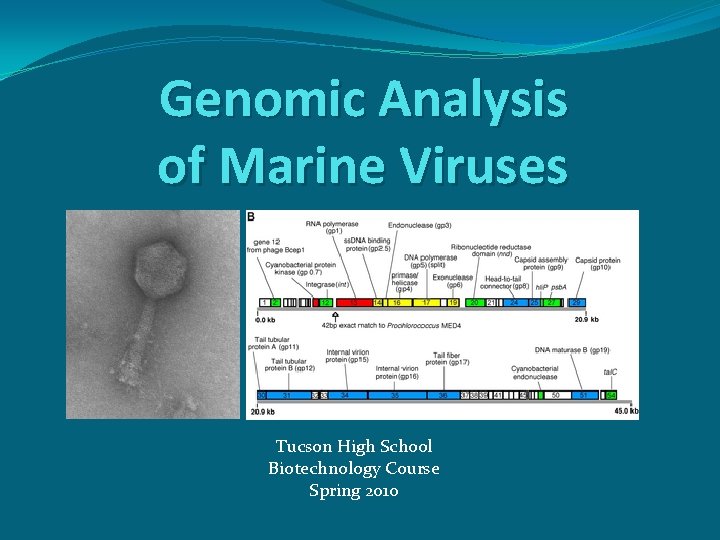
Genomic Analysis of Marine Viruses Tucson High School











- Slides: 33

Genomic Analysis of Marine Viruses Tucson High School Biotechnology Course Spring 2010

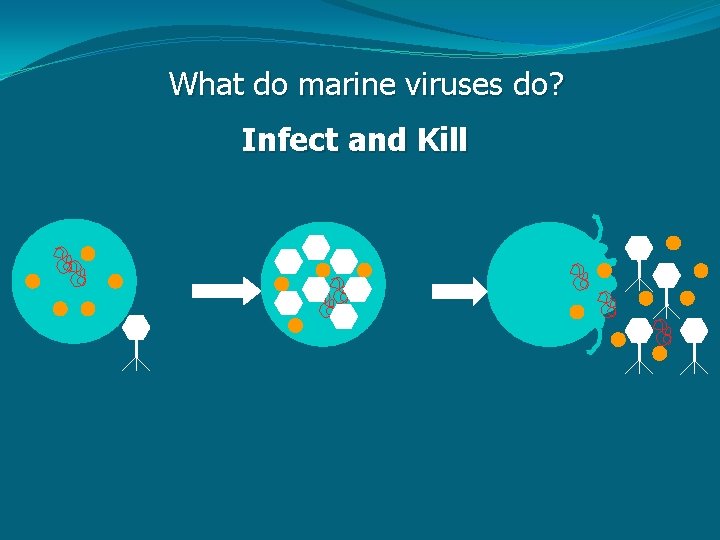
What do marine viruses do? Infect and Kill

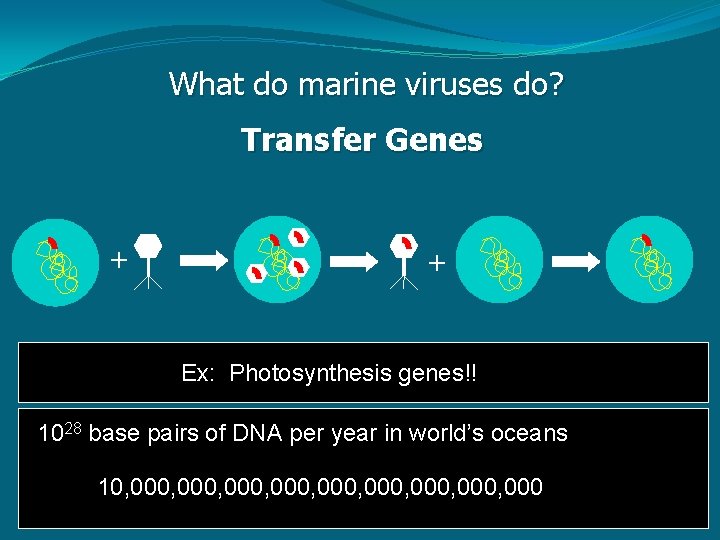
What do marine viruses do? Transfer Genes + + Ex: Photosynthesis genes!! 1028 base pairs of DNA per year in world’s oceans 10, 000, 000, 000

