Infectious Diseases Drug Discovery An Astra Zeneca Perspective




![Increasing [Inh] Rate (RFU/min) Biochemical Confirmation of Inhibition Mode ΔRFU [D-Glu] (μM) [Inhibitor] μM Increasing [Inh] Rate (RFU/min) Biochemical Confirmation of Inhibition Mode ΔRFU [D-Glu] (μM) [Inhibitor] μM](https://slidetodoc.com/presentation_image_h/aaa9364f9f1d2d6fe345f0fbd75ed995/image-31.jpg)






- Slides: 60

Infectious Diseases Drug Discovery: An Astra. Zeneca Perspective Tomas Lundqvist GSC LG-DECS Astra. Zeneca R&D Mölndal Stewart L. Fisher Infection Discovery Astra. Zeneca R&D Boston

Astra. Zeneca R&D Boston History • • AZ’s newest research facility Construction initiated August 1998 (Astra) Building completed March 2000 (Astra. Zeneca) Three Research Areas – Infection Discovery (Global Center) – Oncology – Discovery Informatics • Building expansion completed 2003 – Increased resourcing for Oncology • Approximately 450 employees • Expansion underway: – $100 mil investment in capital (buildings) – Increased resource for Infection Research

Why Focus on Infectious Disease? Ø Medical Need Ø Business Opportunity Ø Social Responsibility

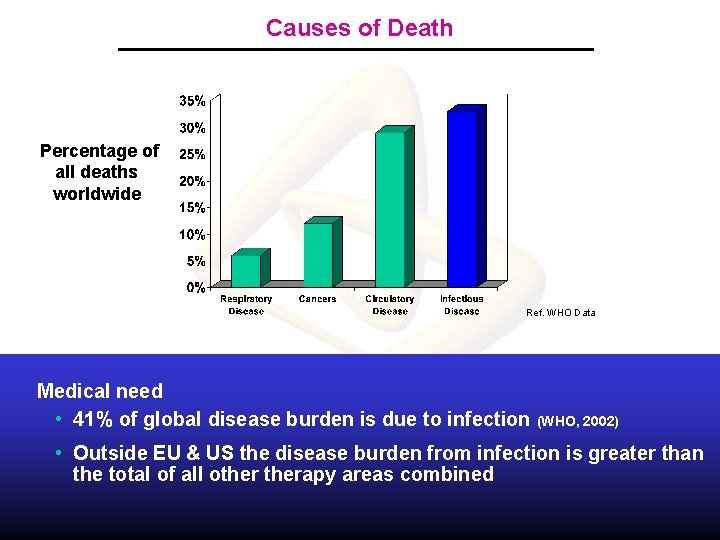
Causes of Death Percentage of all deaths worldwide Ref. WHO Data Medical need • 41% of global disease burden is due to infection (WHO, 2002) • Outside EU & US the disease burden from infection is greater than the total of all otherapy areas combined

A Major Issue for All

The Golden Age & Today The Golden Age of Antibiotic Discovery was very brief, mid 1930 s- early 1960 s penicillin, cephalosporin, streptomycin, erythromycin, tetracycline, vancomycin ØThe pipeline for new antibacterials is drying up ØResistance to antibacterials continues to rise ØThere is a clear & present danger of import to both individual patients and the public health

Target Based Approaches • 1990’s: Dominant lead generation approach – – “Genomic era” Combinatorial/parallel chemistry = large compound libraries Automated screening technologies provided economy of scale Structural approaches most amenable to bacterial targets • Soluble • High yield overproduction/purification • 2000 -present – Approach seen as “not delivering the pipeline” – Many reasons for “failure” • • Poor compound libraries (not as clean as envisioned) Difficult to choose the “druggable” targets Enzyme inhibition ≠ antimicrobial activity (efflux) Sufficient patience in the industry?

Cell Based Approaches • 1990’s: Diminished activity due to target-based approaches – Hit followup appeared “messy” relative to target based – Identification of novel antibiotics increasingly difficult – Major efforts in combinatorial biosynthesis • Genetic manipulation of natural product producers • 2000 -present – renewed interest – Less faith in target based approaches (e. g. lessons from GSK Fab. I) – Improvements in genomic technologies allows facile hit followup • Regulated gene libraries • Target identification via resistance gene mapping – Automated screening technologies affords novel approaches – Approach amenable to pathways and difficult targets

“Look Back” Programs • Revisiting past discoveries, finding new value – – • Ramoplanin, Tiacumicin B – value of C. difficile in 1980 s? Daptomycin – value of MRSA in 1980’s Advances in chemistry make intractable scaffolds amenable – – – ADEPs Anisomycin Moiramide

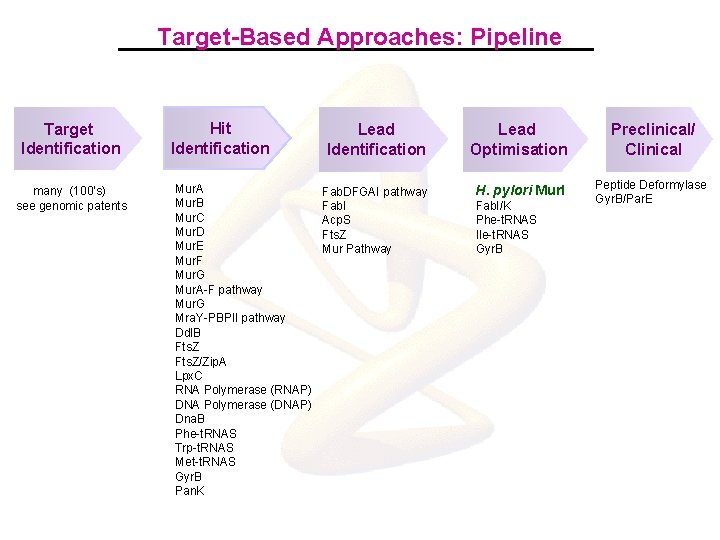
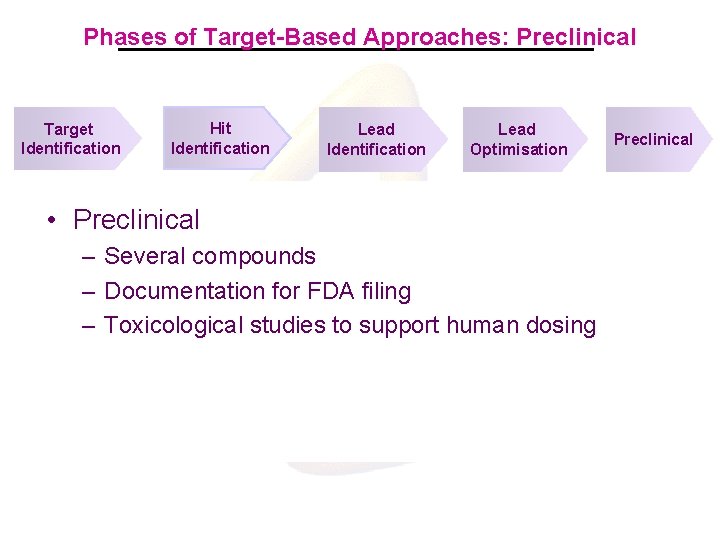
Target-Based Approaches: Pipeline Target Identification many (100’s) see genomic patents Hit Identification Mur. A Mur. B Mur. C Mur. D Mur. E Mur. F Mur. G Mur. A-F pathway Mur. G Mra. Y-PBPII pathway Ddl. B Fts. Z/Zip. A Lpx. C RNA Polymerase (RNAP) DNA Polymerase (DNAP) Dna. B Phe-t. RNAS Trp-t. RNAS Met-t. RNAS Gyr. B Pan. K Lead Identification Lead Optimisation Preclinical/ Clinical Fab. DFGAI pathway Fab. I Acp. S Fts. Z Mur Pathway H. pylori Mur. I Peptide Deformylase Gyr. B/Par. E Fab. I/K Phe-t. RNAS Ile-t. RNAS Gyr. B

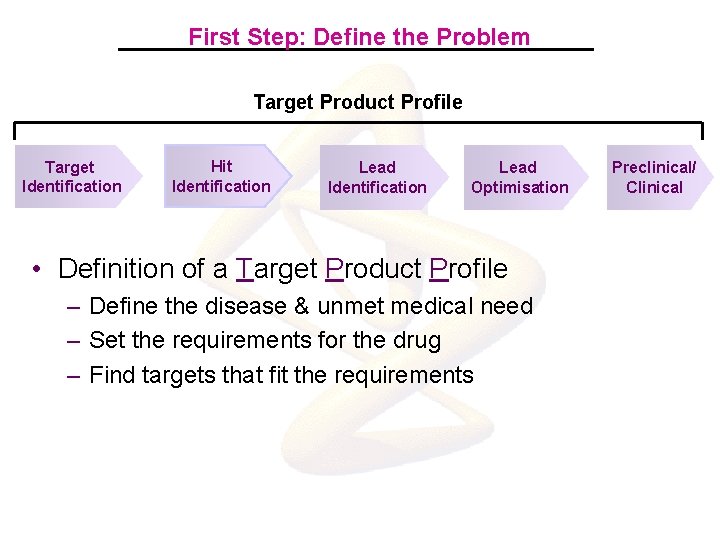
First Step: Define the Problem Target Product Profile Target Identification Hit Identification Lead Optimisation • Definition of a Target Product Profile – Define the disease & unmet medical need – Set the requirements for the drug – Find targets that fit the requirements Preclinical/ Clinical

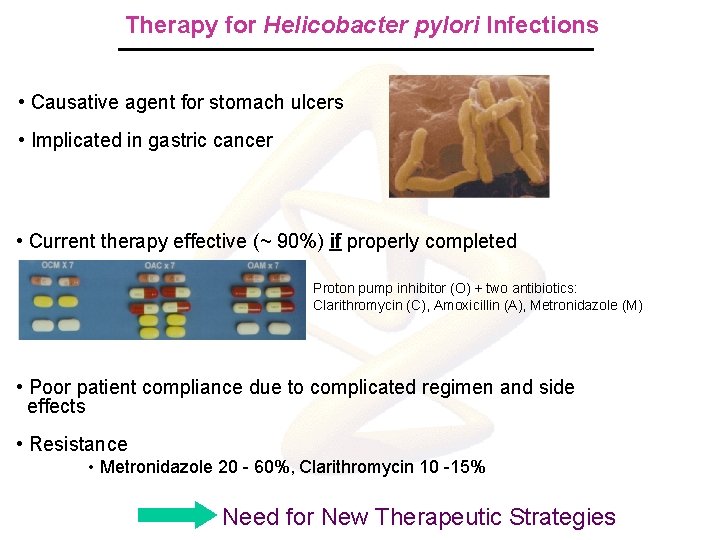
Therapy for Helicobacter pylori Infections • Causative agent for stomach ulcers • Implicated in gastric cancer • Current therapy effective (~ 90%) if properly completed Proton pump inhibitor (O) + two antibiotics: Clarithromycin (C), Amoxicillin (A), Metronidazole (M) • Poor patient compliance due to complicated regimen and side effects • Resistance • Metronidazole 20 - 60%, Clarithromycin 10 -15% Need for New Therapeutic Strategies

