Genetics of Axis Specification in Drosophila Anterior Posterior














- Slides: 35

Genetics of Axis Specification in Drosophila: Anterior. Posterior Axis Determination Gilbert - Chapter 9

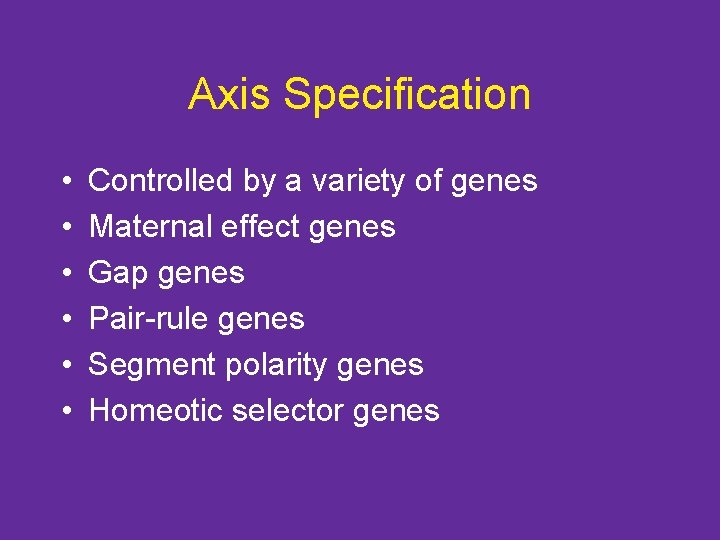
Axis Specification • • • Controlled by a variety of genes Maternal effect genes Gap genes Pair-rule genes Segment polarity genes Homeotic selector genes


Genetic Screen for Genes involved in Drosophila Development • Nusslien-Volhard, Wieschaus • Fed mutagens to Drosophila • Then breed until mutation is homozygous recessive • Examined embryos for patterning defects • Used embryonic cuticles to do screens – Looked at pattern of denticles, shapes of segments

Goals of Genetic Screen • Create small mutations in fruit fly genome – Enough to mutate EVERY gene in the genome at least once – Identify EVERY mutation in the genome that affects embryonic development in the fruit fly – How many genes are there in fruit flies? – Note - At this time the D. melanogaster genome was not sequenced

How was the genetic screen performed? • Feed adult fruit flies with a mutagen – EMS - causes high mutation rate in offspring – Point mutations, short deletions, rearrangements – Can cause defective proteins, absence of proteins • THEN - breed the flies until the mutation is homozygous, and look for ANY embryo that has abnormalities • TENS OF THOUSANDS OF EMBRYOS ARE EXAMINED!!!

The wild-type body is segmented and each segment has a unique identity and thus produces distinctive structures -The genetic screen looked for changes in this body plan

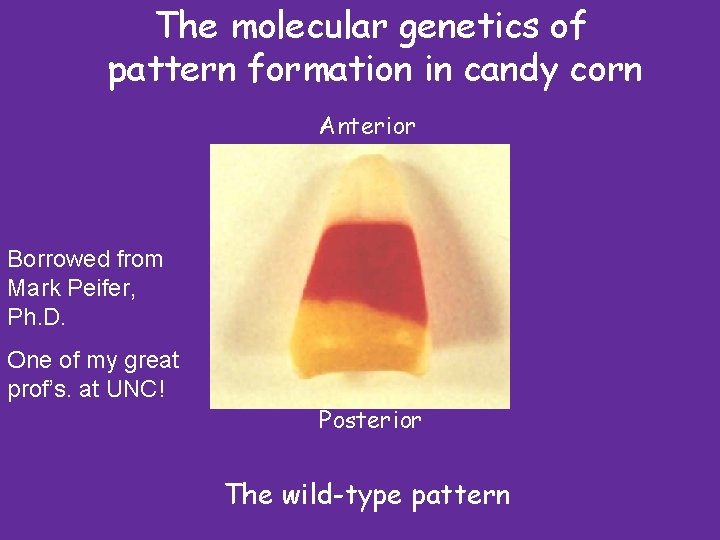
The molecular genetics of pattern formation in candy corn Anterior Borrowed from Mark Peifer, Ph. D. One of my great prof’s. at UNC! Posterior The wild-type pattern