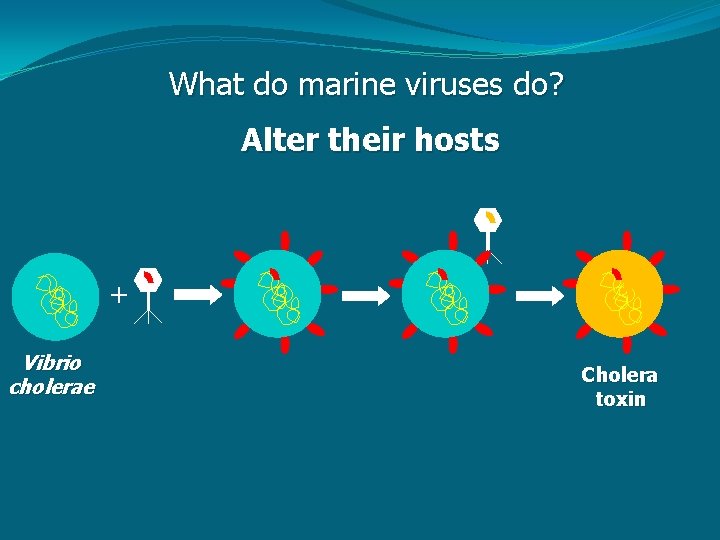
What do marine viruses do? Alter their hosts + Vibrio cholerae Cholera toxin

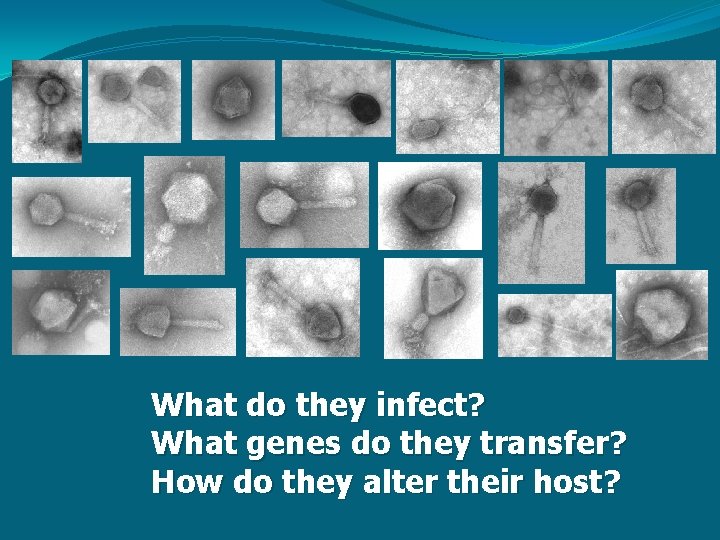
What do they infect? What genes do they transfer? How do they alter their host?

We need to use genetics… UNIVERSAL genes Bacteria have 16 S gene Eukaryotes have 18 S gene NO universal gene for viruses!!

So we use CONCERVED genes

How will WE use genetics? … to find out what type of virus we have. psb. A Myovirus Podovirus X X X DNA pol g 23 X

PCR Forward primer DNA pol Reverse primer standard psb. A DNA g 23 pol

How will WE use genetics? … to find out what type of virus we have. psb. A Myovirus Podovirus X X X DNA pol g 23 X

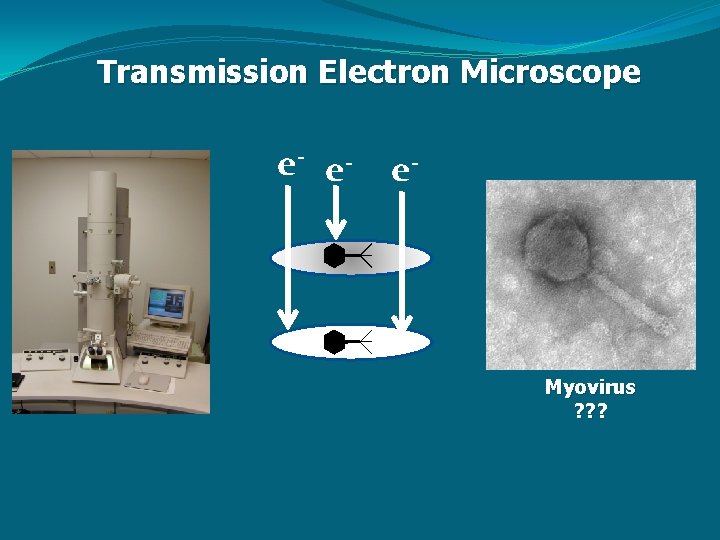
Transmission Electron Microscope e- e- e- Myovirus ? ? ?