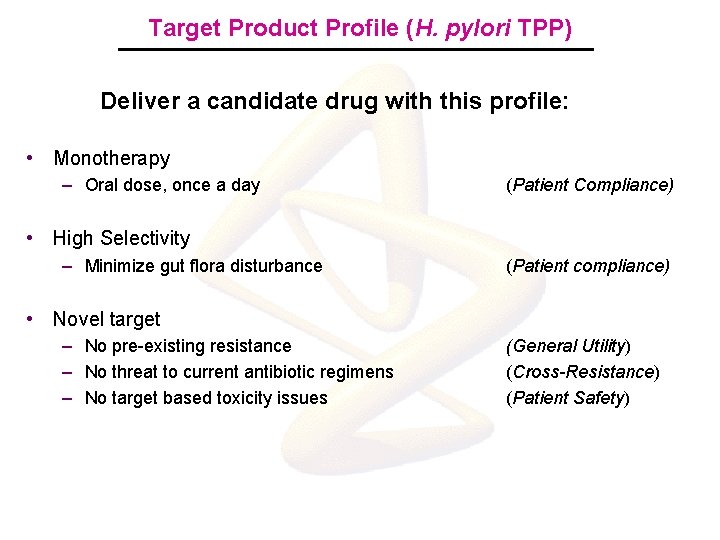
Target Product Profile (H. pylori TPP) Deliver a candidate drug with this profile: • Monotherapy – Oral dose, once a day (Patient Compliance) • High Selectivity – Minimize gut flora disturbance (Patient compliance) • Novel target – No pre-existing resistance – No threat to current antibiotic regimens – No target based toxicity issues (General Utility) (Cross-Resistance) (Patient Safety)

Phases of Target-Based Approach: Target Identification • Target Identification – – Genomics-based selection Validation of essentiality in relevant organisms Cloning and expression of target proteins Production of target proteins

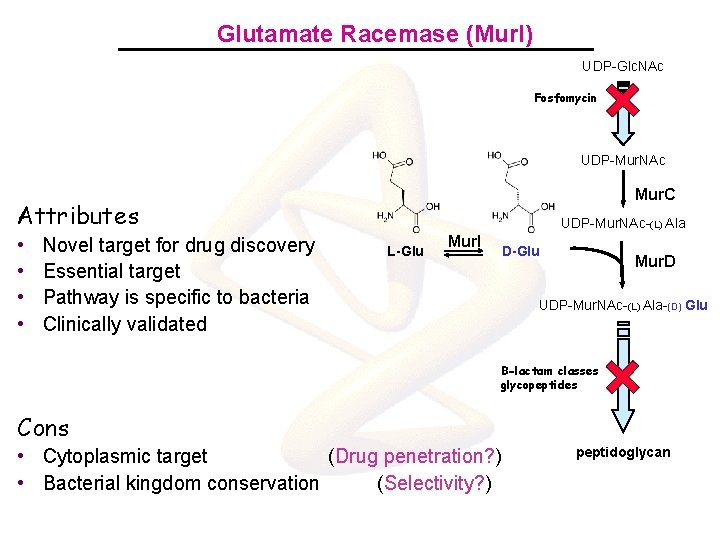
Glutamate Racemase (Mur. I) UDP-Glc. NAc Fosfomycin UDP-Mur. NAc Mur. C Attributes • • Novel target for drug discovery Essential target Pathway is specific to bacteria Clinically validated UDP-Mur. NAc-(L) Ala L-Glu Mur. I D-Glu Mur. D UDP-Mur. NAc-(L) Ala-(D) Glu B-lactam classes glycopeptides Cons • Cytoplasmic target (Drug penetration? ) • Bacterial kingdom conservation (Selectivity? ) peptidoglycan

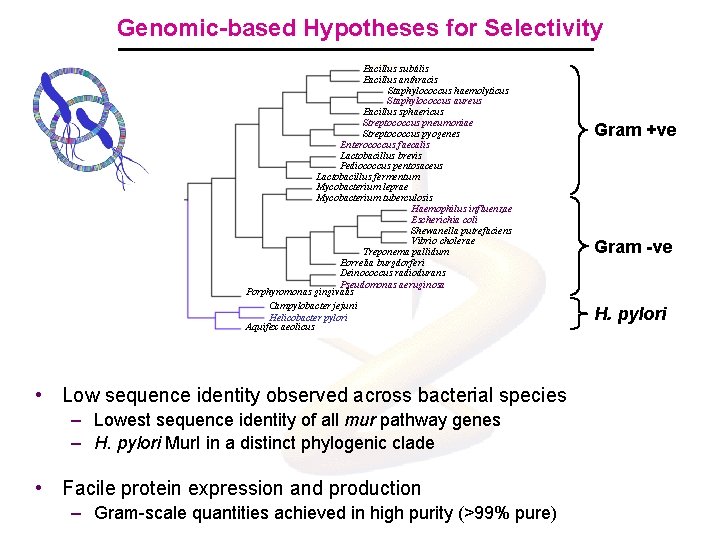
Genomic-based Hypotheses for Selectivity Bacillus subtilis Bacillus anthracis Staphylococcus haemolyticus Staphylococcus aureus Bacillus sphaericus Streptococcus pneumoniae Streptococcus pyogenes Enterococcus faecalis Lactobacillus brevis Pediococcus pentosaceus Lactobacillus fermentum Mycobacterium leprae Mycobacterium tuberculosis Haemophilus influenzae Escherichia coli Shewanella putrefaciens Vibrio cholerae Treponema pallidum Borrelia burgdorferi Deinococcus radiodurans Pseudomonas aeruginosa Porphyromonas gingivalis Campylobacter jejuni Helicobacter pylori Aquifex aeolicus • Low sequence identity observed across bacterial species – Lowest sequence identity of all mur pathway genes – H. pylori Mur. I in a distinct phylogenic clade • Facile protein expression and production – Gram-scale quantities achieved in high purity (>99% pure) Gram +ve Gram -ve H. pylori

Phases of Target-Based Approach: Hit Identification Target Identification Hit Identification • Hit Identification – Biophysical and biochemical characterization of targets – Development of primary assay and secondary assays for evaluation of hits – Kinetic mechanism studies for enzyme targets – Screening (e. g. HTS, virtual) and chem-informatic analysis – Limited SAR generation

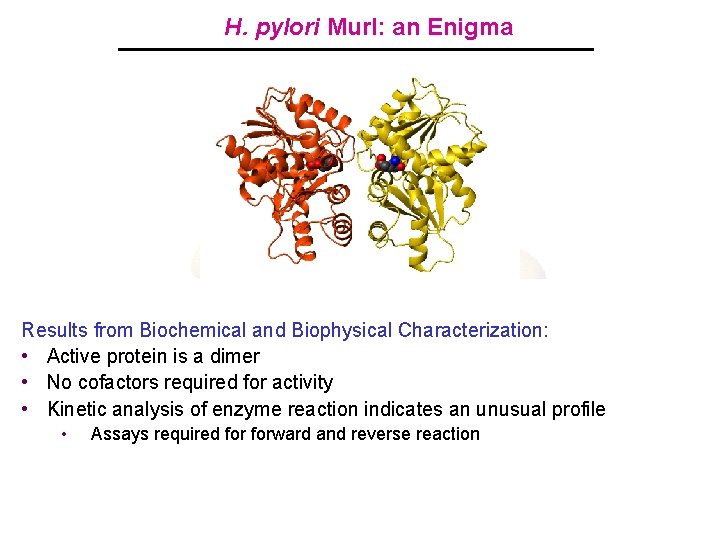
H. pylori Mur. I: an Enigma • Novel Enzyme Crystal Structure Solved – 1998 Results from Biochemical and Biophysical Characterization: Crystal Structure Features • • Active protein is a dimer Dimericrequired enzyme for activity • No –cofactors – Active sitesofoccluded solvent • Kinetic analysis enzymefrom reaction indicates an unusual profile Selective binding D-Glu and reverse reaction • – Assays required for of forward

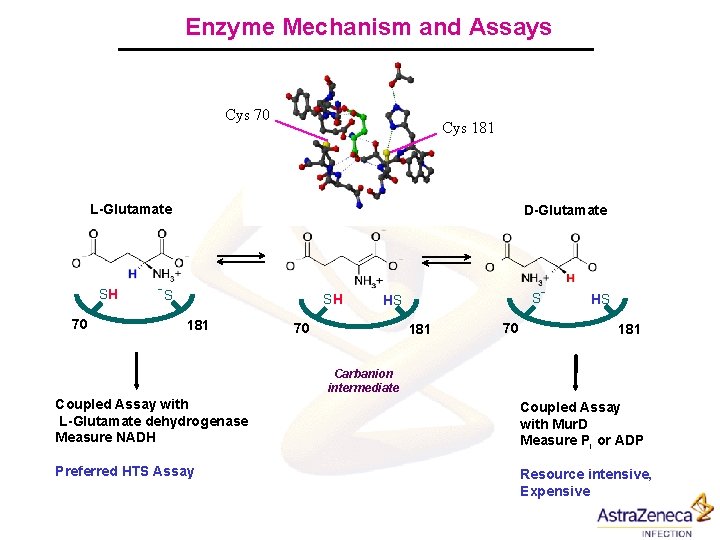
Enzyme Mechanism and Assays Cys 70 Cys 181 L-Glutamate SH 70 D-Glutamate -S SH 181 S HS 70 181 70 - HS 181 Carbanion intermediate Coupled Assay with L-Glutamate dehydrogenase Measure NADH Coupled Assay with Mur. D Measure Pi or ADP Preferred HTS Assay Resource intensive, Expensive