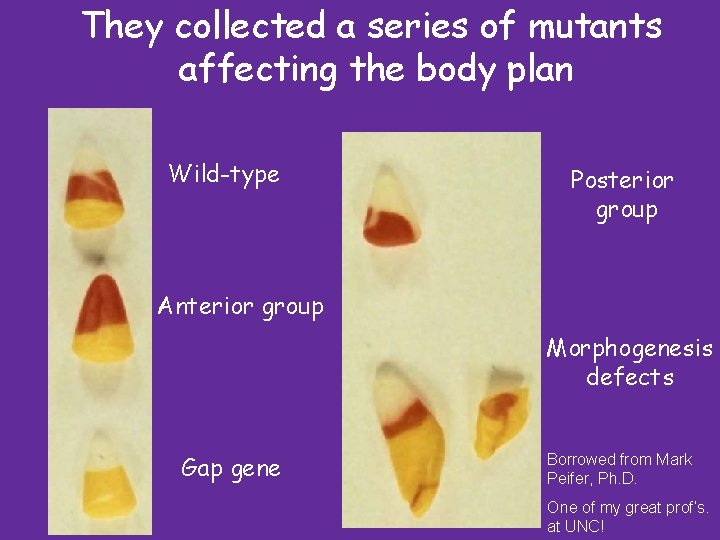
They collected a series of mutants affecting the body plan Wild-type Posterior group Anterior group Morphogenesis defects Gap gene Borrowed from Mark Peifer, Ph. D. One of my great prof’s. at UNC!

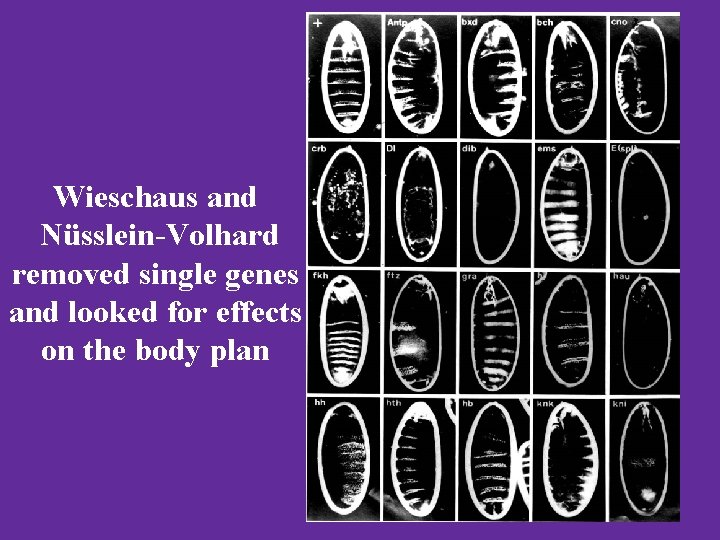
Wieschaus and Nüsslein-Volhard removed single genes and looked for effects on the body plan

Genes involved in embryogenesis • Genes controlling embryonic development are either –maternal-effect genes • m. RNA or protein already deposited in the egg – zygotic genes • Transcribed from nucleus of zygote

Maternal effect genes: Anterior - Posterior Polarity • Placed into developing oocyte by maternal cells – Nurse cells that surround the egg • Several genes were discovered • What phenotype would you be looking for in your embryos? ?

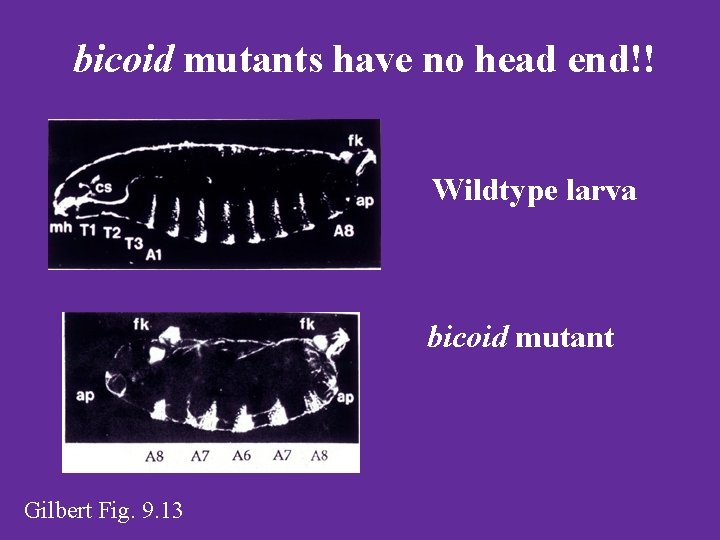
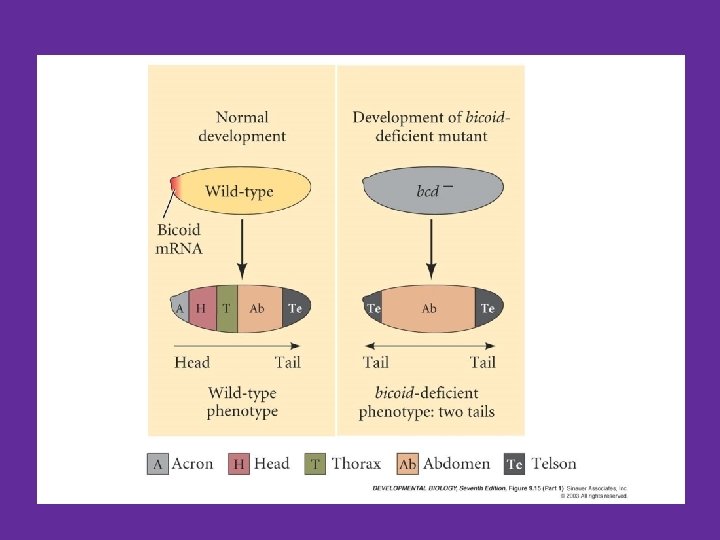
bicoid mutants have no head end!! Wildtype larva bicoid mutant Gilbert Fig. 9. 13