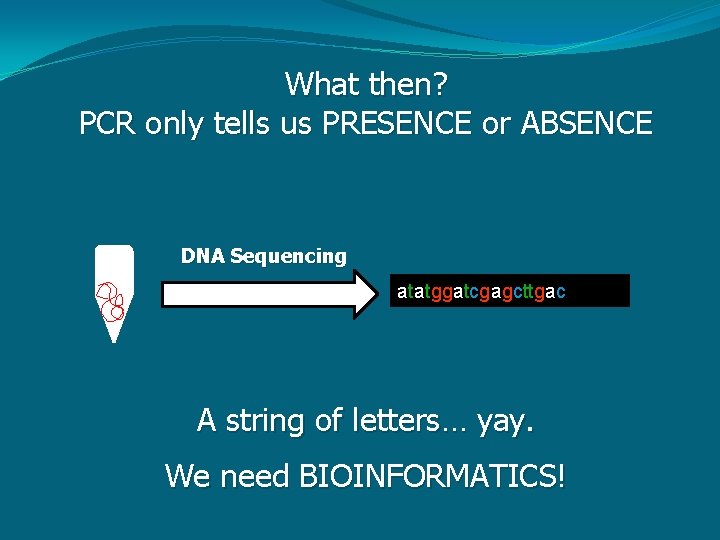
What then? PCR only tells us PRESENCE or ABSENCE DNA Sequencing atatggatcgagcttgac A string of letters… yay. We need BIOINFORMATICS!

Bioinformatics and Genomics Bonnie Hurwitz Graduate student TMPL

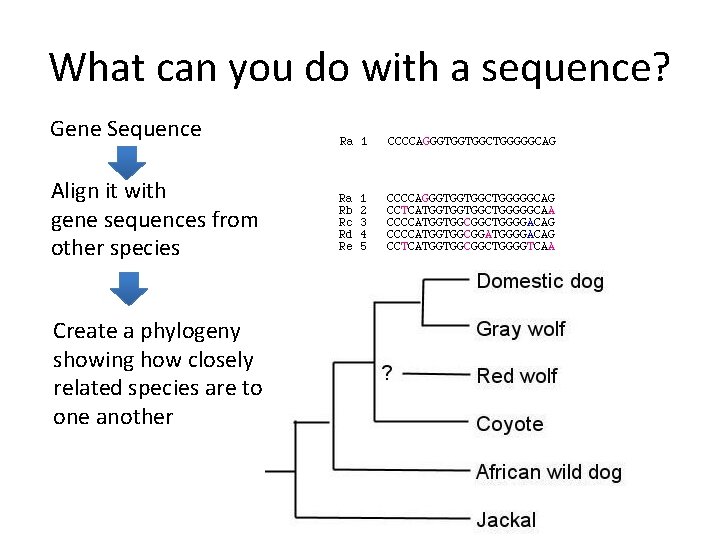
What can you do with a sequence? Gene Sequence Align it with gene sequences from other species Create a phylogeny showing how closely related species are to one another

