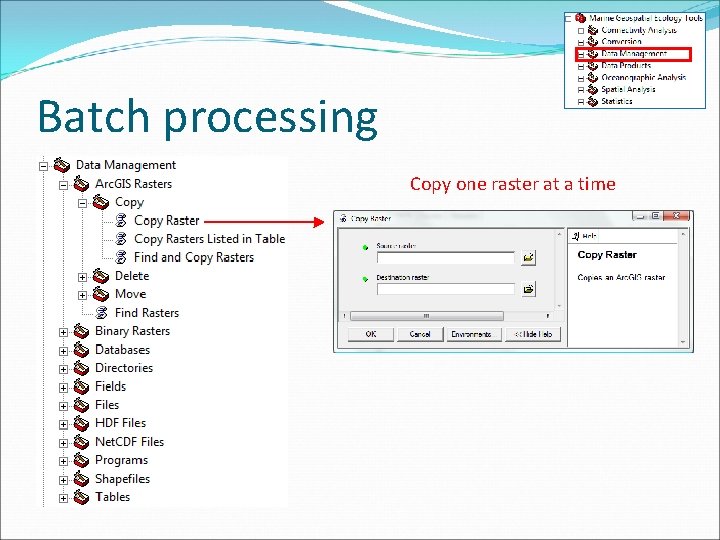
Batch processing Copy one raster at a time

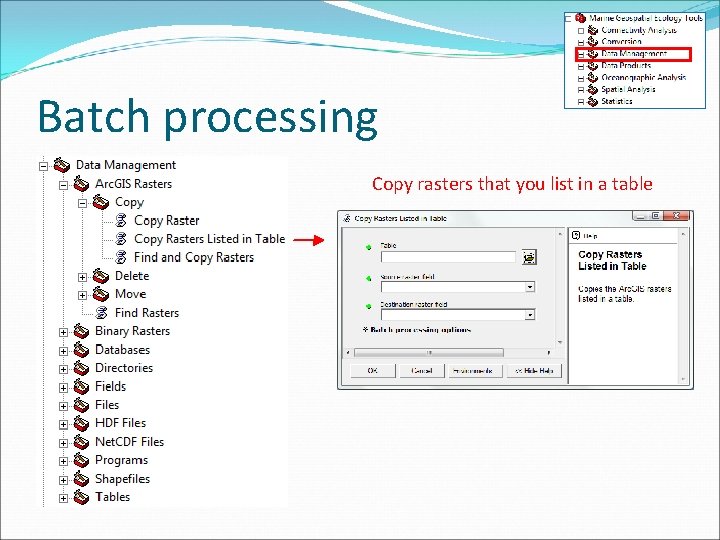
Batch processing Copy rasters that you list in a table

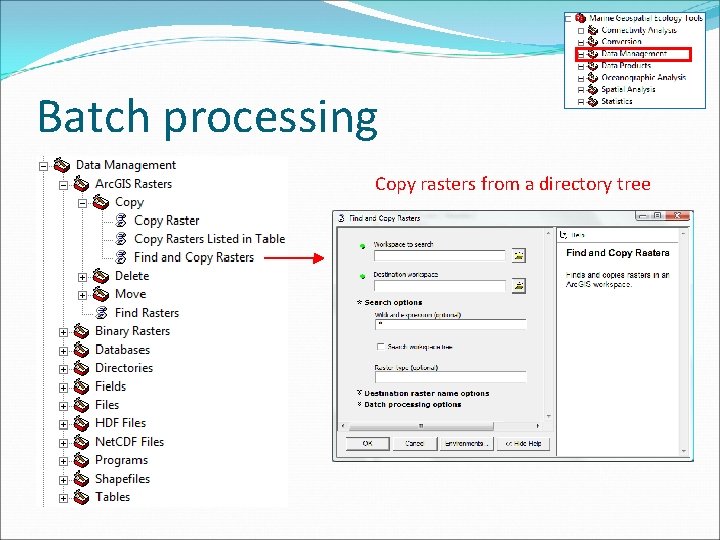
Batch processing Copy rasters from a directory tree