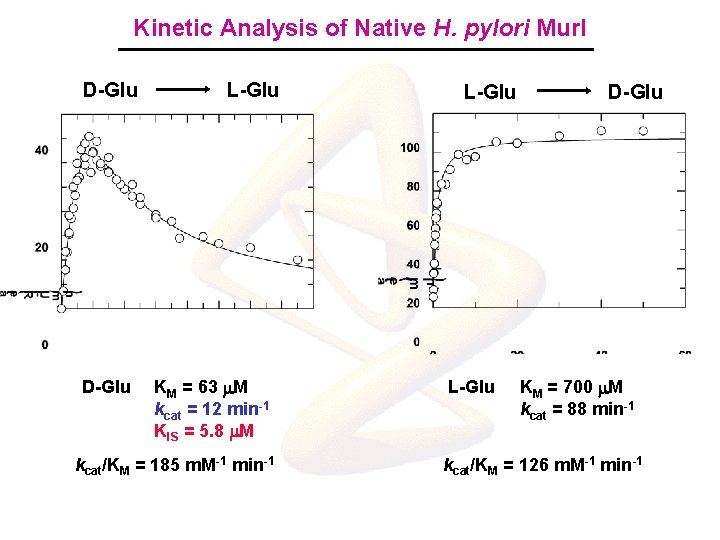
Kinetic Analysis of Native H. pylori Mur. I D-Glu L-Glu KM = 63 m. M kcat = 12 min-1 KIS = 5. 8 m. M kcat/KM = 185 m. M-1 min-1 L-Glu D-Glu KM = 700 m. M kcat = 88 min-1 kcat/KM = 126 m. M-1 min-1

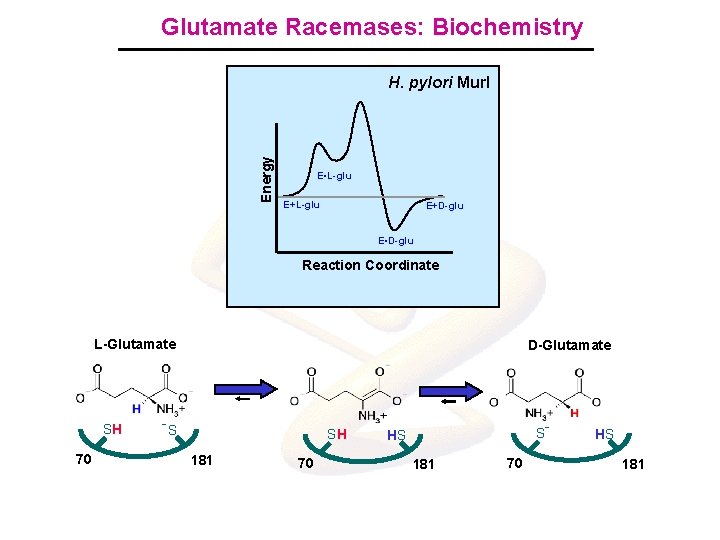
Glutamate Racemases: Biochemistry Energy H. pylori Mur. I E. L-glu E+L-glu E+D-glu E. L-glu E. D-glu. E D-glu Reaction. Coordinate L-Glutamate SH 70 D-Glutamate -S SH 181 70 S HS 181 70 - HS 181

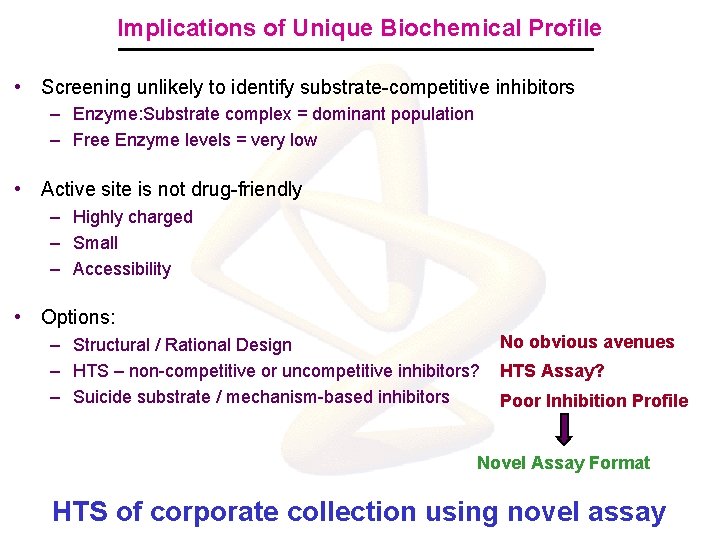
Implications of Unique Biochemical Profile • Screening unlikely to identify substrate-competitive inhibitors – Enzyme: Substrate complex = dominant population – Free Enzyme levels = very low • Active site is not drug-friendly – Highly charged – Small – Accessibility • Options: – Structural / Rational Design – HTS – non-competitive or uncompetitive inhibitors? – Suicide substrate / mechanism-based inhibitors No obvious avenues HTS Assay? Poor Inhibition Profile Novel Assay Format HTS of corporate collection using novel assay

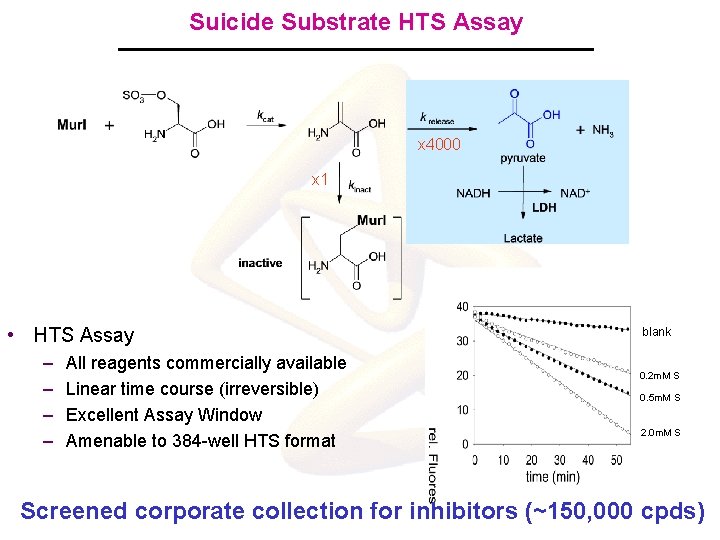
Suicide Substrate HTS Assay x 4000 x 1 • HTS Assay – – All reagents commercially available Linear time course (irreversible) Excellent Assay Window Amenable to 384 -well HTS format blank 0. 2 m. M S 0. 5 m. M S 2. 0 m. M S Screened corporate collection for inhibitors (~150, 000 cpds)

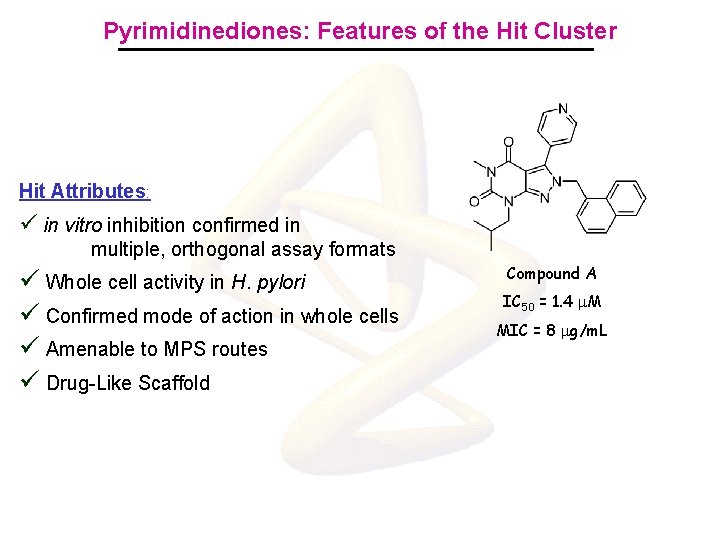
Pyrimidinediones: Features of the Hit Cluster Hit Attributes: ü in vitro inhibition confirmed in multiple, orthogonal assay formats ü Whole cell activity in H. pylori ü Confirmed mode of action in whole cells ü Amenable to MPS routes ü Drug-Like Scaffold Compound A IC 50 = 1. 4 m. M MIC = 8 mg/m. L

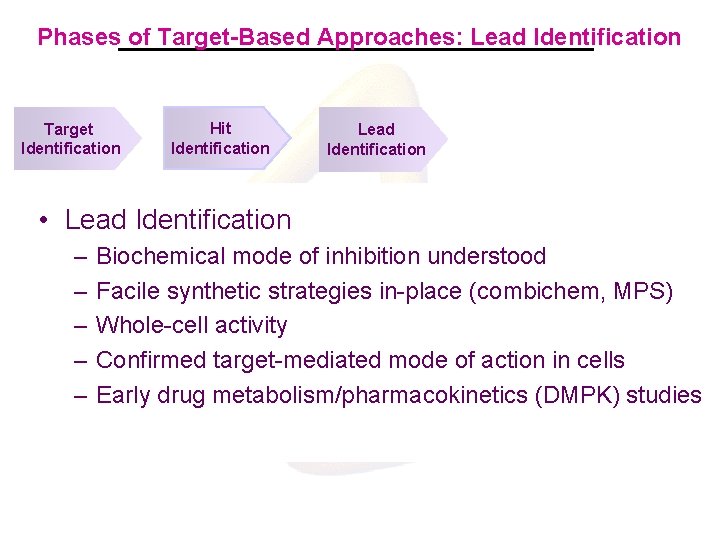
Phases of Target-Based Approaches: Lead Identification Target Identification Hit Identification Lead Identification Hit Identification • • Target Identification Lead Identification Biochemicaland mode of inhibition understood of targets Biophysical biochemical characterization –– Genomics-based selection Facile synthetic in-place (combichem, MPS) Development of strategies primary in HTS assayorganisms and secondary –– Validation of essentiality relevant assays for evaluation of hits – Whole-cell activity – Cloning and expression of target proteins – Kinetic mechanism studies for enzyme targets – Confirmed target-mediated mode of action in cells – Production of target proteins – and chem-informatic analysis – HTS Early. Screening drug metabolism/pharmacokinetics (DMPK) studies – Limited SAR generation