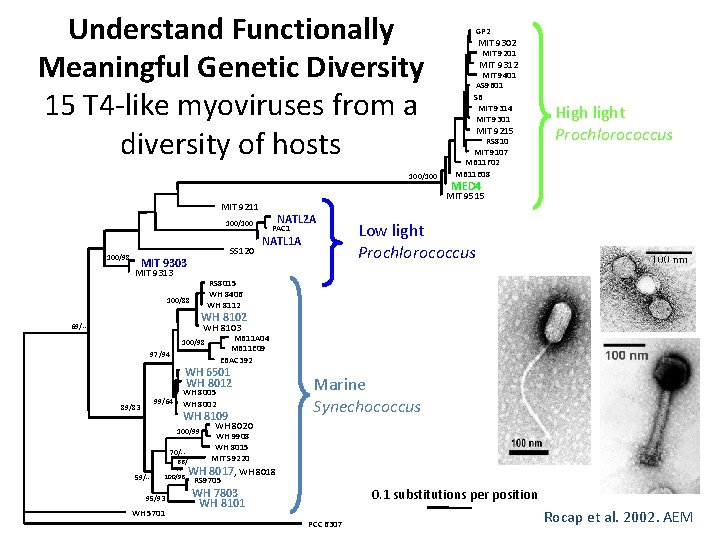
Understand Functionally Meaningful Genetic Diversity 15 T 4 -like myoviruses from a diversity of hosts 100/100 NATL 2 A 100/100 SS 120 MIT 9303 MIT 9313 MIT 9302 MIT 9201 MIT 9312 MIT 9401 AS 9601 SB MIT 9314 MIT 9301 MIT 9215 RS 810 MIT 9107 MB 11 F 02 MB 11 E 08 High light Prochlorococcus MED 4 MIT 9515 MIT 9211 100/98 GP 2 PAC 1 NATL 1 A Low light Prochlorococcus RS 8015 WH 8406 WH 8112 100/88 WH 8102 69/-- WH 8103 MB 11 A 04 MB 11 E 09 EBAC 392 100/98 97/94 WH 6501 WH 8012 99/64 89/83 WH 8005 WH 8002 WH 8109 100/99 70/-- 59/-- 66/ -100/98 95/93 WH 5701 Marine Synechococcus WH 8020 WH 9908 WH 8015 MIT S 9220 WH 8017, WH 8018 RS 9705 WH 7803 WH 8101 0. 1 substitutions per position PCC 6307 Rocap et al. 2002. AEM

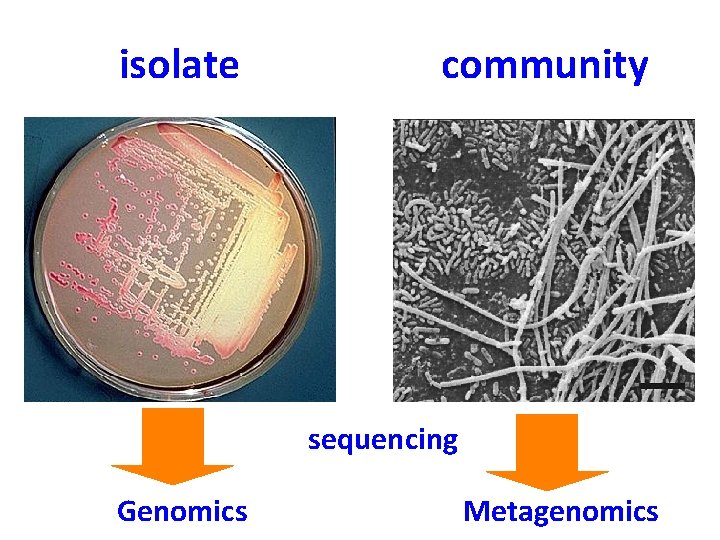
What can you do with a lot of sequences? What is a (meta)genome?

isolate community sequencing Genomics Metagenomics

Genome assembly

Genome assembly

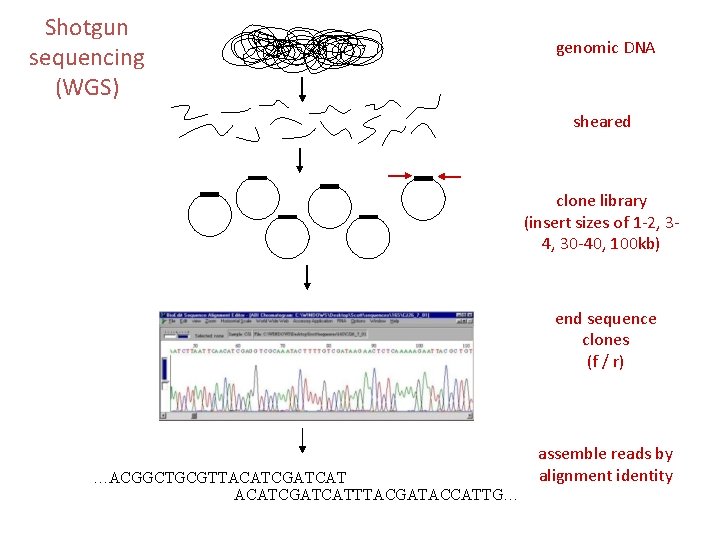
Shotgun sequencing (WGS) genomic DNA sheared clone library (insert sizes of 1 -2, 34, 30 -40, 100 kb) end sequence clones (f / r) …ACGGCTGCGTTACATCGATCATTTACGATACCATTG… assemble reads by alignment identity

