K 812 30 Oct 07 LQT 9 and




- Slides: 22

K 812 30 Oct 07 LQT 9 and 1

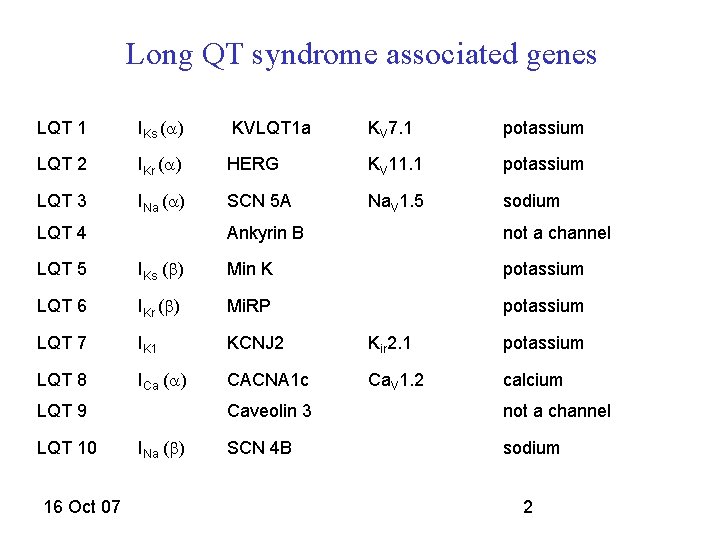
Long QT syndrome associated genes LQT 1 IKs (a) KVLQT 1 a KV 7. 1 potassium LQT 2 IKr (a) HERG KV 11. 1 potassium LQT 3 INa (a) SCN 5 A Na. V 1. 5 sodium LQT 4 Ankyrin B not a channel LQT 5 IKs (b) Min K potassium LQT 6 IKr (b) Mi. RP potassium LQT 7 IK 1 KCNJ 2 Kir 2. 1 potassium LQT 8 ICa (a) CACNA 1 c Ca. V 1. 2 calcium LQT 9 LQT 10 16 Oct 07 INa (b) Caveolin 3 not a channel SCN 4 B sodium 2

Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Circulation 114: 2104 -12, 2006

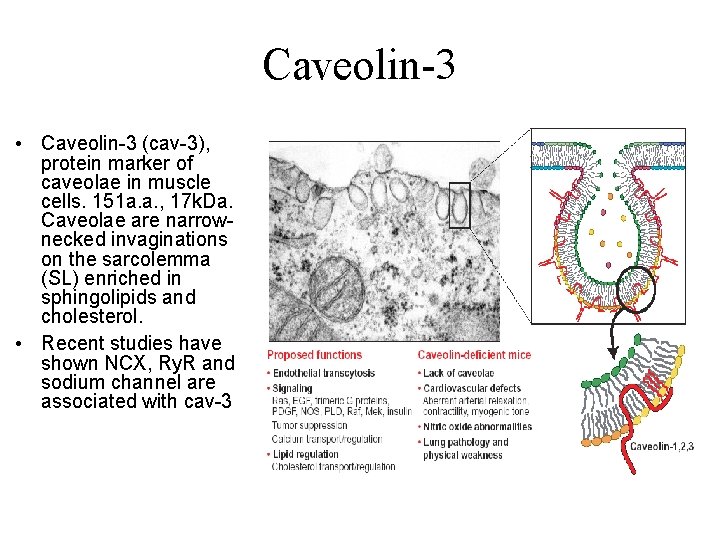
Caveolin-3 • Caveolin-3 (cav-3), protein marker of caveolae in muscle cells. 151 a. a. , 17 k. Da. Caveolae are narrownecked invaginations on the sarcolemma (SL) enriched in sphingolipids and cholesterol. • Recent studies have shown NCX, Ry. R and sodium channel are associated with cav-3 Parton R. G. Science