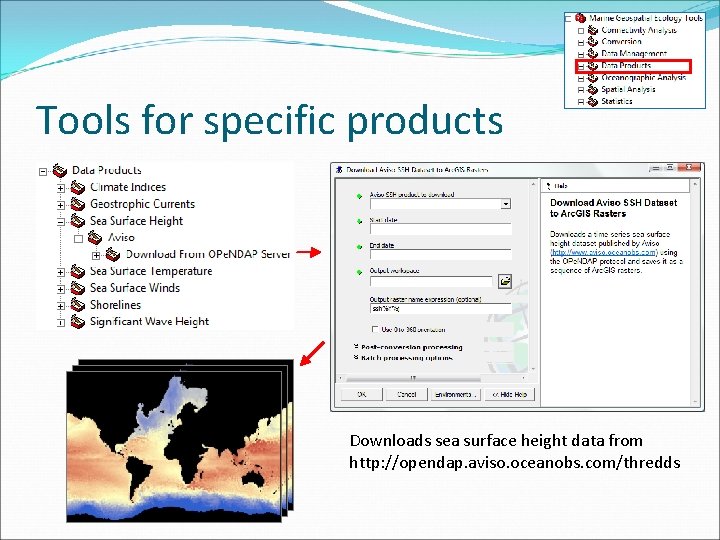
Tools for specific products Downloads sea surface height data from http: //opendap. aviso. oceanobs. com/thredds

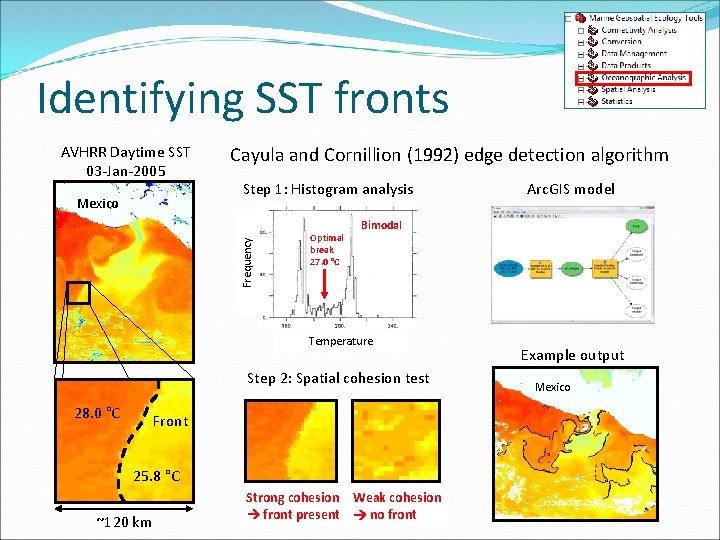
Identifying SST fronts AVHRR Daytime SST 03 -Jan-2005 Step 1: Histogram analysis Frequency Mexico Cayula and Cornillion (1992) edge detection algorithm Optimal break 27. 0 °C Bimodal Temperature Step 2: Spatial cohesion test 28. 0 °C Front 25. 8 °C ~120 km Arc. GIS model Strong cohesion Weak cohesion front present no front Example output Mexico

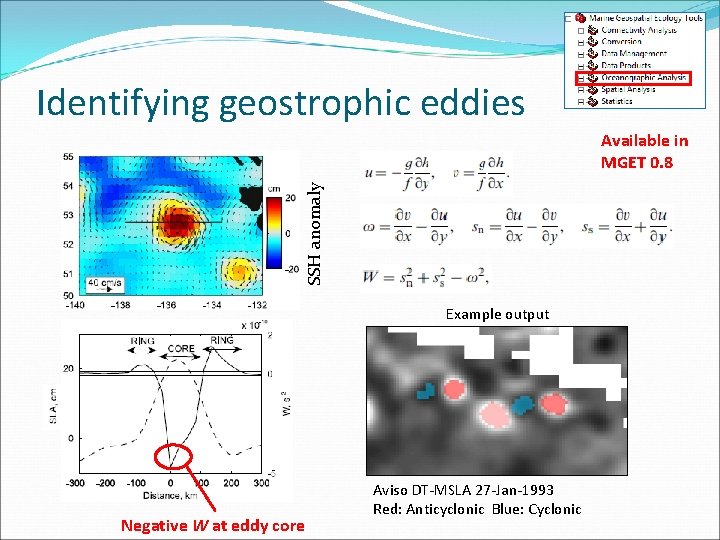
Identifying geostrophic eddies SSH anomaly Available in MGET 0. 8 Example output Negative W at eddy core Aviso DT-MSLA 27 -Jan-1993 Red: Anticyclonic Blue: Cyclonic