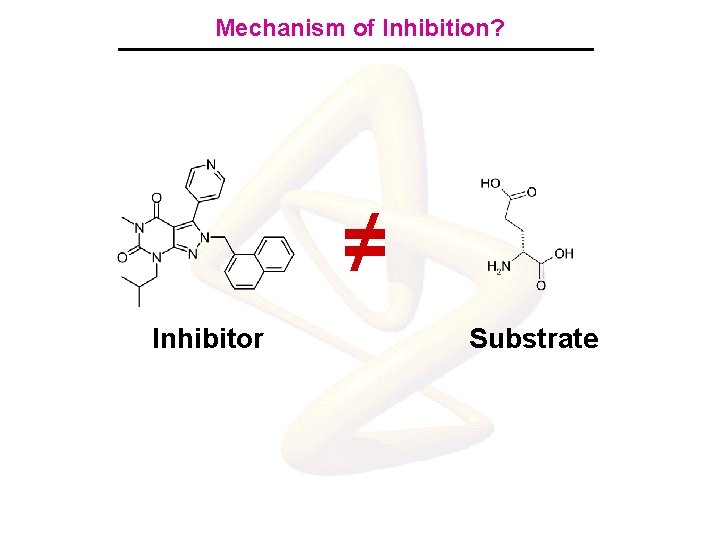
Mechanism of Inhibition? ≠ Inhibitor Substrate

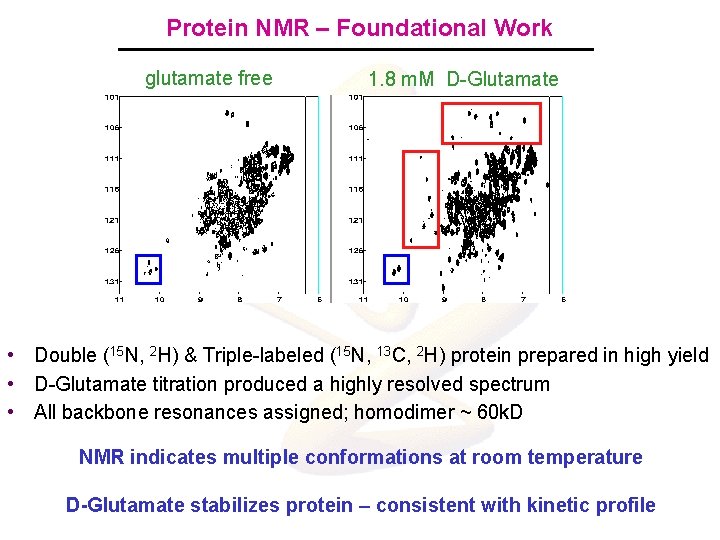
Protein NMR – Foundational Work glutamate free 1. 8 m. M D-Glutamate • Double (15 N, 2 H) & Triple-labeled (15 N, 13 C, 2 H) protein prepared in high yield • D-Glutamate titration produced a highly resolved spectrum • All backbone resonances assigned; homodimer ~ 60 k. D NMR indicates multiple conformations at room temperature D-Glutamate stabilizes protein – consistent with kinetic profile

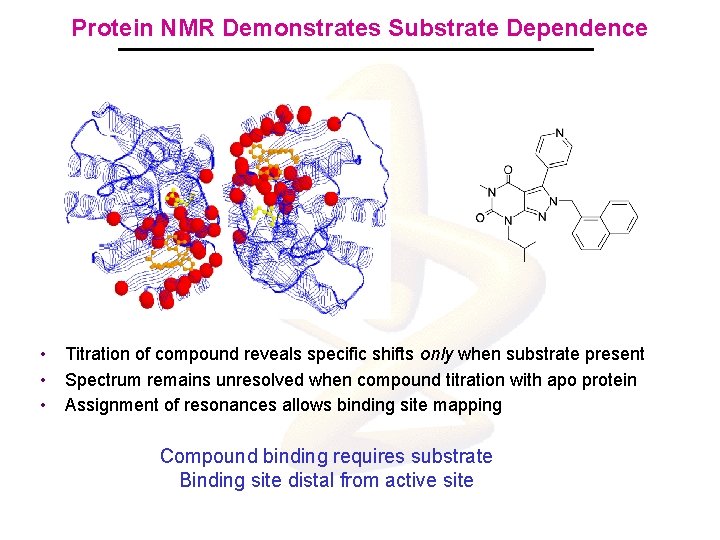
Protein NMR Demonstrates Substrate Dependence Black = D-Glu + Mur. I Red = D-Glu + Mur. I + Inh • • • Titration of compound reveals specific shifts only when substrate present Spectrum remains unresolved when compound titration with apo protein Assignment of resonances allows binding site mapping Compound binding requires substrate Binding site distal from active site

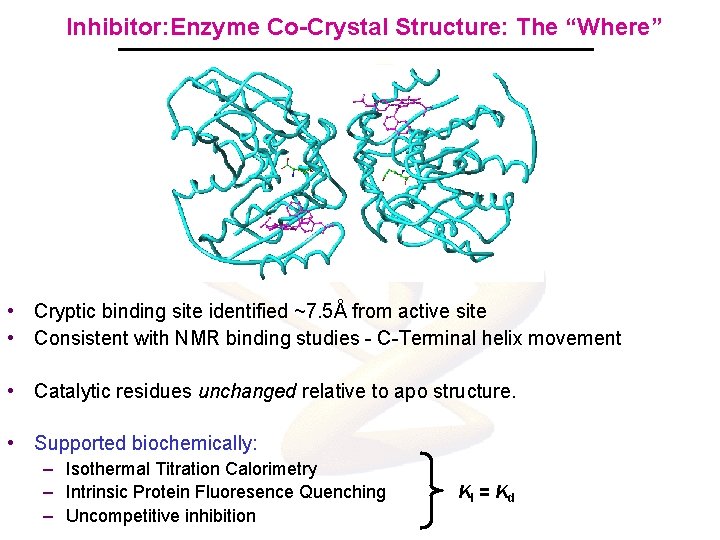
Inhibitor: Enzyme Co-Crystal Structure: The “Where” • Cryptic binding site identified ~7. 5Å from active site • Consistent with NMR binding studies - C-Terminal helix movement • Catalytic residues unchanged relative to apo structure. • Supported biochemically: – Isothermal Titration Calorimetry – Intrinsic Protein Fluoresence Quenching – Uncompetitive inhibition KI = Kd

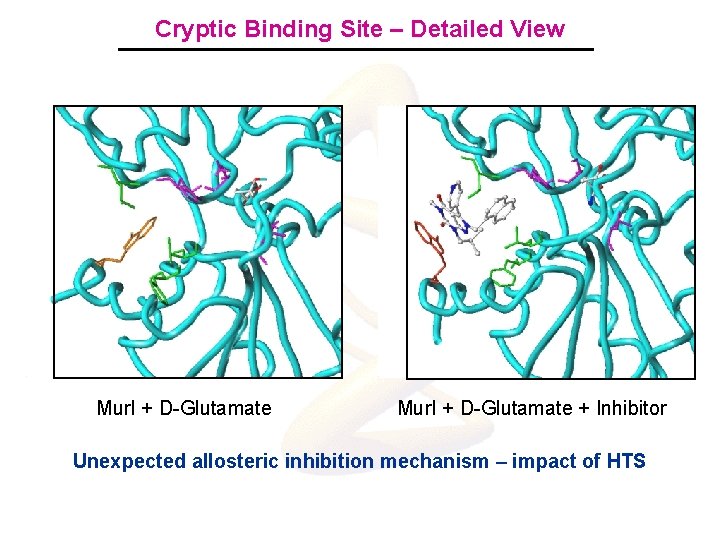
Cryptic Binding Site – Detailed View Mur. I + D-Glutamate + Inhibitor Unexpected allosteric inhibition mechanism – impact of HTS
![Increasing Inh Rate RFUmin Biochemical Confirmation of Inhibition Mode ΔRFU DGlu μM Inhibitor μM Increasing [Inh] Rate (RFU/min) Biochemical Confirmation of Inhibition Mode ΔRFU [D-Glu] (μM) [Inhibitor] μM](https://slidetodoc.com/presentation_image_h/aaa9364f9f1d2d6fe345f0fbd75ed995/image-31.jpg)
Increasing [Inh] Rate (RFU/min) Biochemical Confirmation of Inhibition Mode ΔRFU [D-Glu] (μM) [Inhibitor] μM • Binding mode confirmed in multiple formats: – Intrinsic Protein Fluorescence Quenching – Isothermal Titration Calorimetry • Kinetic Mechanism Consistent with Uncompetitive Inhibition KI = IC 50

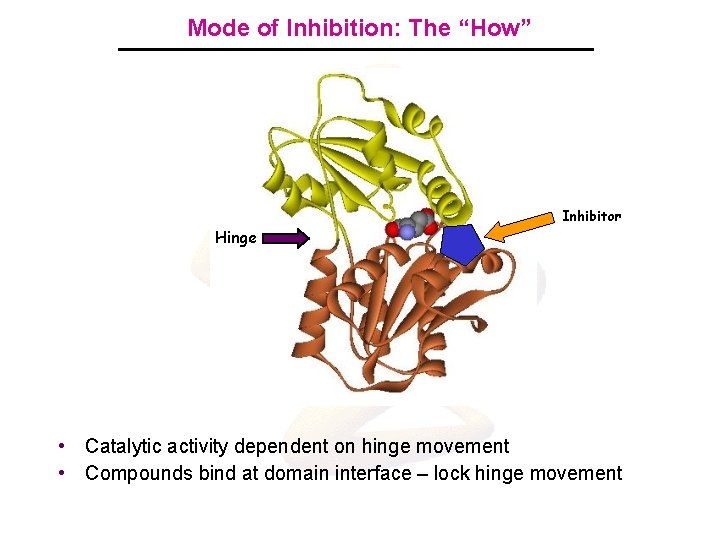
Mode of Inhibition: The “How” Inhibitor Hinge • Catalytic activity dependent on hinge movement • Compounds bind at domain interface – lock hinge movement