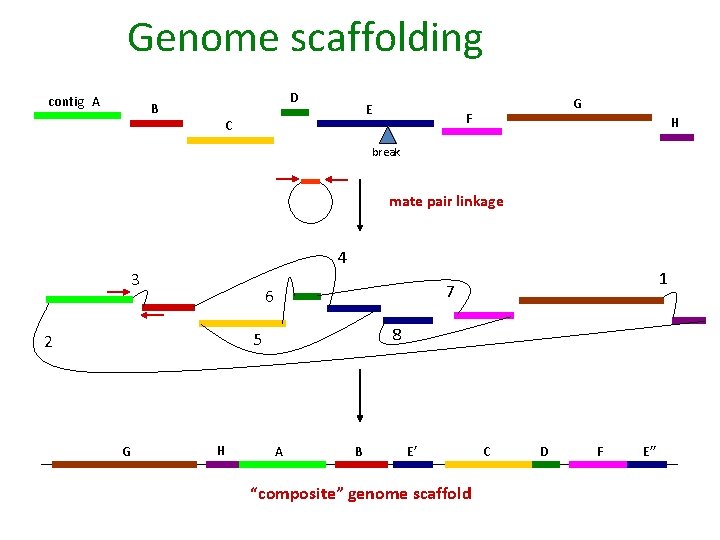
Genome scaffolding contig A B D E C G F H break mate pair linkage 4 3 7 6 8 5 2 G H 1 A B E’ “composite” genome scaffold C D F E’’