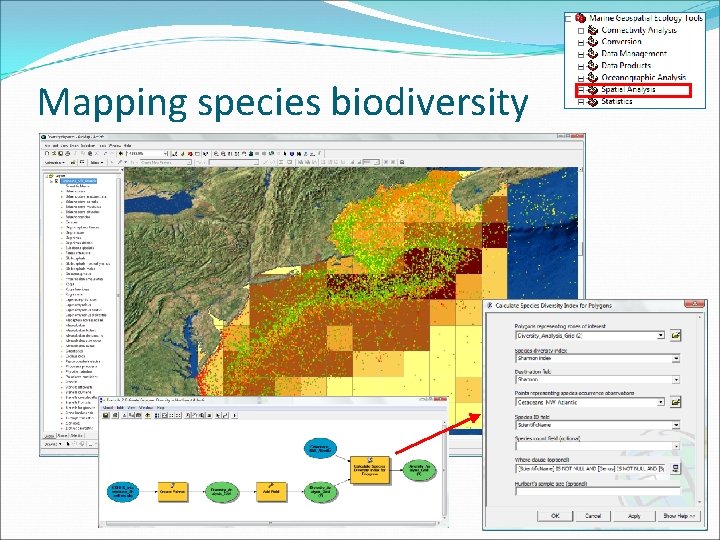
Mapping species biodiversity

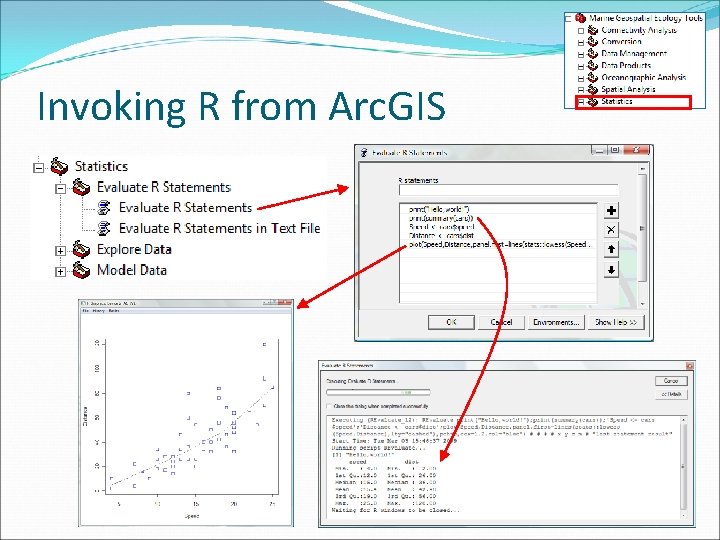
Invoking R from Arc. GIS

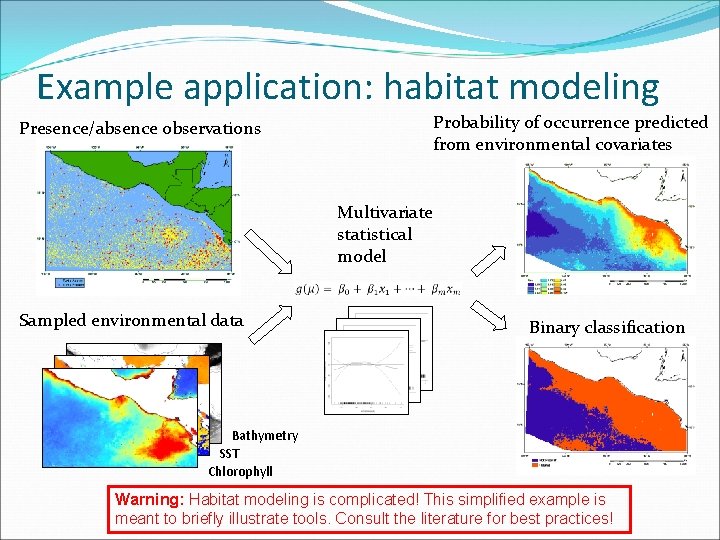
Example application: habitat modeling Probability of occurrence predicted from environmental covariates Presence/absence observations Multivariate statistical model Sampled environmental data Binary classification Bathymetry SST Chlorophyll Warning: Habitat modeling is complicated! This simplified example is meant to briefly illustrate tools. Consult the literature for best practices!