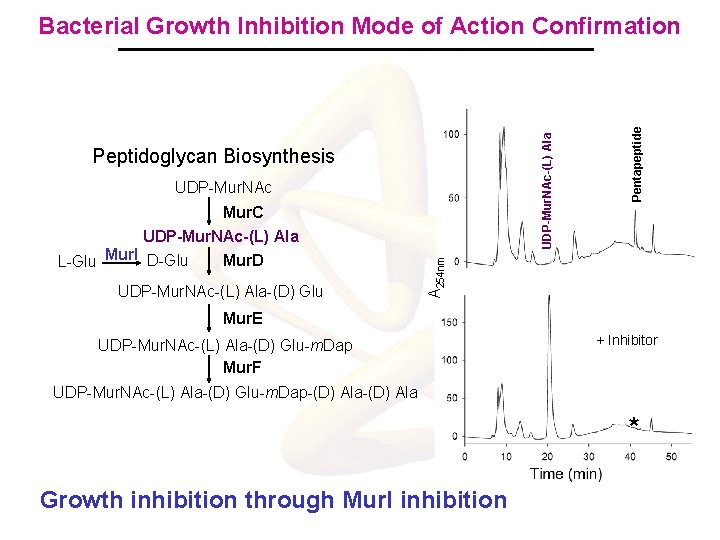
Peptidoglycan Biosynthesis UDP-Mur. NAc L-Glu Mur. I D-Glu Mur. D UDP-Mur. NAc-(L) Ala-(D) Glu A 254 nm Mur. C UDP-Mur. NAc-(L) Ala Pentapeptide UDP-Mur. NAc-(L) Ala Bacterial Growth Inhibition Mode of Action Confirmation Mur. E UDP-Mur. NAc-(L) Ala-(D) Glu-m. Dap Mur. F + Inhibitor UDP-Mur. NAc-(L) Ala-(D) Glu-m. Dap-(D) Ala * Growth inhibition through Mur. I inhibition

Phases of Target-Based Approaches: Lead Optimization Target Identification Hit Identification • Lead Optimization Hit Identification • Target. Identification Lead Optimization – Biophysical Focus on analogs of central scaffold(s) Facile synthetic (combichem, MPS) and strategies biochemical characterization of targets – Genomics-based selection in-place Activity in animal disease-state model Biochemical mode of inhibition understood Development of primary HTS assay and secondary –– Validation of essentiality in relevant organisms for evaluation of hits – assays Assess potential for resistance Whole-cell activity – Cloning and expression of target proteins – Kinetic mechanism studies for enzyme targets in vivo DMPK studies for human dosing estimation – Confirmed target-mediated mode of action in cells – Production of target proteins – Screening and studies chem-informatic analysis in vitro toxicological – HTS Early drug metabolism/pharmacokinetics (DMPK) studies – Limited generation Scale up. SAR synthesis; process chemistry

Trojan Horse or Goldmine? Can we improve potency? What is the potential for resistance? Can we achieve the desired selectivity margin?

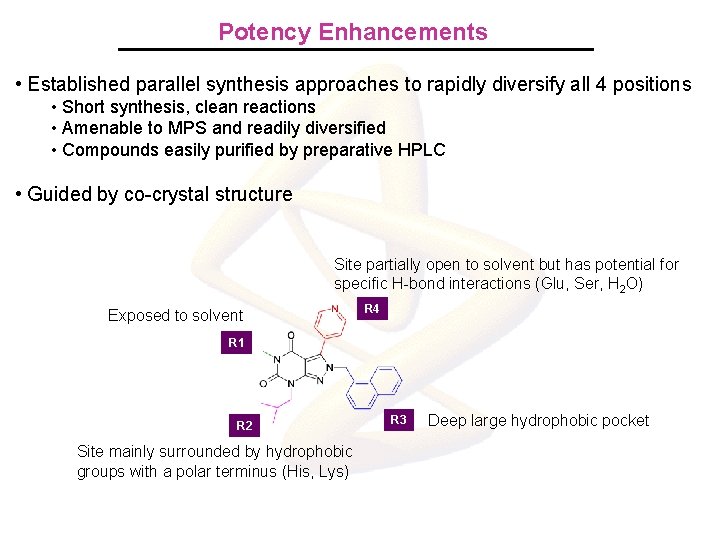
Potency Enhancements • Established parallel synthesis approaches to rapidly diversify all 4 positions • Short synthesis, clean reactions • Amenable to MPS and readily diversified • Compounds easily purified by preparative HPLC • Guided by co-crystal structure Site partially open to solvent but has potential for specific H-bond interactions (Glu, Ser, H 2 O) Exposed to solvent R 4 R 1 R 2 Site mainly surrounded by hydrophobic groups with a polar terminus (His, Lys) R 3 Deep large hydrophobic pocket

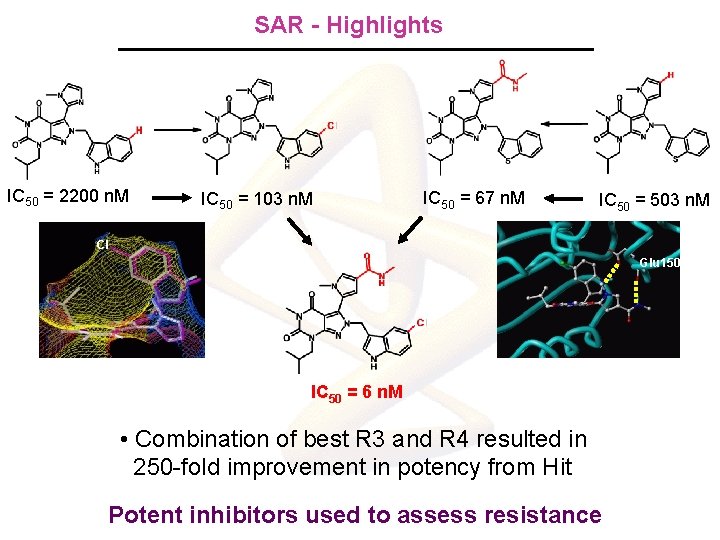
SAR - Highlights IC 50 = 2200 n. M IC 50 = 103 n. M IC 50 = 67 n. M IC 50 = 503 n. M Cl Glu 150 IC 50 = 6 n. M • Combination of best R 3 and R 4 resulted in 250 -fold improvement in potency from Hit Potent inhibitors used to assess resistance

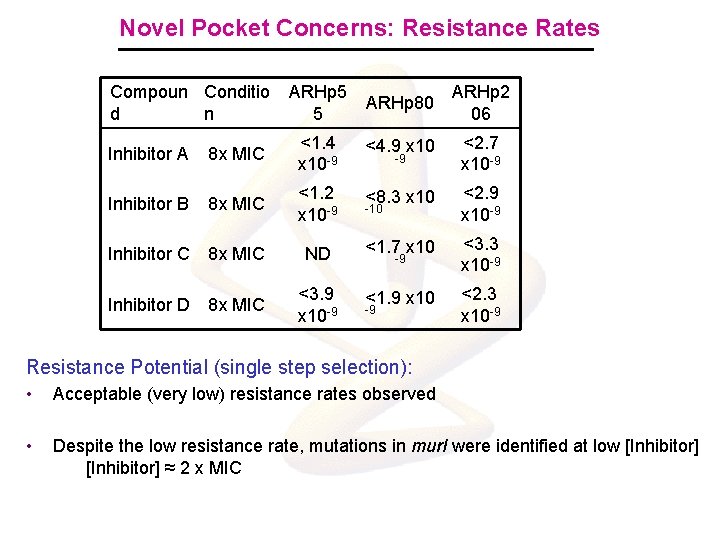
Novel Pocket Concerns: Resistance Rates Compoun Conditio d n ARHp 5 5 ARHp 80 ARHp 2 06 Inhibitor A 8 x MIC <1. 4 x 10 -9 <4. 9 x 10 <2. 7 x 10 -9 Inhibitor B 8 x MIC <1. 2 x 10 -9 <8. 3 x 10 <2. 9 x 10 -9 Inhibitor C 8 x MIC ND <1. 7 x 10 <3. 3 x 10 -9 Inhibitor D 8 x MIC <3. 9 x 10 -9 <1. 9 x 10 <2. 3 x 10 -9 -9 -10 -9 -9 Resistance Potential (single step selection): • Acceptable (very low) resistance rates observed • Despite the low resistance rate, mutations in mur. I were identified at low [Inhibitor] ≈ 2 x MIC

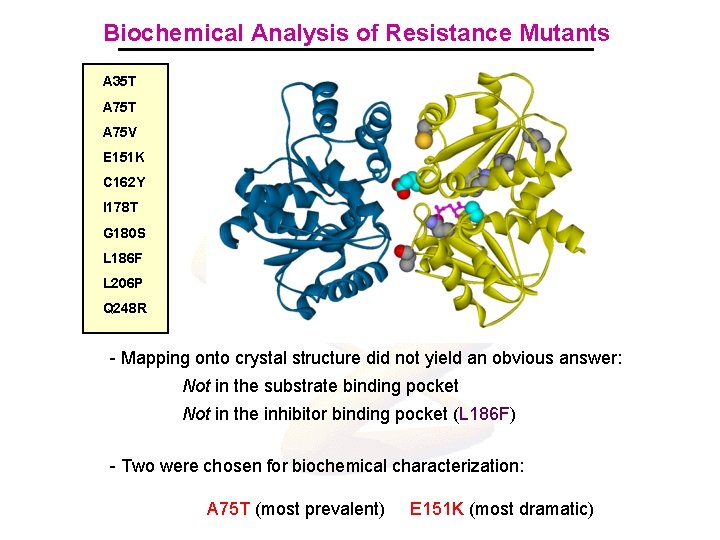
Biochemical Analysis of Resistance Mutants A 35 T A 75 V E 151 K C 162 Y I 178 T G 180 S L 186 F L 206 P Q 248 R - Mapping onto crystal structure did not yield an obvious answer: Not in the substrate binding pocket Not in the inhibitor binding pocket (L 186 F) - Two were chosen for biochemical characterization: A 75 T (most prevalent) E 151 K (most dramatic)