Bicoid Mutant • Lacks anterior structures, posterior structures are duplicated • Leads to lots of Molecular Biology questions! – How can we find this gene and its sequence? – What does the phenotype tell us about the function of bicoid? – What is the bicoid protein like in this mutant?

Bicoid and Nanos • Bicoid m. RNA – Concentrated in the future anterior end of the ovum by the nurse cells – Nurse cells are ovary cells of the mother – The m. RNA is held in place by a network of microtubules • Nanos m. RNA – Tethered to the cytoskeleton at the future posterior end of the egg


Bicoid and Specification of the Anterior pole • Bicoid appears to be essential for the formation of anterior structures • Further evidence – Bicoid m. RNA is localized to the anterior end of the oocyte – As bicoid m. RNA gets translated, a gradient of bicoid is created from A to P • More concentrated at most Anterior end

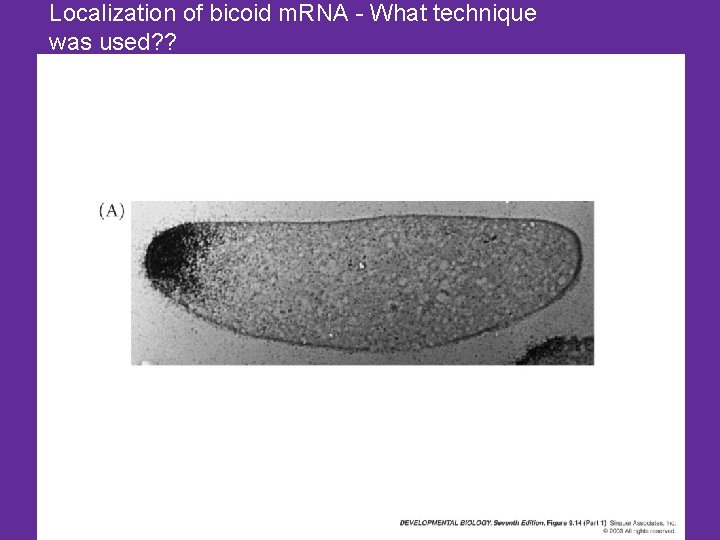
Localization of bicoid m. RNA - What technique was used? ?

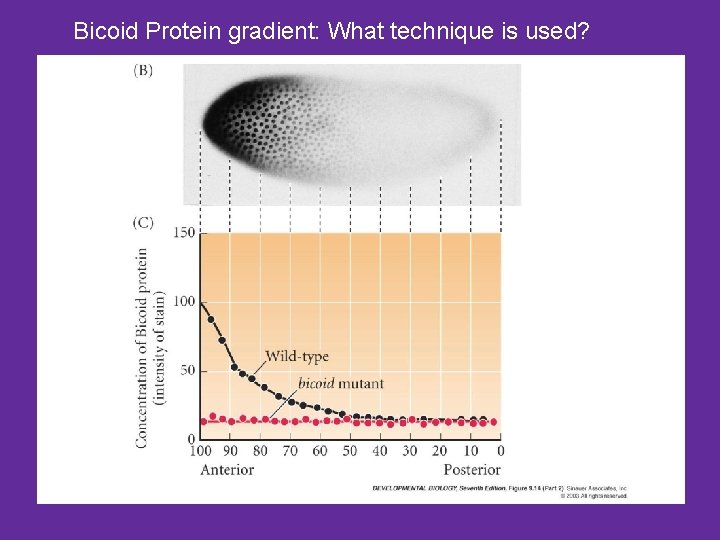
Bicoid Protein gradient: What technique is used?