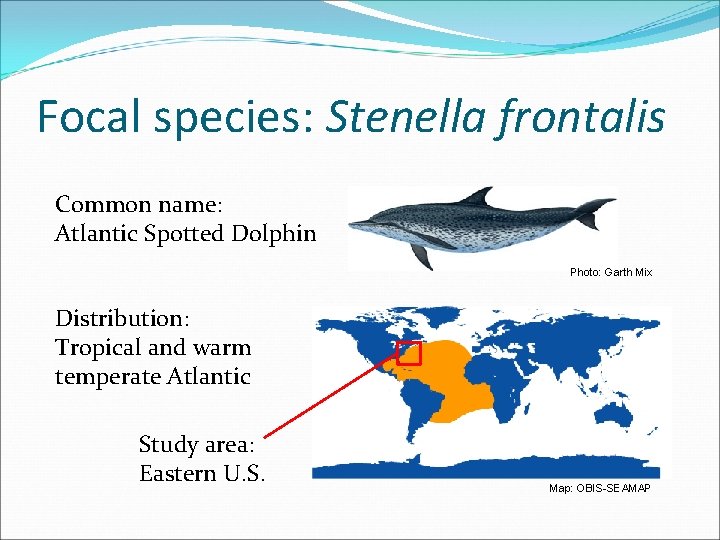
Focal species: Stenella frontalis Common name: Atlantic Spotted Dolphin Photo: Garth Mix Distribution: Tropical and warm temperate Atlantic Study area: Eastern U. S. Map: OBIS-SEAMAP

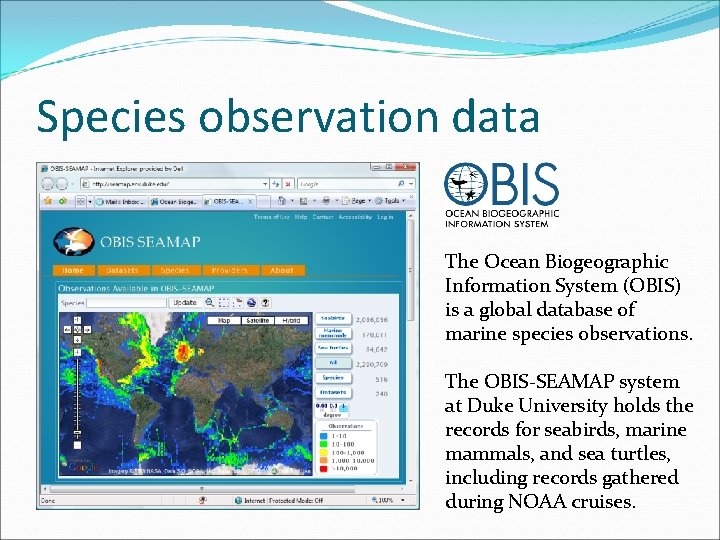
Species observation data The Ocean Biogeographic Information System (OBIS) is a global database of marine species observations. The OBIS-SEAMAP system at Duke University holds the records for seabirds, marine mammals, and sea turtles, including records gathered during NOAA cruises.

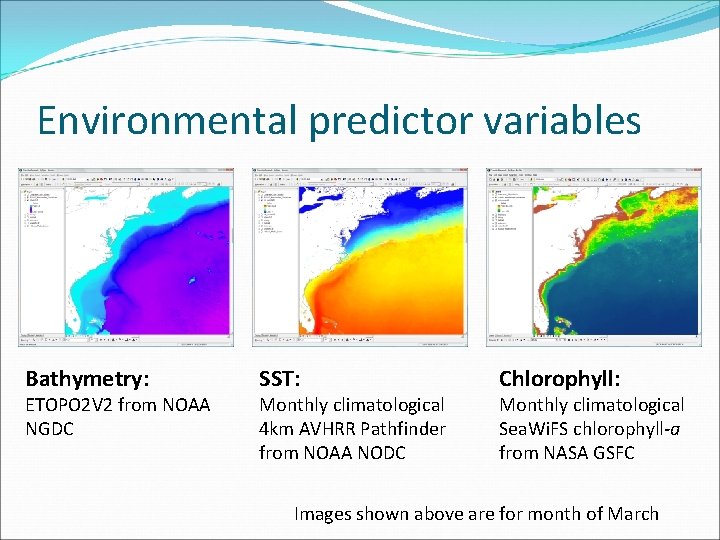
Environmental predictor variables Bathymetry: ETOPO 2 V 2 from NOAA NGDC SST: Monthly climatological 4 km AVHRR Pathfinder from NOAA NODC Chlorophyll: Monthly climatological Sea. Wi. FS chlorophyll-a from NASA GSFC Images shown above are for month of March