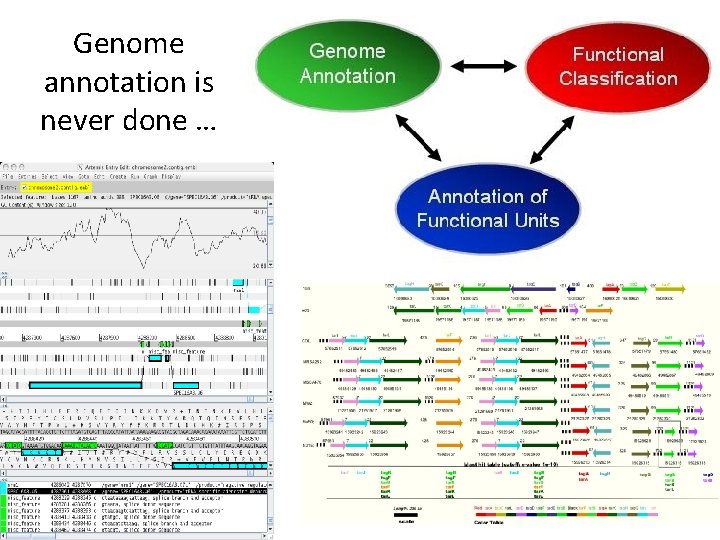
Genome annotation is never done …

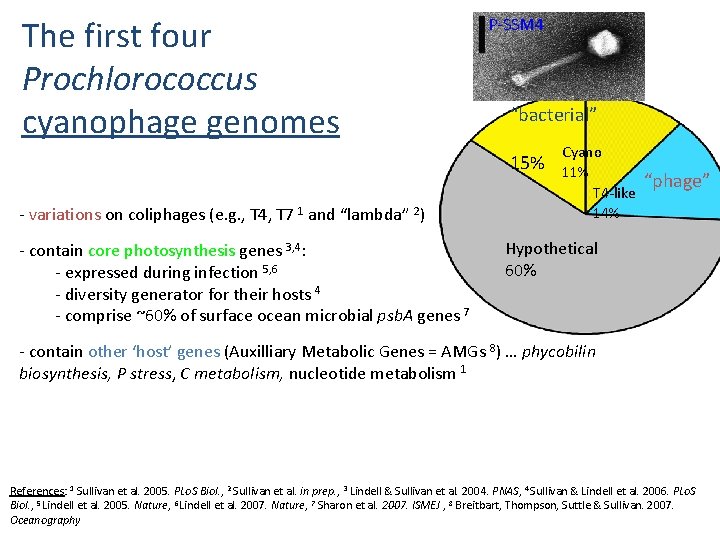
The first four Prochlorococcus cyanophage genomes P-SSM 4 “bacterial” 15% - variations on coliphages (e. g. , T 4, T 7 1 and “lambda” 2) - contain core photosynthesis genes 3, 4: - expressed during infection 5, 6 - diversity generator for their hosts 4 - comprise ~60% of surface ocean microbial psb. A genes 7 Cyano 11% T 4 -like 14% “phage” Hypothetical 60% - contain other ‘host’ genes (Auxilliary Metabolic Genes = AMGs 8) … phycobilin biosynthesis, P stress, C metabolism, nucleotide metabolism 1 References: 1 Sullivan et al. 2005. PLo. S Biol. , 2 Sullivan et al. in prep. , 3 Lindell & Sullivan et al. 2004. PNAS, 4 Sullivan & Lindell et al. 2006. PLo. S Biol. , 5 Lindell et al. 2005. Nature, 6 Lindell et al. 2007. Nature, 7 Sharon et al. 2007. ISMEJ , 8 Breitbart, Thompson, Suttle & Sullivan. 2007. Oceanography

Metagenome assembly

Metagenome assembly

Metagenome assembly

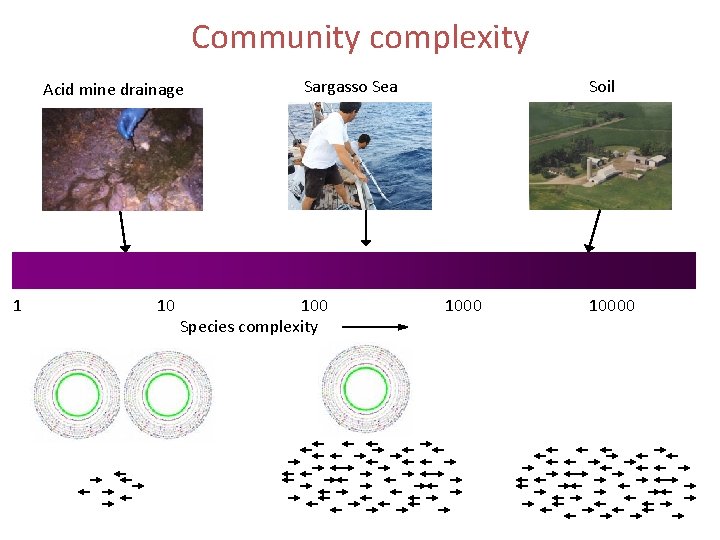
Community complexity Acid mine drainage 1 10 Sargasso Sea 100 Species complexity Soil 10000

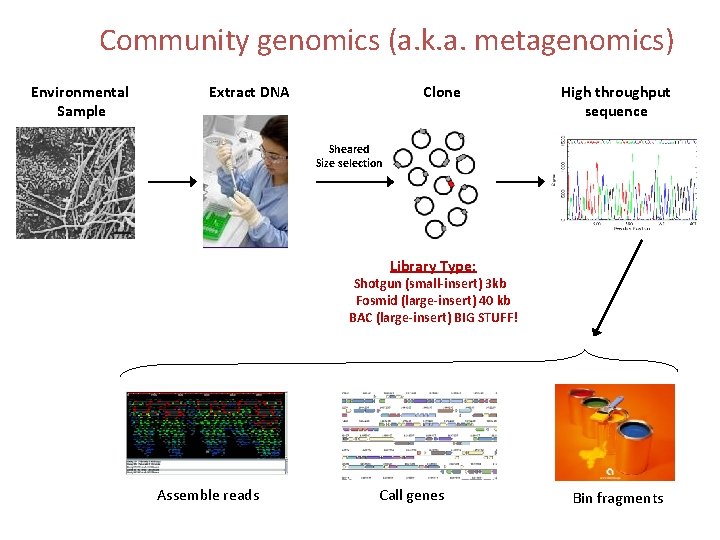
Community genomics (a. k. a. metagenomics) Environmental Sample Extract DNA Clone High throughput sequence Sheared Size selection Library Type: Shotgun (small-insert) 3 kb Fosmid (large-insert) 40 kb BAC (large-insert) BIG STUFF! Assemble reads Call genes Bin fragments