Bicoid - More evidence • Bicoid m. RNA (wild-type) can rescue the Bicoid mutant phenotype – Inject bicoid m. RNA into anterior end of bicoid mutant embryo • Injection of WT bicoid m. RNA anywhere into the early embryo turns that area into a head end!



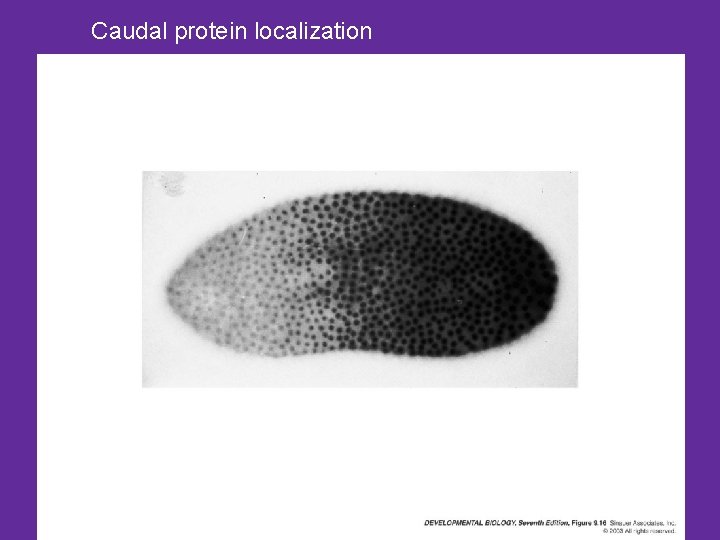
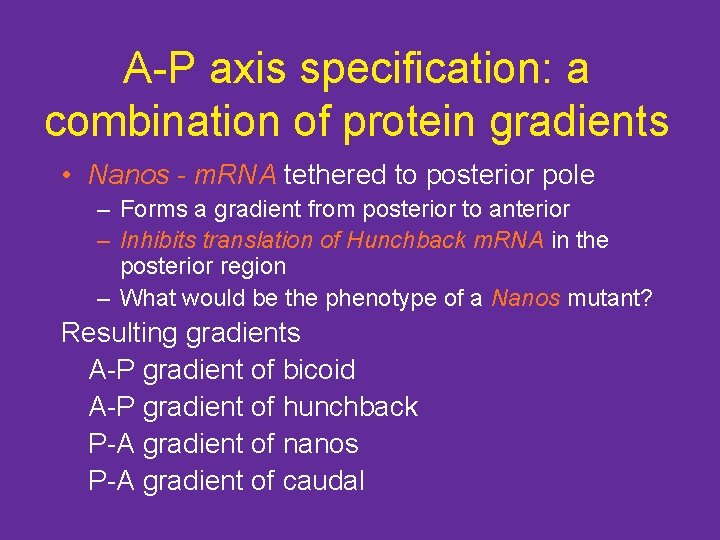
What does bicoid do, and how? • Bicoid protein is arranged in a gradient • It represses the molecules that control posterior identity – Represses translation of a molecule that helps specify the posterior end - Caudal – Caudal m. RNA is found in entire embryo – Caudal protein is found at posterior end

Caudal protein localization

What does bicoid do, and how? • Bicoid protein is a transcription factor • Enters nuclei, activates downstream gene expression – Hunchback - essential for anterior pole formation • Hunchback activates transcription of further head-specific gene products (swallow, exuperantia, buttonhead, orthodenticle)



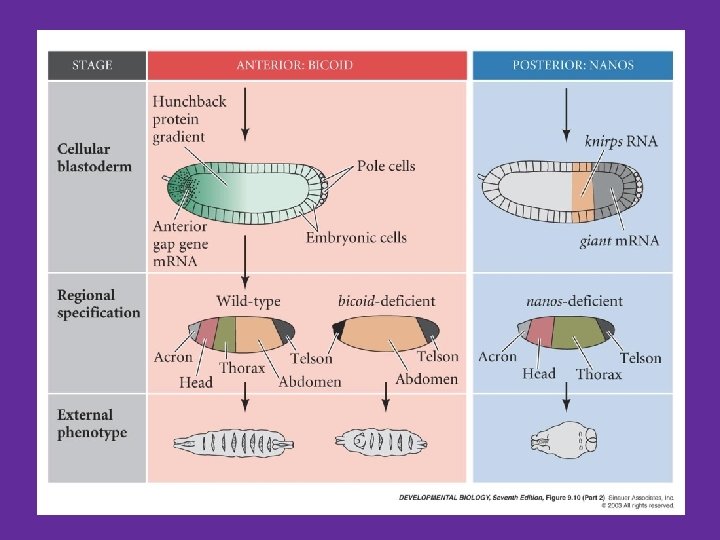
A-P axis specification: a combination of protein gradients • Nanos - m. RNA tethered to posterior pole – Forms a gradient from posterior to anterior – Inhibits translation of Hunchback m. RNA in the posterior region – What would be the phenotype of a Nanos mutant? Resulting gradients A-P gradient of bicoid A-P gradient of hunchback P-A gradient of nanos P-A gradient of caudal