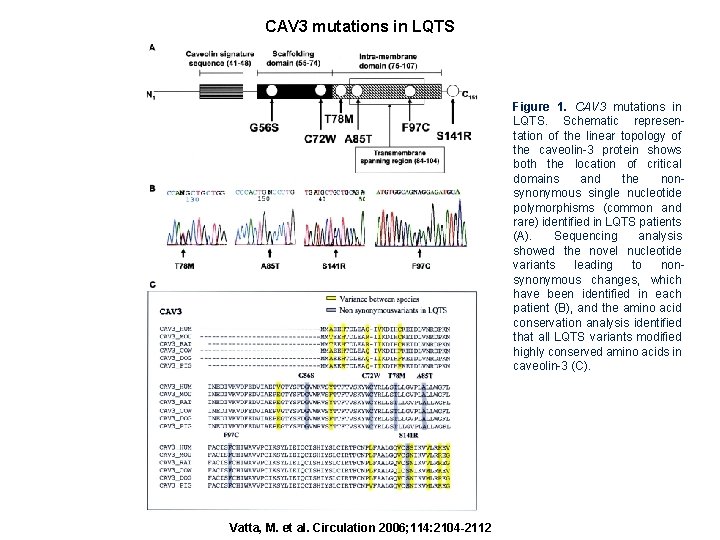
CAV 3 mutations in LQTS Figure 1. CAV 3 mutations in LQTS. Schematic representation of the linear topology of the caveolin-3 protein shows both the location of critical domains and the nonsynonymous single nucleotide polymorphisms (common and rare) identified in LQTS patients (A). Sequencing analysis showed the novel nucleotide variants leading to nonsynonymous changes, which have been identified in each patient (B), and the amino acid conservation analysis identified that all LQTS variants modified highly conserved amino acids in caveolin-3 (C). Vatta, M. et al. Circulation 2006; 114: 2104 -2112

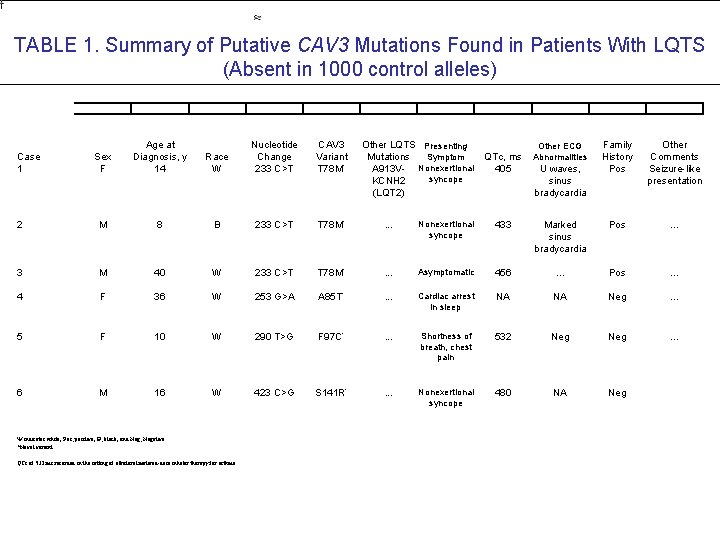
TABLE 1. Summary of Putative CAV 3 Mutations Found in Patients With LQTS (Absent in 1000 control alleles) Sex F Age at Diagnosis, y 14 Race W Nucleotide Change 233 C>T CAV 3 Variant T 78 M* 2 M 8 B 233 C>T T 78 M* . . . Nonexertional syncope 433 3 M 40 W 233 C>T T 78 M* . . . Asymptomatic 4 F 36 W 253 G>A A 85 T* . . . 5 F 10 W 290 T>G F 97 C* 6 M 16 W 423 C>G S 141 R* Case 1 W indicates white; Pos, positive; B, black; and Neg, Negative. *Novel variant. QTc of 532 ms recorded in the setting of albuterol metered-dose inhaler therapy for asthma. Other LQTS Presenting Mutations QTc, ms Symptom A 913 V- Nonexertional 405 syncope KCNH 2 (LQT 2) Family History Pos Other Comments Seizure-like presentation Marked sinus bradycardia Pos . . . 456 . . . Pos . . . Cardiac arrest in sleep NA NA Neg . . . Shortness of breath, chest pain 532 Neg . . . Nonexertional syncope 480 NA Neg Other ECG Abnormalities U waves, sinus bradycardia

ECG of patient with the LQTS-associated mutation F 97 C-CAV 3 Vatta, M. et al. Circulation 2006; 114: 2104 -2112 Copyright © 2006 American Heart Association