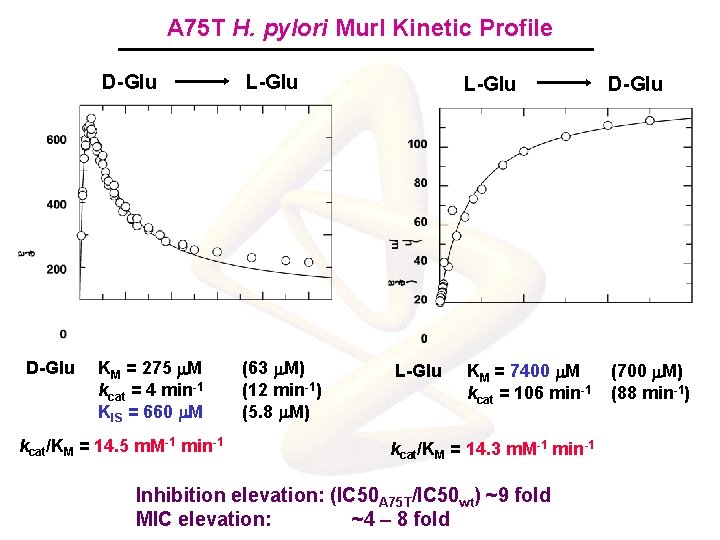
A 75 T H. pylori Mur. I Kinetic Profile D-Glu L-Glu KM = 275 m. M kcat = 4 min-1 KIS = 660 m. M (63 m. M) (12 min-1) (5. 8 m. M) kcat/KM = 14. 5 m. M-1 min-1 L-Glu D-Glu KM = 7400 m. M kcat = 106 min-1 (700 m. M) (88 min-1) kcat/KM = 14. 3 m. M-1 min-1 Inhibition elevation: (IC 50 A 75 T/IC 50 wt) ~9 fold MIC elevation: ~4 – 8 fold

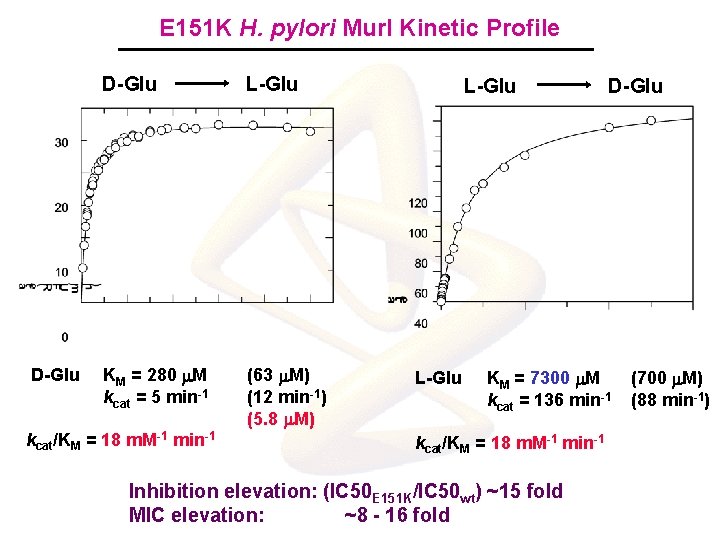
E 151 K H. pylori Mur. I Kinetic Profile D-Glu L-Glu KM = 280 m. M kcat = 5 min-1 (63 m. M) (12 min-1) (5. 8 m. M) kcat/KM = 18 m. M-1 min-1 L-Glu D-Glu KM = 7300 m. M kcat = 136 min-1 kcat/KM = 18 m. M-1 min-1 Inhibition elevation: (IC 50 E 151 K/IC 50 wt) ~15 fold MIC elevation: ~8 - 16 fold (700 m. M) (88 min-1)

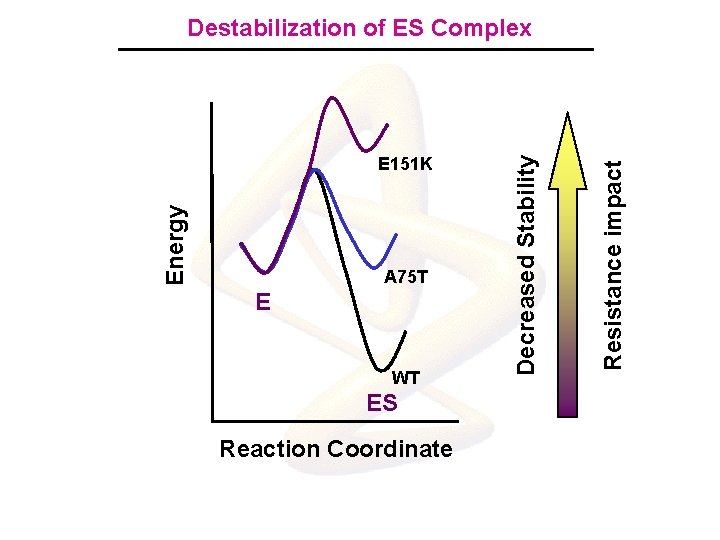
A 75 T E WT ES Reaction Coordinate Resistance impact Energy E 151 K Decreased Stability Destabilization of ES Complex

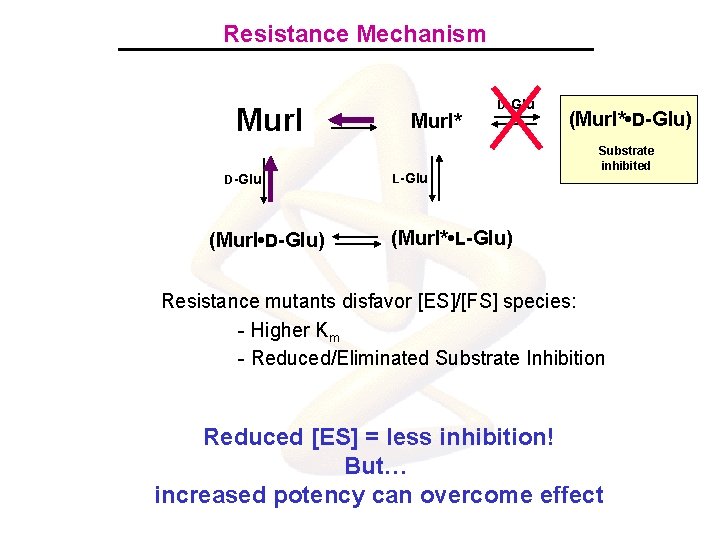
Resistance Mechanism Mur. I D-Glu (Mur. I • D-Glu) Mur. I* D-Glu L-Glu (Mur. I* • D-Glu) Substrate inhibited (Mur. I* • L-Glu) Resistance mutants disfavor [ES]/[FS] species: - Higher Km - Reduced/Eliminated Substrate Inhibition Reduced [ES] = less inhibition! But… increased potency can overcome effect

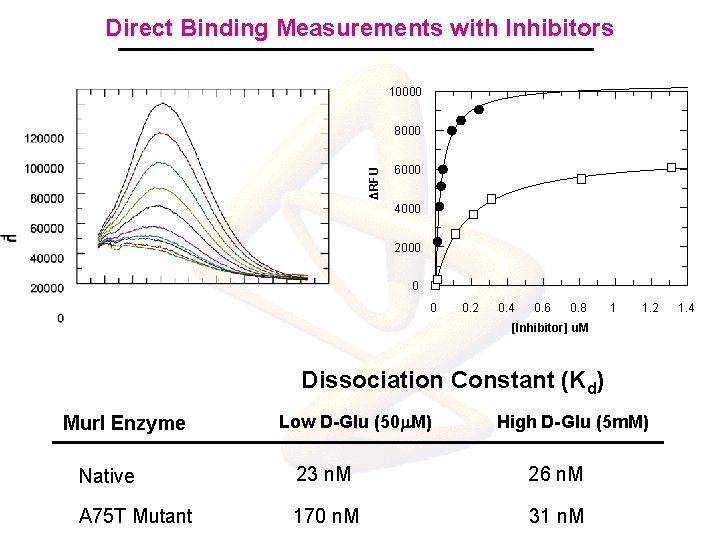
Direct Binding Measurements with Inhibitors 10000 ΔRFU 8000 6000 4000 2000 0 0 0. 2 0. 4 0. 6 0. 8 1 1. 2 [Inhibitor] u. M Dissociation Constant (Kd) Mur. I Enzyme Low D-Glu (50 m. M) High D-Glu (5 m. M) Native 23 n. M 26 n. M A 75 T Mutant 170 n. M 31 n. M 1. 4

Bacterial Selectivity Requirement What about the selectivity profile?

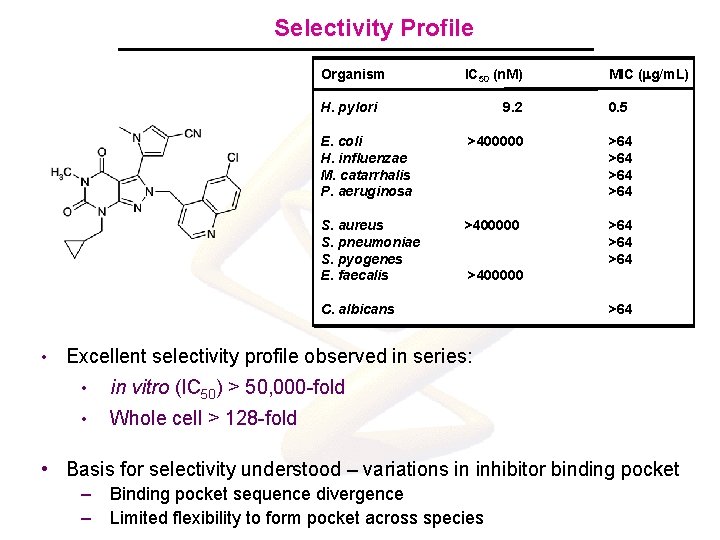
Selectivity Profile Organism IC 50 (n. M) H. pylori 9. 2 0. 5 E. coli H. influenzae M. catarrhalis P. aeruginosa >400000 >64 >64 S. aureus S. pneumoniae S. pyogenes E. faecalis >400000 >64 >64 >400000 C. albicans • MIC (mg/m. L) >64 Excellent selectivity profile observed in series: • in vitro (IC 50) > 50, 000 -fold • Whole cell > 128 -fold • Basis for selectivity understood – variations in inhibitor binding pocket – – Binding pocket sequence divergence Limited flexibility to form pocket across species

Trojan Horse or Goldmine? Can we improve potency? YES! What is the potential for resistance? Low Can we achieve the desired selectivity margin? So, where’s the drug? YES!