Maternal effect genes and AP polarity - summary • Maternal m. RNA’s are tethered to either A or P ends of oocyte • Proteins are translated to create a gradient • Activate or repress embryonic gene expression (hunchback, caudal) • Result is and anterior region and a posterior region

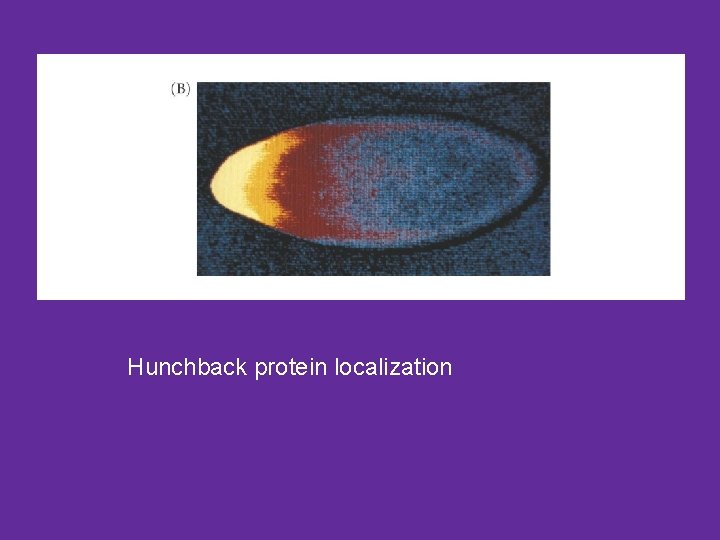
Hunchback protein localization

 Genetics of axis specification in drosophila
Genetics of axis specification in drosophila Anterior posterior axis
Anterior posterior axis Cleavage in bird
Cleavage in bird If lclp is negative number, we set the lclp = 0. why?
If lclp is negative number, we set the lclp = 0. why? Natural variations operations management
Natural variations operations management Four conic sections
Four conic sections Direct axis and quadrature axis
Direct axis and quadrature axis Axis 1 and axis 2 disorders
Axis 1 and axis 2 disorders Axis 1 and axis 2 disorders
Axis 1 and axis 2 disorders Vertical hyperbola equation
Vertical hyperbola equation Dark lamp method
Dark lamp method Anterior posterior pad placement zoll
Anterior posterior pad placement zoll Anterior choroidal artery
Anterior choroidal artery Blood supply abdomen
Blood supply abdomen Difference between anterior and posterior pituitary
Difference between anterior and posterior pituitary Difference between anterior and posterior pituitary
Difference between anterior and posterior pituitary Rutina hombro
Rutina hombro Posterior abdominal wall
Posterior abdominal wall Commissura labiorum
Commissura labiorum Shahab vahdat
Shahab vahdat Cartílago de reichert
Cartílago de reichert Posterior palatal seal area
Posterior palatal seal area Axillary fold
Axillary fold Valvula ileocecal anatomia
Valvula ileocecal anatomia Marine animals without backbone
Marine animals without backbone Fovea palatine
Fovea palatine Cingulum bar rpd
Cingulum bar rpd Dientes posteriores y anteriores
Dientes posteriores y anteriores The anterior and posterior body cavities
The anterior and posterior body cavities Anterior vs posterior abdominal wall
Anterior vs posterior abdominal wall Corpus callosum artery
Corpus callosum artery Anterior posterior palatal bant
Anterior posterior palatal bant Olecranal region
Olecranal region Anterior stroke vs posterior stroke
Anterior stroke vs posterior stroke Anterior vs posterior abdominal wall
Anterior vs posterior abdominal wall Ventral vs dorsal vs anterior and posterior
Ventral vs dorsal vs anterior and posterior