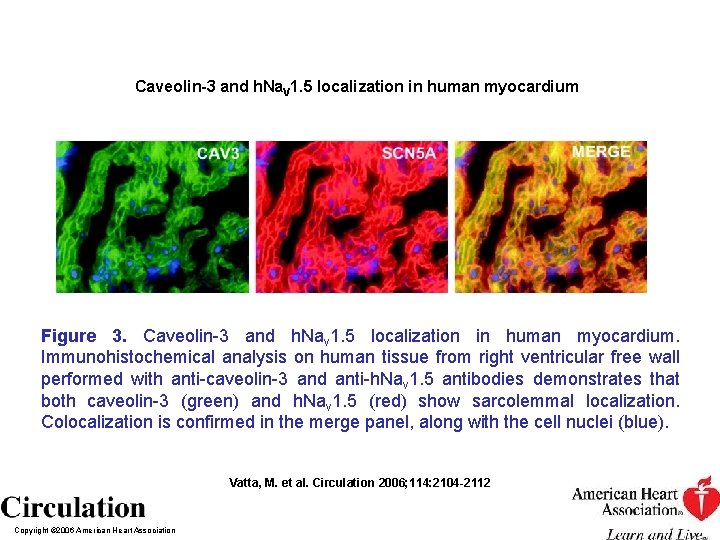
Caveolin-3 and h. Na. V 1. 5 localization in human myocardium Figure 3. Caveolin-3 and h. Nav 1. 5 localization in human myocardium. Immunohistochemical analysis on human tissue from right ventricular free wall performed with anti-caveolin-3 and anti-h. Nav 1. 5 antibodies demonstrates that both caveolin-3 (green) and h. Nav 1. 5 (red) show sarcolemmal localization. Colocalization is confirmed in the merge panel, along with the cell nuclei (blue). Vatta, M. et al. Circulation 2006; 114: 2104 -2112 Copyright © 2006 American Heart Association

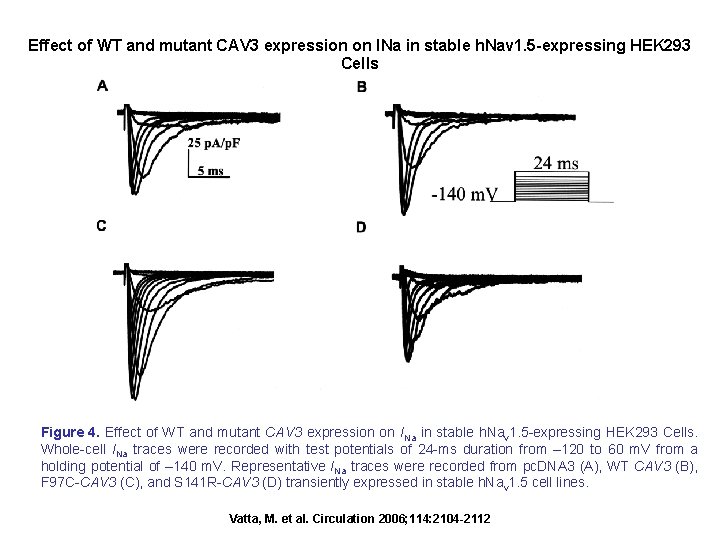
Effect of WT and mutant CAV 3 expression on INa in stable h. Nav 1. 5 -expressing HEK 293 Cells Figure 4. Effect of WT and mutant CAV 3 expression on INa in stable h. Nav 1. 5 -expressing HEK 293 Cells. Whole-cell INa traces were recorded with test potentials of 24 -ms duration from – 120 to 60 m. V from a holding potential of – 140 m. V. Representative INa traces were recorded from pc. DNA 3 (A), WT CAV 3 (B), F 97 C-CAV 3 (C), and S 141 R-CAV 3 (D) transiently expressed in stable h. Nav 1. 5 cell lines. Vatta, M. et al. Circulation 2006; 114: 2104 -2112