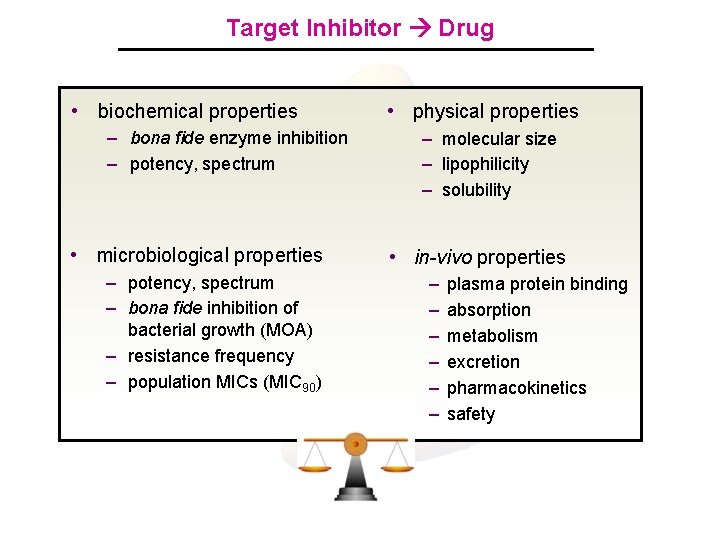
Target Inhibitor Drug • biochemical properties – bona fide enzyme inhibition – potency, spectrum • microbiological properties – potency, spectrum – bona fide inhibition of bacterial growth (MOA) – resistance frequency – population MICs (MIC 90) • physical properties – molecular size – lipophilicity – solubility • in-vivo properties – – – plasma protein binding absorption metabolism excretion pharmacokinetics safety

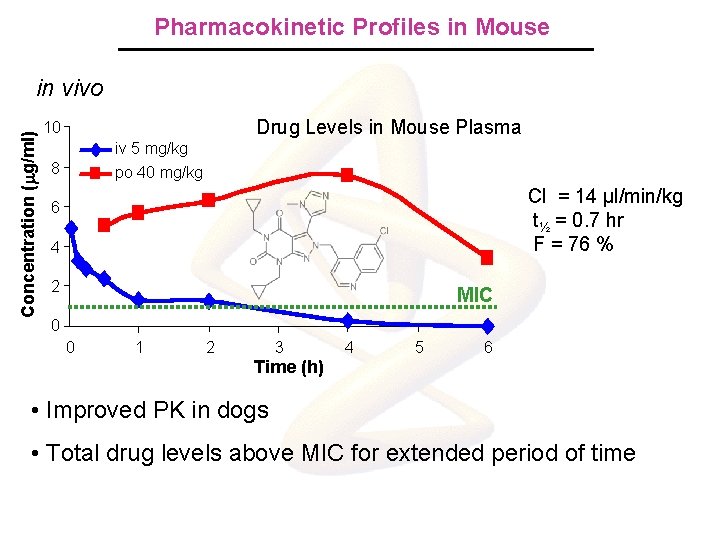
Pharmacokinetic Profiles in Mouse Concentration (mg/ml) in vivo Drug Levels in Mouse Plasma 10 iv 5 mg/kg po 40 mg/kg 8 Cl = 14 µl/min/kg t½ = 0. 7 hr F = 76 % 6 4 2 MIC 0 0 1 2 3 4 5 6 Time (h) • Improved PK in dogs • Total drug levels above MIC for extended period of time

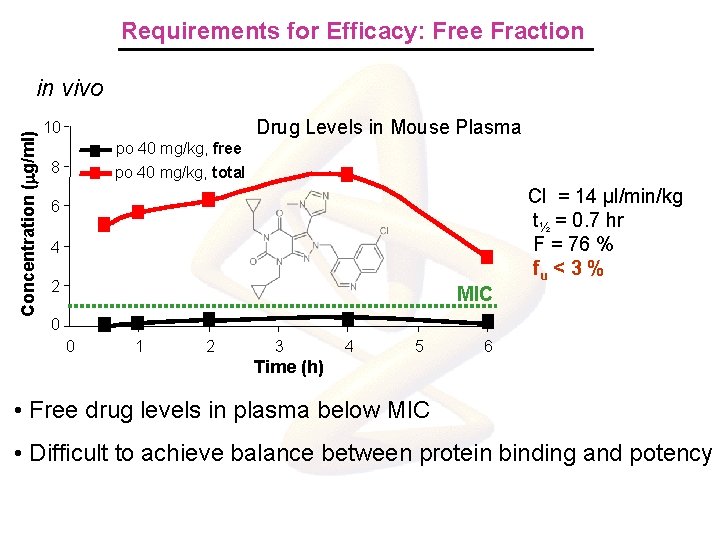
Requirements for Efficacy: Free Fraction Concentration (mg/ml) in vivo Drug Levels in Mouse Plasma 10 po 40 mg/kg, free po 40 mg/kg, total 8 Cl = 14 µl/min/kg t½ = 0. 7 hr F = 76 % fu < 3 % 6 4 2 MIC 0 0 1 2 3 4 5 6 Time (h) • Free drug levels in plasma below MIC • Difficult to achieve balance between protein binding and potency

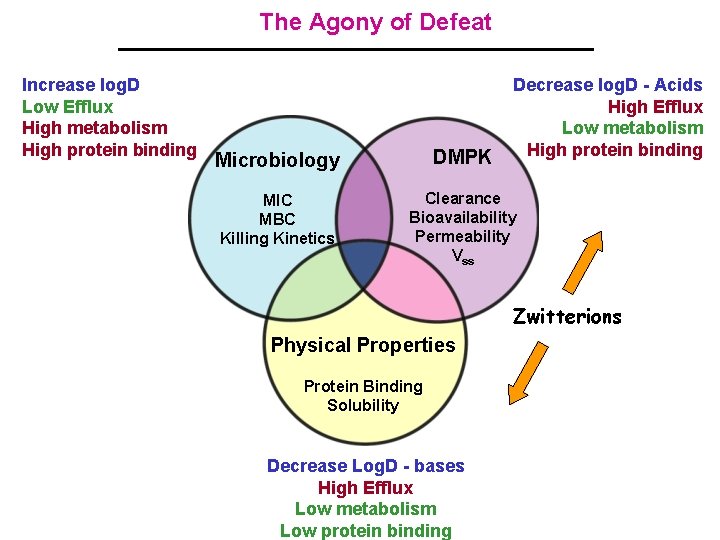
The Agony of Defeat Increase log. D Low Efflux High metabolism High protein binding Decrease log. D - Acids High Efflux Low metabolism High protein binding Microbiology DMPK MIC MBC Killing Kinetics Clearance Bioavailability Permeability Vss Zwitterions Physical Properties Protein Binding Solubility Decrease Log. D - bases High Efflux Low metabolism Low protein binding

Phases of Target-Based Approaches: Preclinical Target Identification Hit Identification • Lead Optimization • Preclinical Hit Identification • Target. Identification Lead Optimisation Preclinical – Biophysical Focus oncompounds analogs of central scaffold(s) Several Facile synthetic (combichem, MPS) and strategies biochemical characterization of targets – –Genomics-based selection in-place Activity in animal disease-state model Documentation for filing Biochemical mode of. FDA inhibition understood Development of primary HTS assay and secondary –––Validation of essentiality in relevant organisms for evaluation of hits ––assays in vivo DMPK studies for human dosing estimation Toxicological studies to support human Whole-cell activity – Cloning and expression of target proteinsdosing – Kinetic mechanism studies enzyme targets in vitro toxicological studiesfor – Confirmed target-mediated mode of action in cells – Production of target proteins – and process chem-informatic analysis Scale up synthesis; chemistry – HTS Early. Screening drug metabolism/pharmacokinetics (DMPK) studies – Limited SAR generation

Thoughts • Mur. I Specific: – Essentiality & target conservation may be insufficient to gauge potential – Niche opportunities may be more tractable than broad spectrum • General: – Understand the target: • Mechanistic studies can clarify appropriate strategies for Hit ID • Evaluate the physiological context of in vitro data • Structural studies are integral – HTS can provide novelty – with luck and persistence – Don’t be satisfied with your best lead series – keep looking!

More reading

Acknowledgments • AZ Boston • AZ Mölndal Richard Alm Barbara Arsenault April Blodgett Ken Coleman Boudewijn de. Jonge Joe Eyermann Ning Gao Madhu Gowravaram Lena Grosser Pamela Hill Janette Jones Thomas Keating Amy Kutschke Jim Loch Larry Mac. Pherson Cynthia Mascolo Marshall Morningstar Brian Noonan Olga Rivin Maria Uria-Nickelsen Jonny Yang Mark Zambrowski Beth Andrews Greg Basarab Gloria Breault Janelle Comita Gejing Deng Tatyana Friedman Bolin Geng Oluyinka Green Laurel Hajec Sussie Hopkins Camil Joubran Gunther Kern Stephania Livchak Kathleen Mc. Cormack John Manchester Scott Mills Trevor Newton Linda Otterson Mike Rooney Jim Whiteaker Wei Yang Marie Andersen Tomas Lundqvist Rutger Folmer Yafeng Xue Nan Albertson Bo Xu Mark Divers Christer Cederberg John Primeau Trevor Trust Mark Wuonola Paul Manning Gautam Sanyal Peter Webborn

Supporting Slides

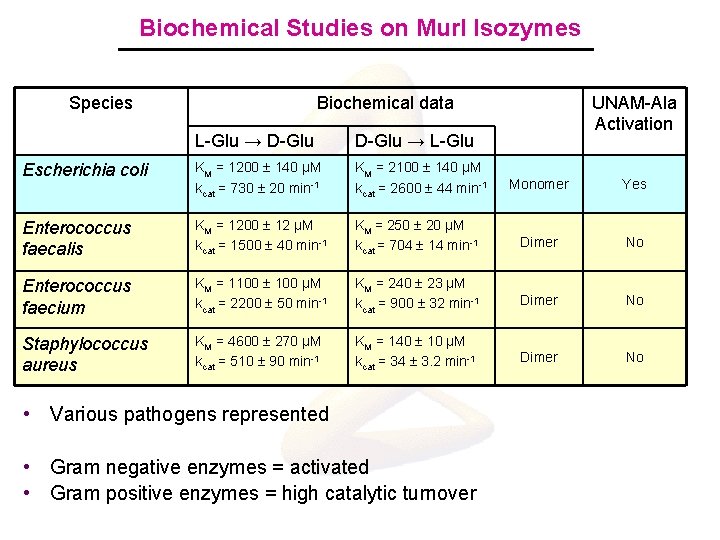
Biochemical Studies on Mur. I Isozymes Species Biochemical data L-Glu → D-Glu → L-Glu Escherichia coli KM = 1200 140 μM kcat = 730 20 min-1 KM = 2100 140 μM kcat = 2600 44 min-1 Enterococcus faecalis KM = 1200 12 μM kcat = 1500 40 min-1 Enterococcus faecium Staphylococcus aureus UNAM-Ala Activation Monomer Yes KM = 250 20 μM kcat = 704 14 min-1 Dimer No KM = 1100 μM kcat = 2200 50 min-1 KM = 240 23 μM kcat = 900 32 min-1 Dimer No KM = 4600 270 μM kcat = 510 90 min-1 KM = 140 10 μM kcat = 34 3. 2 min-1 Dimer No • Various pathogens represented • Gram negative enzymes = activated • Gram positive enzymes = high catalytic turnover