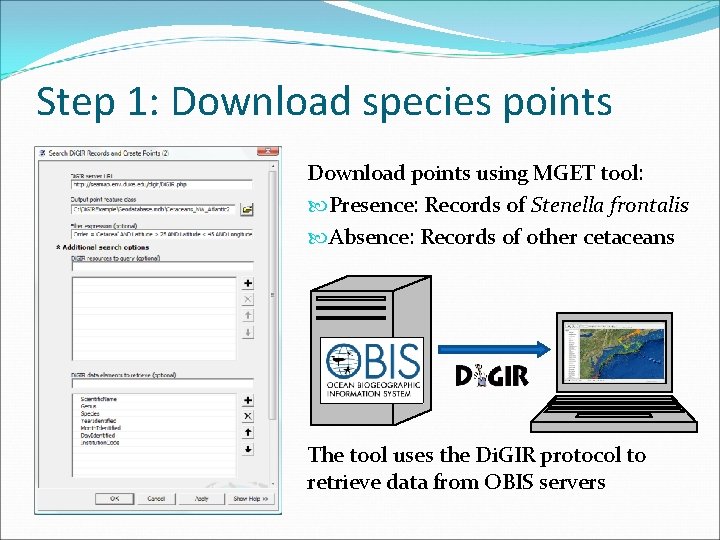
Step 1: Download species points Download points using MGET tool: Presence: Records of Stenella frontalis Absence: Records of other cetaceans The tool uses the Di. GIR protocol to retrieve data from OBIS servers