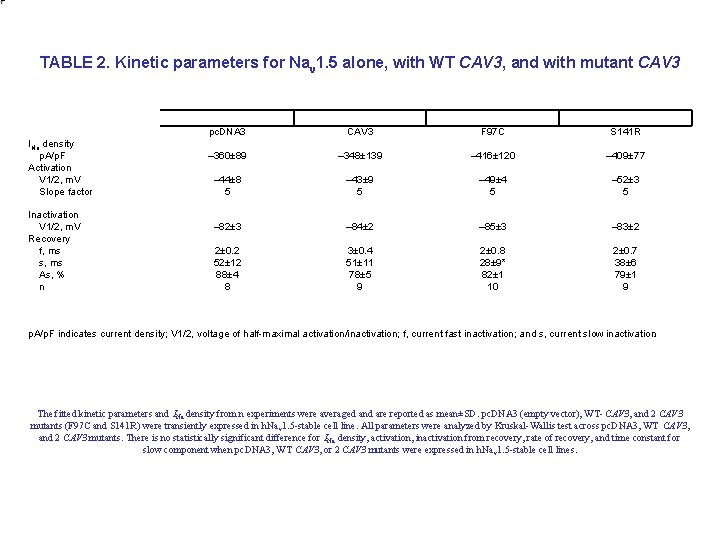
TABLE 2. Kinetic parameters for Nav 1. 5 alone, with WT CAV 3, and with mutant CAV 3 INa density p. A/p. F Activation V 1/2, m. V Slope factor Inactivation V 1/2, m. V Recovery f, ms s, ms As, % n pc. DNA 3 CAV 3 F 97 C S 141 R – 360± 89 – 348± 139 – 416± 120 – 409± 77 – 44± 8 5 – 43± 9 5 – 49± 4 5 – 52± 3 5 – 82± 3 – 84± 2 – 85± 3 – 83± 2 2± 0. 2 52± 12 88± 4 8 3± 0. 4 51± 11 78± 5 9 2± 0. 8 28± 9* 82± 1 10 2± 0. 7 38± 6 79± 1 9 p. A/p. F indicates current density; V 1/2, voltage of half-maximal activation/inactivation; f, current fast inactivation; and s, current slow inactivation. The fitted kinetic parameters and INa density from n experiments were averaged and are reported as mean±SD. pc. DNA 3 (empty vector), WT- CAV 3, and 2 CAV 3 mutants (F 97 C and S 141 R) were transiently expressed in h. Na v 1. 5 -stable cell line. All parameters were analyzed by Kruskal-Wallis test across pc. DNA 3, WT CAV 3, and 2 CAV 3 mutants. There is no statistically significant difference for INa density, activation, inactivation from recovery, rate of recovery, and time constant for slow component when pc. DNA 3, WT CAV 3, or 2 CAV 3 mutants were expressed in h. Na v 1. 5 -stable cell lines.

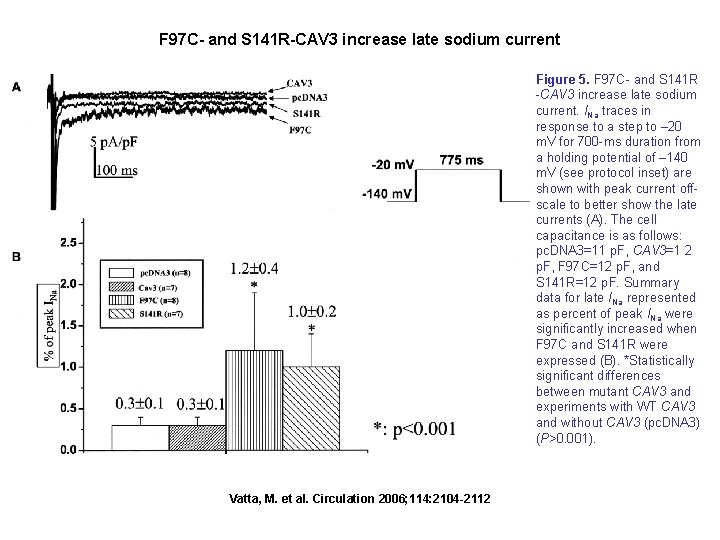
F 97 C- and S 141 R-CAV 3 increase late sodium current Figure 5. F 97 C- and S 141 R -CAV 3 increase late sodium current. INa traces in response to a step to – 20 m. V for 700 -ms duration from a holding potential of – 140 m. V (see protocol inset) are shown with peak current offscale to better show the late currents (A). The cell capacitance is as follows: pc. DNA 3=11 p. F, CAV 3=1 2 p. F, F 97 C=12 p. F, and S 141 R=12 p. F. Summary data for late INa represented as percent of peak INa were significantly increased when F 97 C and S 141 R were expressed (B). *Statistically significant differences between mutant CAV 3 and experiments with WT CAV 3 and without CAV 3 (pc. DNA 3) (P>0. 001). Vatta, M. et al. Circulation 2006; 114: 2104 -2112