What to do with the data? EGTs = Environmental Gene Tags Predict ORFs (genes) in sequence data Assign a function to ORFs Compare relative abundance across habitats

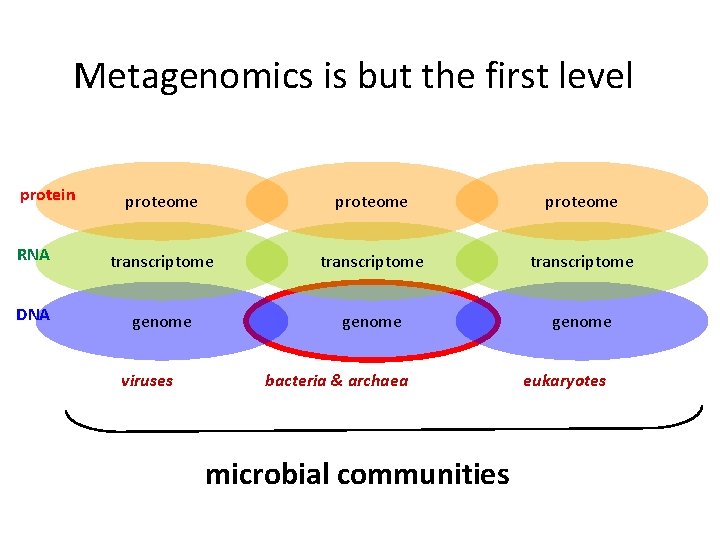
Metagenomics is but the first level protein proteome RNA transcriptome DNA genome viruses bacteria & archaea microbial communities eukaryotes

Summary • The smallest but arguably most important ocean inhabitants are microbes and phages • Using metagenomics to sequence previously undetectable microbes and phages has expanded our knowledge of the oceans’ ecosystems • Looking a genes in genomes can give us an idea of the potential function and role these organisms play in ocean ecology • Looking at gene expression can tell us which genes are playing an active role in the ecosystem and who the major players are

Our goals • Assemble and annotate a phage genome – Next Tuesday and Thursday • Build a gene phylogeny and determine what phage you have based on it’s relationship to other phages – April 6 th

higher trophic levels grazers phytoplankton Dissolved bacteria viral lysis
 Unlike lytic viruses lysogenic viruses do not
Unlike lytic viruses lysogenic viruses do not Watch?v=dhlvcdeg0yg
Watch?v=dhlvcdeg0yg Tucson high magnet school
Tucson high magnet school Principle of genomic equivalence
Principle of genomic equivalence Genomic england
Genomic england Genomic england
Genomic england Genomic england
Genomic england Genomic instability
Genomic instability Genomic
Genomic Genomic imprinting definition
Genomic imprinting definition Genomic signal processing
Genomic signal processing Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic equivalence definition
Genomic equivalence definition Kino r
Kino r Tucson water quality
Tucson water quality Tucson modern streetcar
Tucson modern streetcar Tucson flood 1983
Tucson flood 1983 Endotipos
Endotipos Dysgraphia testing tucson
Dysgraphia testing tucson Tucson tce contamination map
Tucson tce contamination map Weptac tucson 2019
Weptac tucson 2019 Dr warneke tucson az
Dr warneke tucson az Tucson audubon mason center
Tucson audubon mason center Thqta
Thqta Why are viruses considered nonliving?
Why are viruses considered nonliving? Bacteriophage characteristics
Bacteriophage characteristics General characteristics of viruses
General characteristics of viruses Viruses
Viruses Lysogenic viruses do not
Lysogenic viruses do not Section 19-3 diseases caused by bacteria and viruses
Section 19-3 diseases caused by bacteria and viruses Cultivation of viruses
Cultivation of viruses Viral inoculation in embryonated egg
Viral inoculation in embryonated egg Egrette chapter 21
Egrette chapter 21 Nonliving particle that replicates inside a living cell
Nonliving particle that replicates inside a living cell