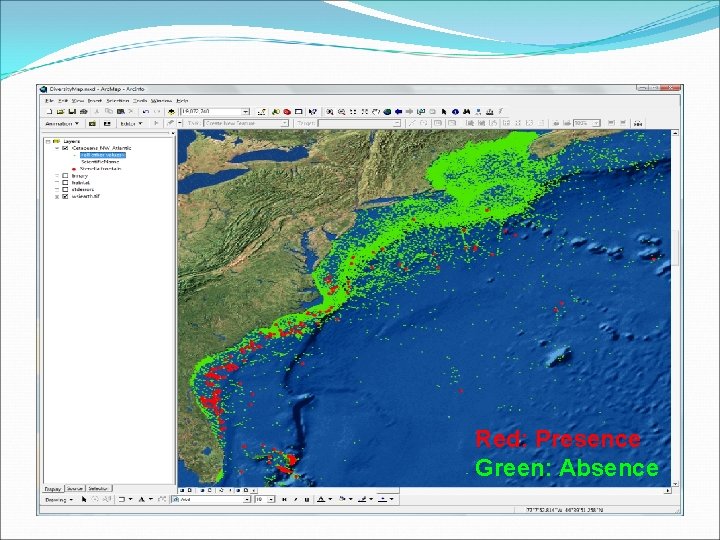
Red: Presence Green: Absence

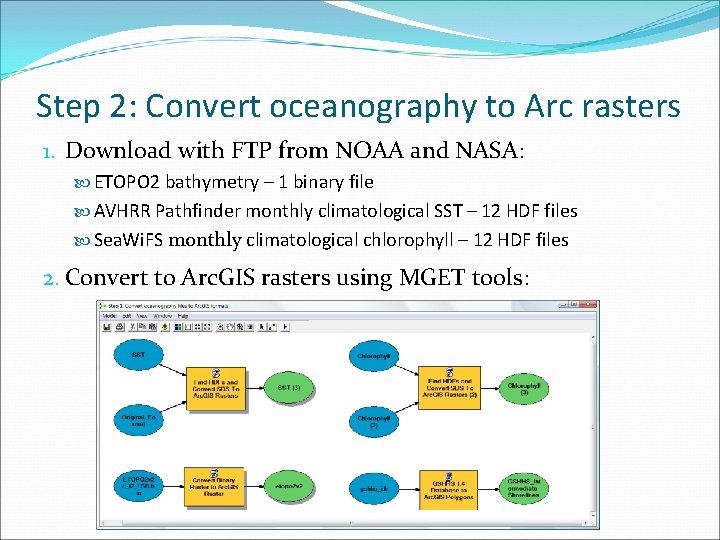
Step 2: Convert oceanography to Arc rasters 1. Download with FTP from NOAA and NASA: ETOPO 2 bathymetry – 1 binary file AVHRR Pathfinder monthly climatological SST – 12 HDF files Sea. Wi. FS monthly climatological chlorophyll – 12 HDF files 2. Convert to Arc. GIS rasters using MGET tools:

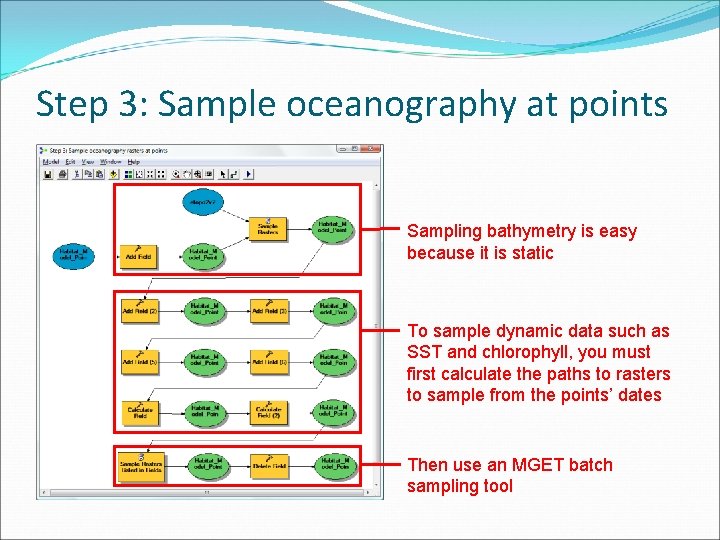
Step 3: Sample oceanography at points • Need to sample rasters and populate fields • Must sample SST and chlorophyll by date

Step 3: Sample oceanography at points Sampling bathymetry is easy because it is static To sample dynamic data such as SST and chlorophyll, you must first calculate the paths to rasters to sample from the points’ dates Then use an MGET batch sampling tool

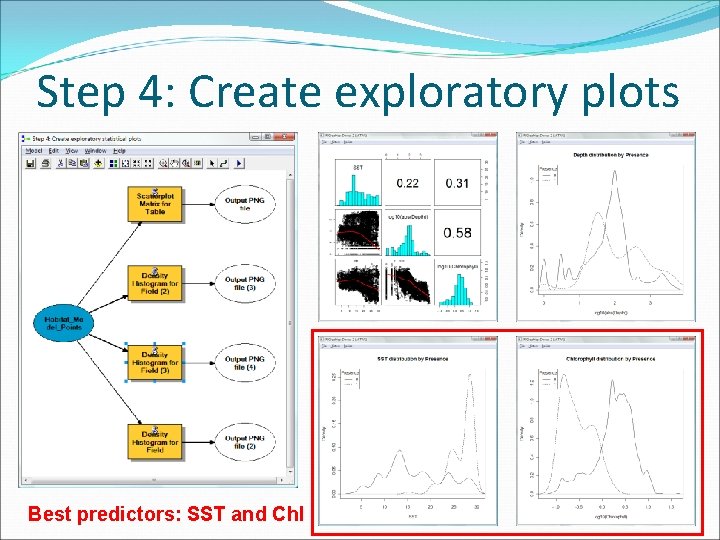
Step 4: Create exploratory plots Best predictors: SST and Chl