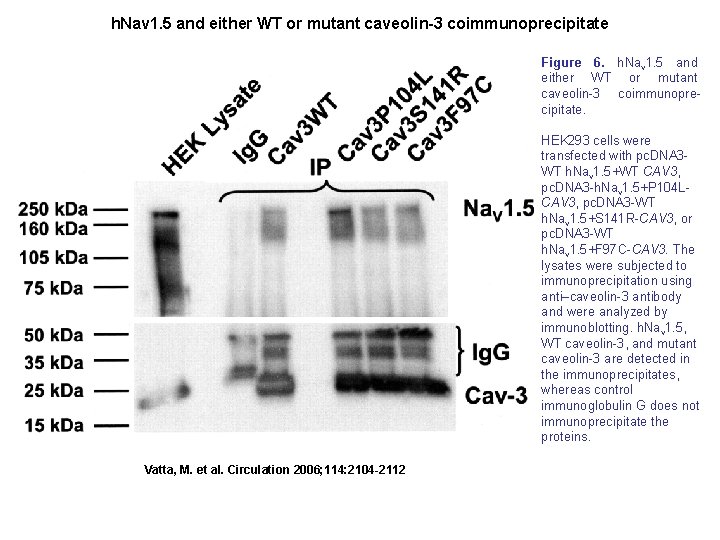
h. Nav 1. 5 and either WT or mutant caveolin-3 coimmunoprecipitate Figure 6. h. Nav 1. 5 and either WT or mutant caveolin-3 coimmunoprecipitate. HEK 293 cells were transfected with pc. DNA 3 WT h. Nav 1. 5+WT CAV 3, pc. DNA 3 -h. Nav 1. 5+P 104 LCAV 3, pc. DNA 3 -WT h. Nav 1. 5+S 141 R-CAV 3, or pc. DNA 3 -WT h. Nav 1. 5+F 97 C-CAV 3. The lysates were subjected to immunoprecipitation using anti–caveolin-3 antibody and were analyzed by immunoblotting. h. Nav 1. 5, WT caveolin-3, and mutant caveolin-3 are detected in the immunoprecipitates, whereas control immunoglobulin G does not immunoprecipitate the proteins. Vatta, M. et al. Circulation 2006; 114: 2104 -2112

Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ 1 gene Moss AJ, Shimizu W, Wilde AA, Towbin JA, Zareba W, Robinson JL, Qi M, Vincent GM, Ackerman MJ, Kaufman ES, Hofman N, Seth R, Kamakura S, Miyamoto Y, Goldenberg I, Andrews ML, Mc. Nitt S Circulation 115: 2481 -9, 2007