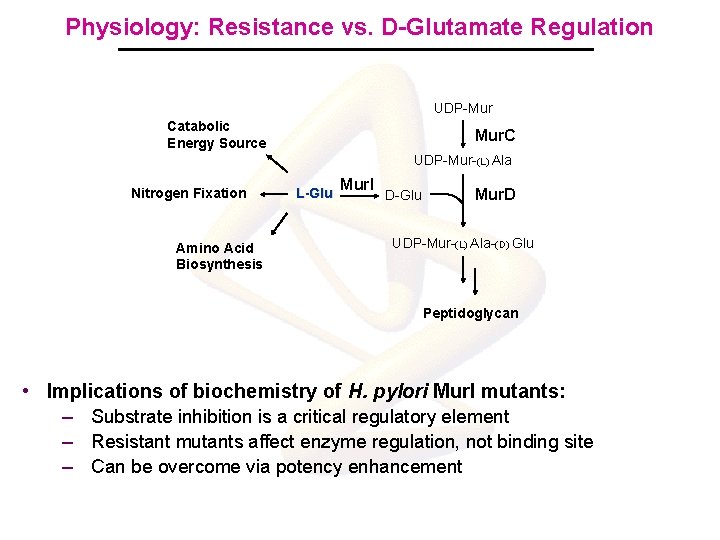
Physiology: Resistance vs. D-Glutamate Regulation UDP-Mur Catabolic Energy Source Mur. C UDP-Mur-(L) Ala Nitrogen Fixation Amino Acid Biosynthesis L-Glu Mur. I D-Glu Mur. D UDP-Mur-(L) Ala-(D) Glu Peptidoglycan • Implications of biochemistry of H. pylori Mur. I mutants: – Substrate inhibition is a critical regulatory element – Resistant mutants affect enzyme regulation, not binding site – Can be overcome via potency enhancement

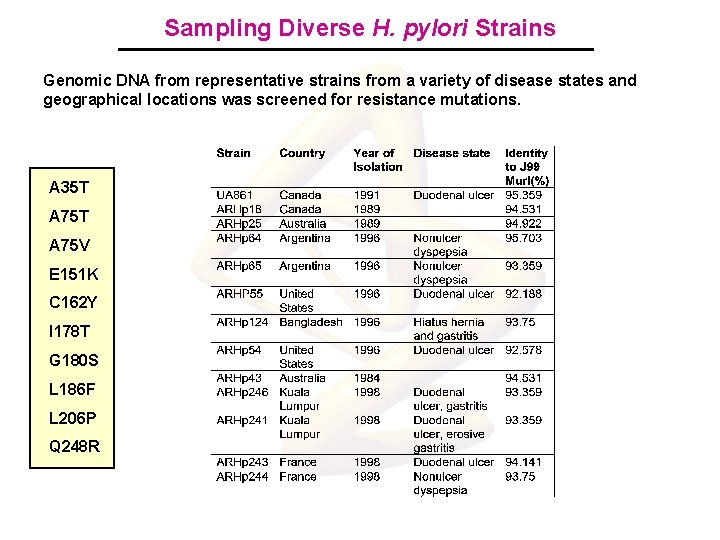
Sampling Diverse H. pylori Strains Genomic DNA from representative strains from a variety of disease states and geographical locations was screened for resistance mutations. A 35 T A 75 V E 151 K C 162 Y I 178 T G 180 S L 186 F L 206 P Q 248 R

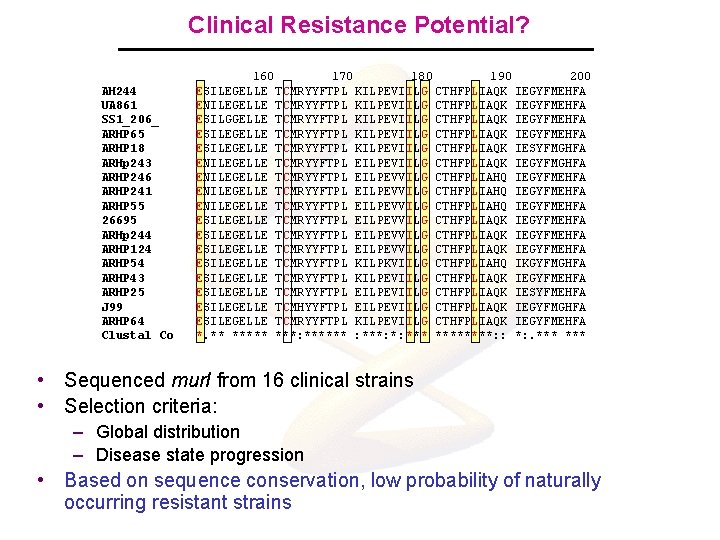
Clinical Resistance Potential? AH 244 UA 861 SS 1_206_ ARHP 65 ARHP 18 ARHp 243 ARHP 246 ARHP 241 ARHP 55 26695 ARHp 244 ARHP 124 ARHP 54 ARHP 43 ARHP 25 J 99 ARHP 64 Clustal Co 160 170 180 190 200 ESILEGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IEGYFMEHFA ENILEGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IEGYFMEHFA ESILGGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IEGYFMEHFA ESILEGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IESYFMGHFA ENILEGELLE TCMRYYFTPL EILPEVIILG CTHFPLIAQK IEGYFMGHFA ENILEGELLE TCMRYYFTPL EILPEVVILG CTHFPLIAHQ IEGYFMEHFA ESILEGELLE TCMRYYFTPL EILPEVVILG CTHFPLIAQK IEGYFMEHFA ESILEGELLE TCMRYYFTPL KILPKVIILG CTHFPLIAHQ IKGYFMGHFA ESILEGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IEGYFMEHFA ESILEGELLE TCMRYYFTPL EILPEVIILG CTHFPLIAQK IESYFMEHFA ESILEGELLE TCMHYYFTPL EILPEVIILG CTHFPLIAQK IEGYFMGHFA ESILEGELLE TCMRYYFTPL KILPEVIILG CTHFPLIAQK IEGYFMEHFA *. ** ***: ****** : ***: *: ********: : *: . *** • Sequenced mur. I from 16 clinical strains • Selection criteria: – Global distribution – Disease state progression • Based on sequence conservation, low probability of naturally occurring resistant strains
 Renjas
Renjas Astra schults
Astra schults Certain infectious and parasitic diseases
Certain infectious and parasitic diseases Emerging infectious diseases
Emerging infectious diseases Rastra zeneca
Rastra zeneca Drug discovery process
Drug discovery process Nature reviews drug discovery
Nature reviews drug discovery Exhausted drug
Exhausted drug Astra aukšmuksta
Astra aukšmuksta Astra
Astra Contoh kasus manajemen risiko bank
Contoh kasus manajemen risiko bank Astra aukšmuksta
Astra aukšmuksta Astra aukšmuksta
Astra aukšmuksta Honda sales operation
Honda sales operation Liceul astra pitesti
Liceul astra pitesti Astra zenec
Astra zenec Per aspera ad astra interpretacja
Per aspera ad astra interpretacja Apa yang dimaksud dengan pancagatra
Apa yang dimaksud dengan pancagatra Sammu pikkus
Sammu pikkus Ad astra dalba
Ad astra dalba Ethical hacking definition
Ethical hacking definition One point perspective staircase
One point perspective staircase Silo perspective vs business process perspective
Silo perspective vs business process perspective Infectious mononucleosis
Infectious mononucleosis Stages of infection
Stages of infection Infectious stunting syndrome
Infectious stunting syndrome Stridor
Stridor Infectious nucleic acid
Infectious nucleic acid Hennepin county infectious disease manual
Hennepin county infectious disease manual Chapter 26 infectious disease prevention and control
Chapter 26 infectious disease prevention and control Blood smear
Blood smear Infectious waste definition
Infectious waste definition Skeleton hand whmis
Skeleton hand whmis Viruses are the smallest infectious agents
Viruses are the smallest infectious agents Stages of infectious disease
Stages of infectious disease Infectious disease
Infectious disease Infectious disease quality controls
Infectious disease quality controls Noncellular infectious protein particles are called
Noncellular infectious protein particles are called Infectious canine hepatitis in dogs
Infectious canine hepatitis in dogs Infectious canine hepatitis in dogs
Infectious canine hepatitis in dogs Chapter 21 mental health diseases and disorders
Chapter 21 mental health diseases and disorders Milady nail diseases and disorders
Milady nail diseases and disorders Albugo eye
Albugo eye Section 19-3 diseases caused by bacteria and viruses
Section 19-3 diseases caused by bacteria and viruses Non communicable diseases infographic
Non communicable diseases infographic Major nutritional deficiency diseases in emergencies
Major nutritional deficiency diseases in emergencies Sphyx medical term
Sphyx medical term Protein deficiency diseases
Protein deficiency diseases Columbian exchange restaurant names
Columbian exchange restaurant names Diseases spread by columbian exchange
Diseases spread by columbian exchange Global alliance against chronic respiratory diseases
Global alliance against chronic respiratory diseases Helminthic diseases
Helminthic diseases Pain transmission
Pain transmission Myth and fallacies about non-communicable diseases
Myth and fallacies about non-communicable diseases Public health entomology certificate
Public health entomology certificate Chapter 24 sexually transmitted diseases and hiv/aids
Chapter 24 sexually transmitted diseases and hiv/aids Non common communicable diseases
Non common communicable diseases Vagninitis
Vagninitis Venn diagram of communicable and non-communicable diseases
Venn diagram of communicable and non-communicable diseases Examples of communicable diseases
Examples of communicable diseases Gingival diseases
Gingival diseases