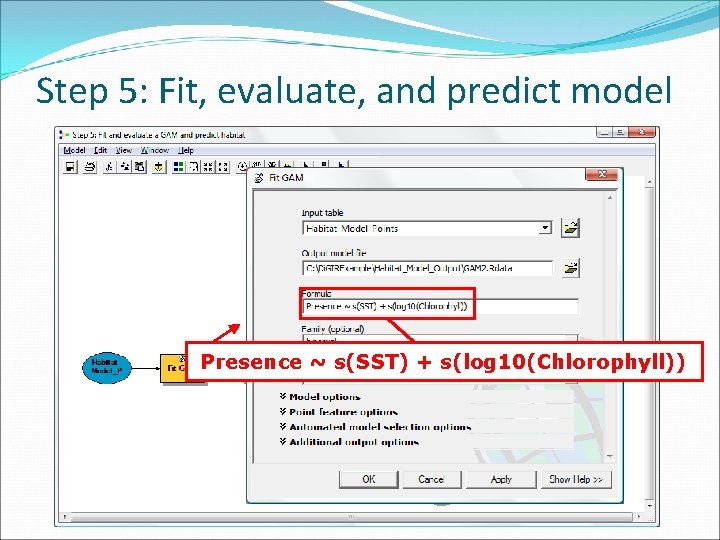
Step 5: Fit, evaluate, and predict model Presence ~ s(SST) + s(log 10(Chlorophyll))

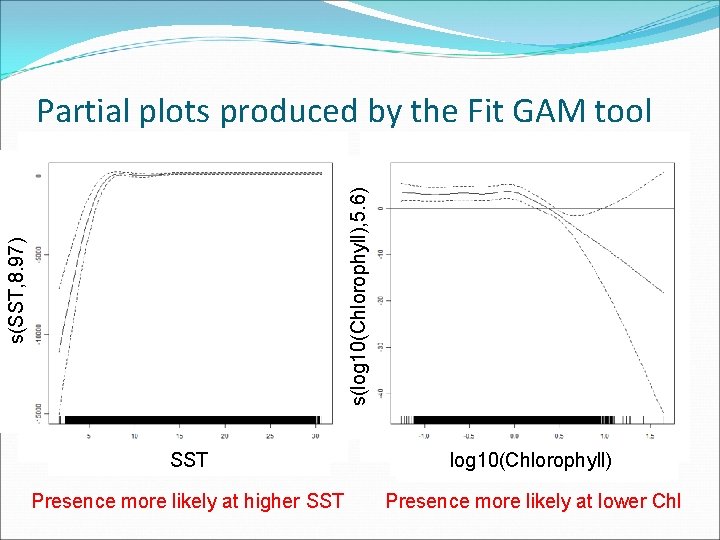
s(SST, 8. 97) s(log 10(Chlorophyll), 5. 6) Partial plots produced by the Fit GAM tool SST log 10(Chlorophyll) SST Presence more likely at higher SST Presence more likely at lower Chl

Plotting a receiver operating characteristic curve

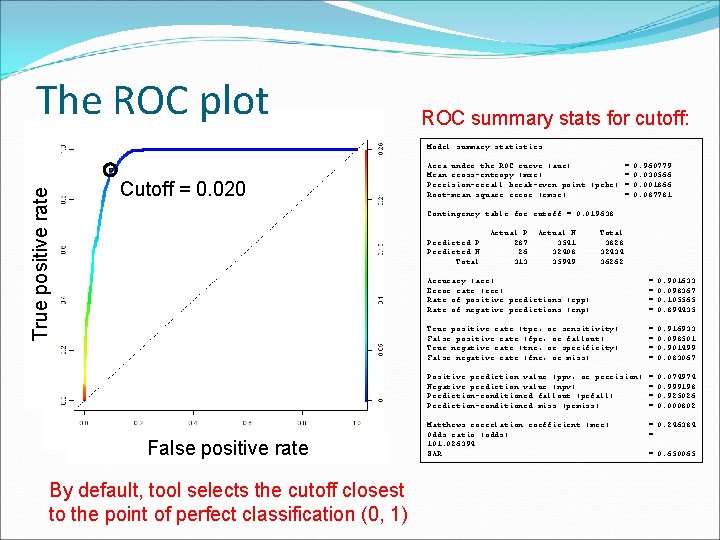
The ROC plot ROC summary stats for cutoff: True positive rate Model summary statistics: Cutoff = 0. 020 Area under the ROC curve (auc) = Mean cross-entropy (mxe) = Precision-recall break-even point (prbe) = Root-mean square error (rmse) = 0. 960779 0. 030566 0. 001866 0. 087781 Contingency table for cutoff = 0. 019638: Predicted P Predicted N Total False positive rate By default, tool selects the cutoff closest to the point of perfect classification (0, 1) Actual P 287 26 313 Actual N 3541 32408 35949 Total 3828 32434 36262 Accuracy (acc) Error rate (err) Rate of positive predictions (rpp) Rate of negative predictions (rnp) = = 0. 901633 0. 098367 0. 105565 0. 894435 True positive rate (tpr, or sensitivity) False positive rate (fpr, or fallout) True negative rate (tnr, or specificity) False negative rate (fnr, or miss) = = 0. 916933 0. 098501 0. 901499 0. 083067 Positive prediction value (ppv, or precision) Negative prediction value (npv) Prediction-conditioned fallout (pcfall) Prediction-conditioned miss (pcmiss) = = 0. 074974 0. 999198 0. 925026 0. 000802 Matthews correlation coefficient (mcc) Odds ratio (odds) 101. 026394 SAR = 0. 246384 = = 0. 650065