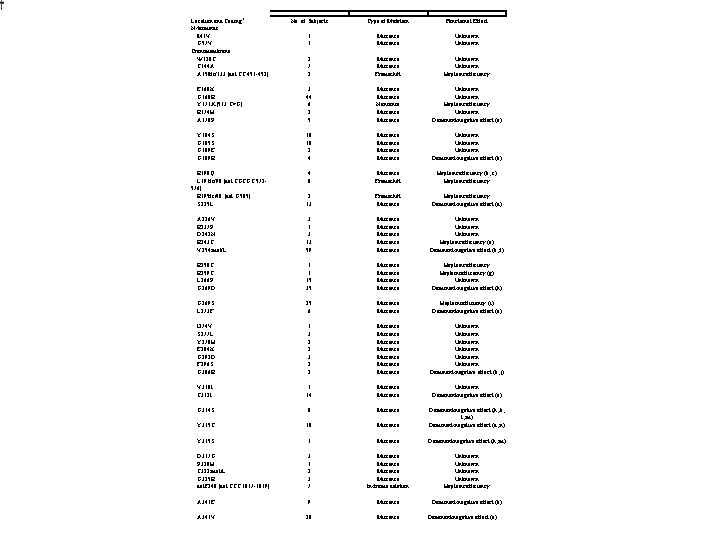
Location and Coding* N-terminus M 1 V G 57 V Transmembrane W 120 C T 144 A A 150 fs/133 [del CT 451 -452] No. of Subjects Type of Mutation Functional Effect 1 1 Missense Unknown 2 7 2 Missense Frameshift Unknown Haploinsufficiency E 160 K G 168 R Y 171 X [513 C>G] R 174 H A 178 P 3 44 6 2 5 Missense Nonsense Missense Unknown Haploinsufficiency Unknown Dominant-negative effect (a) Y 184 S G 185 S G 189 E G 189 R 18 10 2 4 Missense Unknown Dominant-negative effect (b) R 190 Q L 191 fs/90 [del TGCGC 572576] R 195 fs/40 [del G 585] S 225 L 4 8 Missense Frameshift Haploinsufficiency (b, c) Haploinsufficiency 2 13 Frameshift Missense Haploinsufficiency Dominant-negative effect (d) A 226 V R 237 P D 242 N R 243 C V 254 mol/L 3 13 59 Missense Missense Unknown Haploinsufficiency (e) Dominant-negative effect (b, f) R 258 C R 259 C L 266 P G 269 D 1 1 15 35 Missense Haploinsufficiency (g) Unknown Dominant-negative effect (h) G 269 S L 273 F 25 6 Missense Haploinsufficiency (i) Dominant-negative effect (a) I 274 V S 277 L Y 278 H E 284 K G 292 D F 296 S G 306 R 1 3 2 2 Missense Missense Unknown Unknown Dominant-negative effect (b, j) V 310 I T 312 I 1 14 Missense Unknown Dominant-negative effect (a) G 314 S 8 Missense Y 315 C 10 Missense Dominant-negative effect (h, k, l, m) Dominant-negative effect (d, n) Y 315 S 1 Missense Dominant-negative effect (h, m) D 317 G P 320 H T 322 mol/L G 325 R del. F 340 [del CTT 1017 -1019] 3 1 2 3 7 Missense In-frame deletion Unknown Haploinsufficiency A 341 E 9 Missense Dominant-negative effect (b) A 341 V 20 Missense Dominant-negative effect (o)

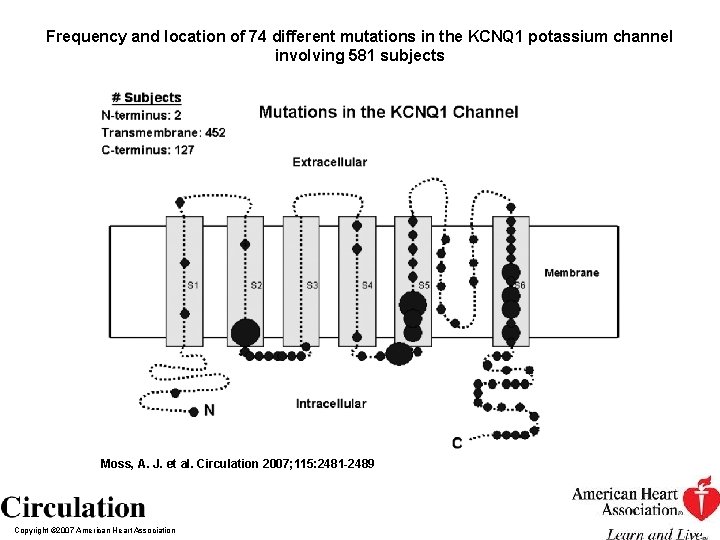
Frequency and location of 74 different mutations in the KCNQ 1 potassium channel involving 581 subjects Moss, A. J. et al. Circulation 2007; 115: 2481 -2489 Copyright © 2007 American Heart Association

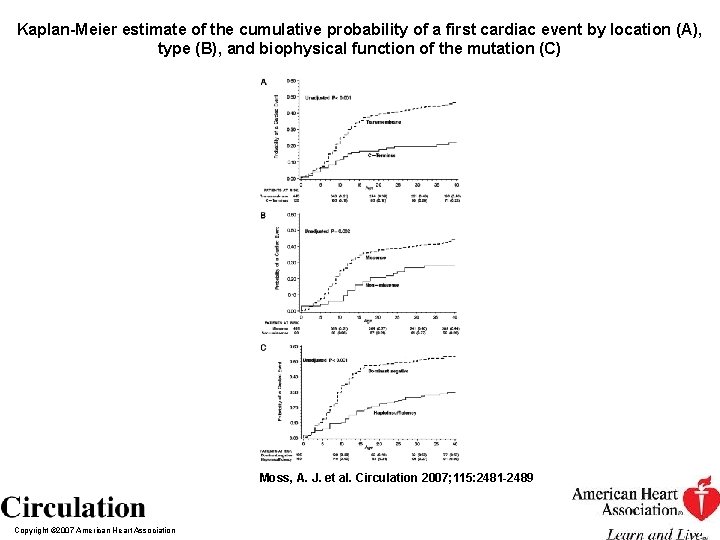
Kaplan-Meier estimate of the cumulative probability of a first cardiac event by location (A), type (B), and biophysical function of the mutation (C) Moss, A. J. et al. Circulation 2007; 115: 2481 -2489 Copyright © 2007 American Heart Association