Predicting presence for oceanographic rasters

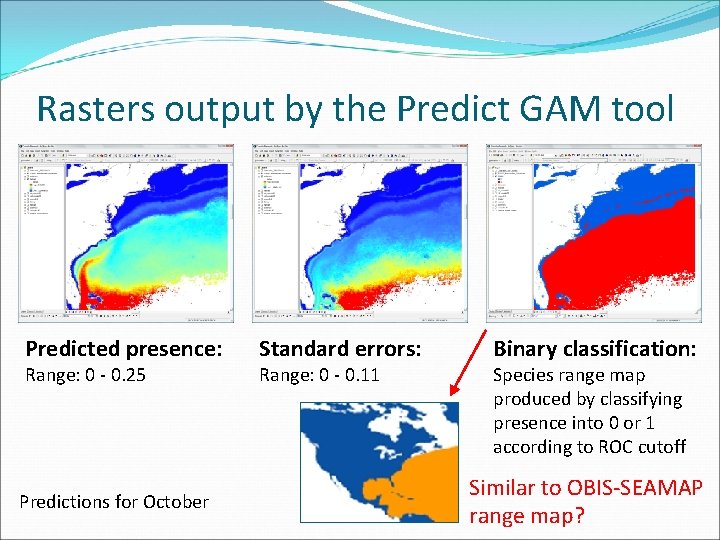
Rasters output by the Predict GAM tool Predicted presence: Range: 0 - 0. 25 Predictions for October Standard errors: Range: 0 - 0. 11 Binary classification: Species range map produced by classifying presence into 0 or 1 according to ROC cutoff Similar to OBIS-SEAMAP range map?

Acknowledgements A special thanks to the many developers of the open source software that MGET is built upon, including: Guido van Rossum and his many collaborators; Mark Hammond; Travis Oliphant and his collaborators; Walter Moreira and Gregory Warnes; Peter Hollemans; David Ullman, Jean-Francois Cayula, and Peter Cornillon; Stephanie Henson; Tobias Sing, Oliver Sander, Niko Beerenwinkel, and Thomas Lengauer; Frank Warmerdam and his collaborators, Howard Butler; Timothy H. Keitt, Roger Bivand, Edzer Pebesma, and Barry Rowlingson; Gerald Evenden; Jeff Whitaker; Roberto De Almeida and his collaborators; Joe Gregorio; David Goodger and his collaborators; Daniel Veillard and his collaborators; Stefan Behnel, Martijn Faassen, and their collaborators; Paul Mc. Guire and his collaborators; Phillip Eby, Bob Ippolito, and their collaborators; Jean-loup Gailly and Mark Adler; the developers of net. CDF; the developers of HDF Thanks to our funders:

For more information Download MGET: http: //code. env. duke. edu/projects/mget Email us: jason. roberts@duke. edu, bbest@duke. edu Learn more about habitat modeling: Guisan, A. , Zimmermann, N. E. (2000) Predictive habitat distribution models in ecology. Ecological Modelling 135, 147– 186. Thanks for attending!