KCNQ 1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin Ghosh S, Nunziato DA, Pitt GS Circ Res. 98: 1048 -54, 2006

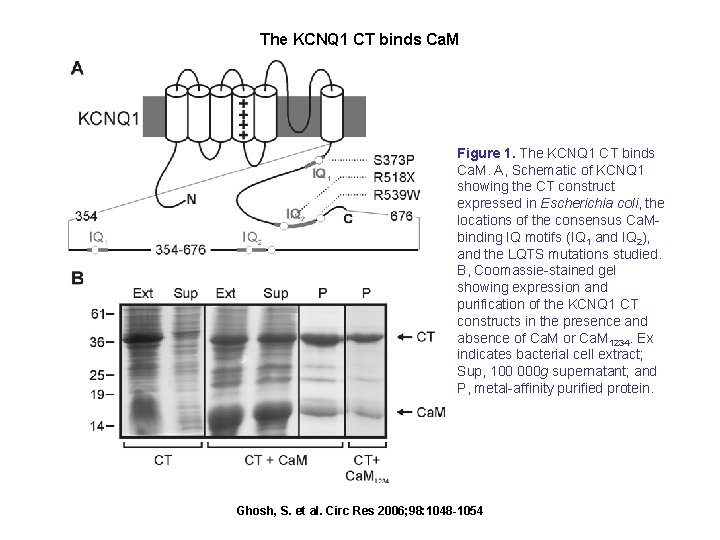
The KCNQ 1 CT binds Ca. M Figure 1. The KCNQ 1 CT binds Ca. M. A, Schematic of KCNQ 1 showing the CT construct expressed in Escherichia coli, the locations of the consensus Ca. Mbinding IQ motifs (IQ 1 and IQ 2), and the LQTS mutations studied. B, Coomassie-stained gel showing expression and purification of the KCNQ 1 CT constructs in the presence and absence of Ca. M or Ca. M 1234. Ex indicates bacterial cell extract; Sup, 100 000 g supernatant; and P, metal-affinity purified protein. Ghosh, S. et al. Circ Res 2006; 98: 1048 -1054

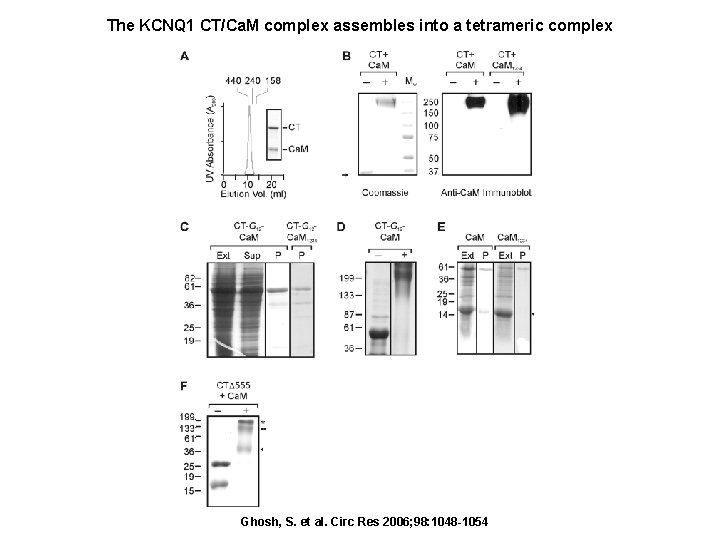
The KCNQ 1 CT/Ca. M complex assembles into a tetrameric complex Ghosh, S. et al. Circ Res 2006; 98: 1048 -1054

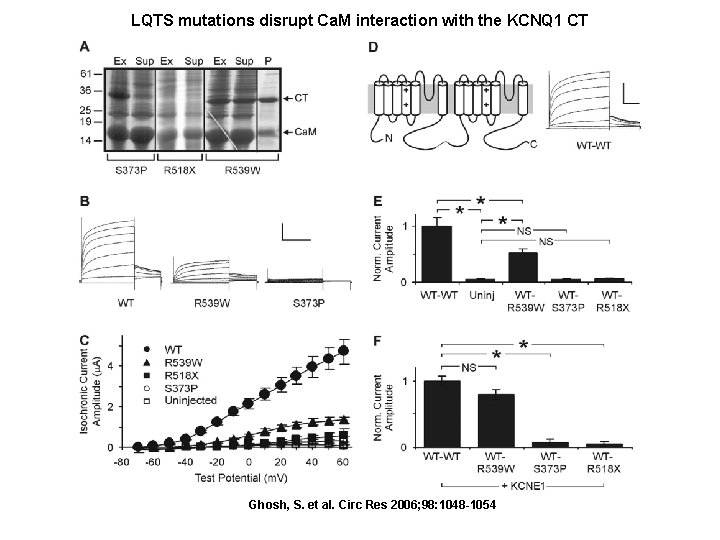
LQTS mutations disrupt Ca. M interaction with the KCNQ 1 CT Ghosh, S. et al. Circ Res 2006; 98: 1048 -1054

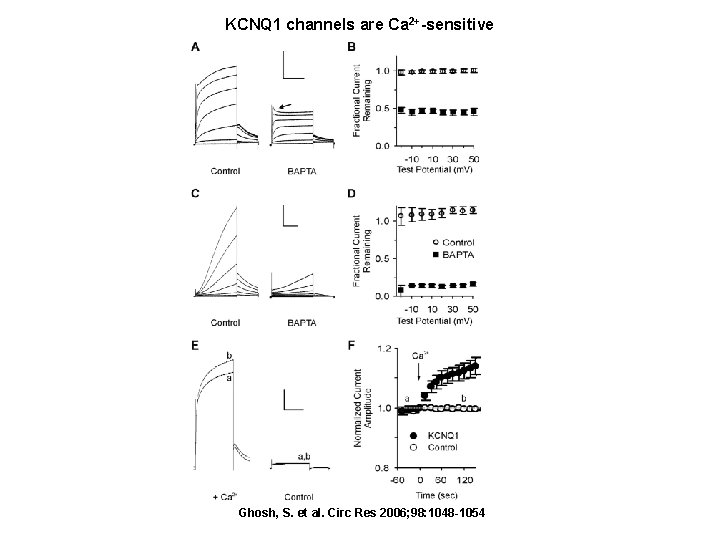
KCNQ 1 channels are Ca 2+-sensitive Ghosh, S. et al. Circ Res 2006; 98: 1048 -1054

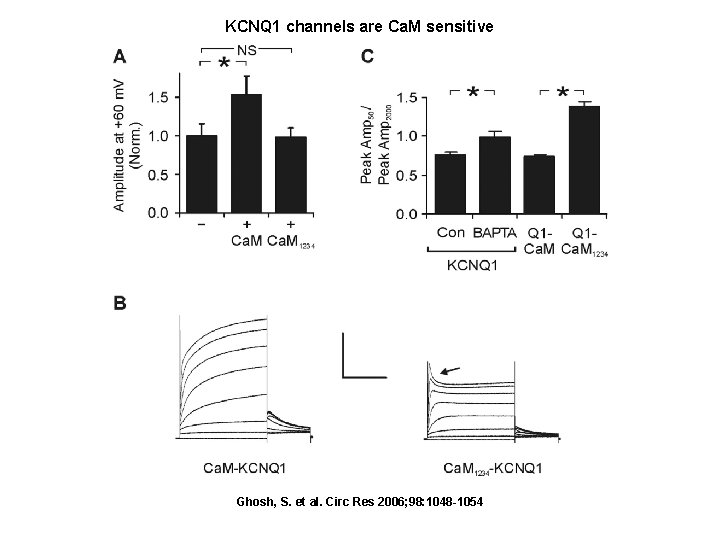
KCNQ 1 channels are Ca. M sensitive Ghosh, S. et al. Circ Res 2006; 98: 1048 -1054