Computational Diagnostics A new research group at the























- Slides: 35

Computational Diagnostics A new research group at the Max Planck Institute for molecular Genetics, Berlin

Will the patient respond to this drug? ?

computational diagnostics A simple solution for simple problems Find all genes that are induced at least x-fold and use them to predict clinical outcomes

computational diagnostics Statistical Modeling Experimental Design, Quality Control, Scaling, Normalization, Dimension Reduction, Predictive Classification, Quantifying the Evidence, Identifying the Evidence

computational diagnostics Computational Infrastructure and more Databases, Automatic Uploading, Standard Analysis Protocols, Analysis Software, Query Language, Understanding the disease, Designing a small Diagnostic Chip

computational diagnostics Clinical Practice Large Patient Databases complemented by expression profiles monitoring the Epidemiology of the disease

Breast Cancer, Expression Profiles and Binary Regression in 7000 Dimensions Rainer Spang, Harry Zuzan, Carrie Blanchette, Erich Huang, Holly Dressman, Jeff Marks, Joe Nevins, Mike West Duke Medical Center & Duke University

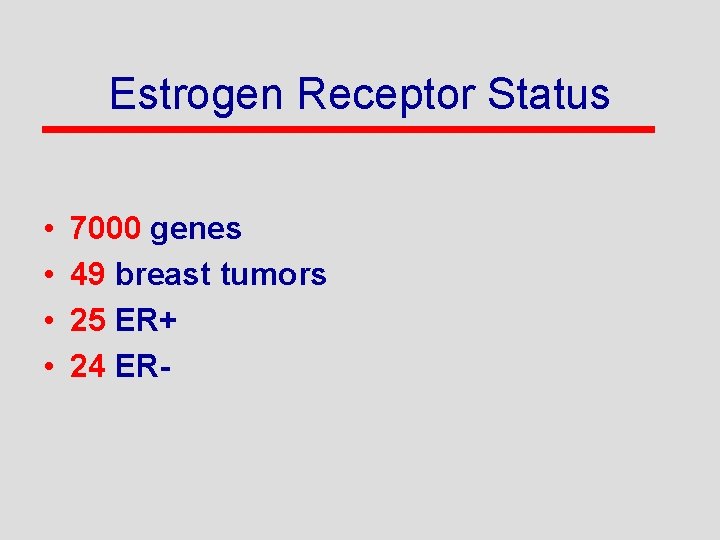
Estrogen Receptor Status • • 7000 genes 49 breast tumors 25 ER+ 24 ER-

Tumor – Chip - 7000 Numbers

We Assume That the Following Steps Are Done: • • • Choosing the patients Doing the surgery Handling the tissues Preparing m. RNA Hybridizing the chips Image analysis Excluding low quality data Normalization Scaling


How Much Evidence Is There? I am 80% sure The probability that I know it the patient has xxx It was a guess given the profile is 0. 8, 1, 0. 5

Given Wanted 89% 7000 Numbers The probability that the tumor is ER+

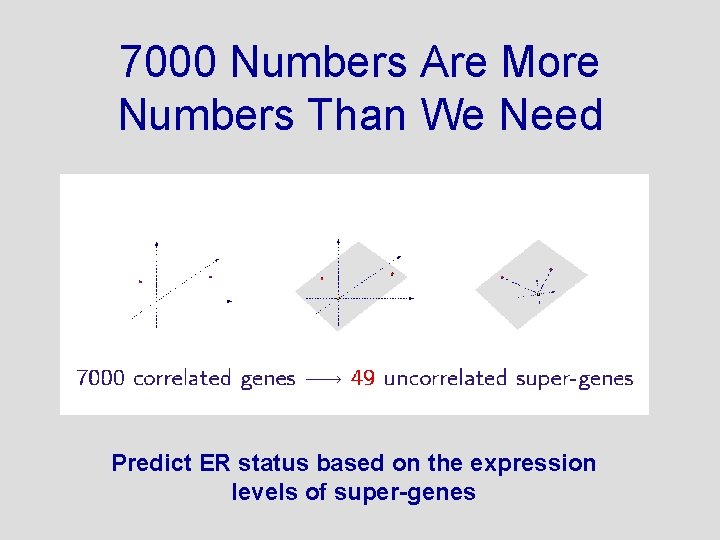
7000 Numbers Are More Numbers Than We Need Predict ER status based on the expression levels of super-genes


Overfitting: We Can Not Identify a Model • There are many different models that assign high probabilities for ER+ tumors and low probabilities for ER- tumors in the training set • For a new patient we find among these models some that support that she is ER+ and others that predict she is ER • ? ? ?

Given the Few Profiles With Known Diagnosis: • The uncertainty on the right model is high • The variance of the model-weights is large • The likelihood landscape is flat • We need additional model assumptions to solve the problem

Informative Priors Likelihood Prior Posterior

If the Prior Is Chosen Badly: • We can not reproduce the diagnosis of the training profiles any more • We still can not identify the model • The diagnosis is driven mostly by the additional assumptions and not by the data

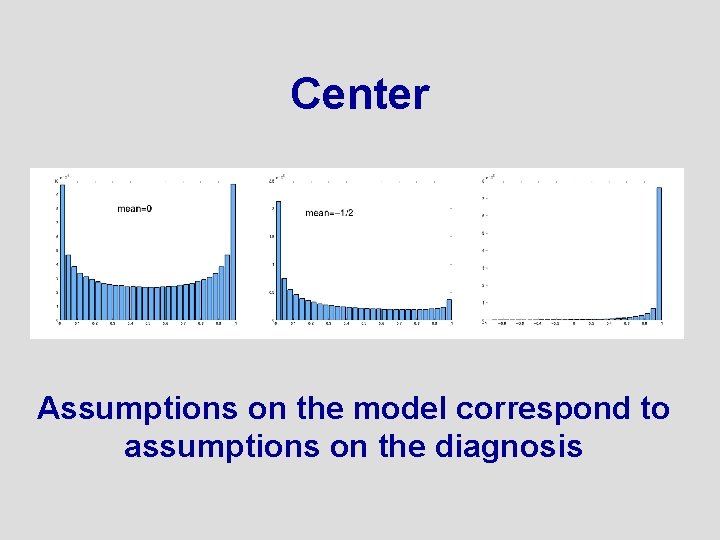
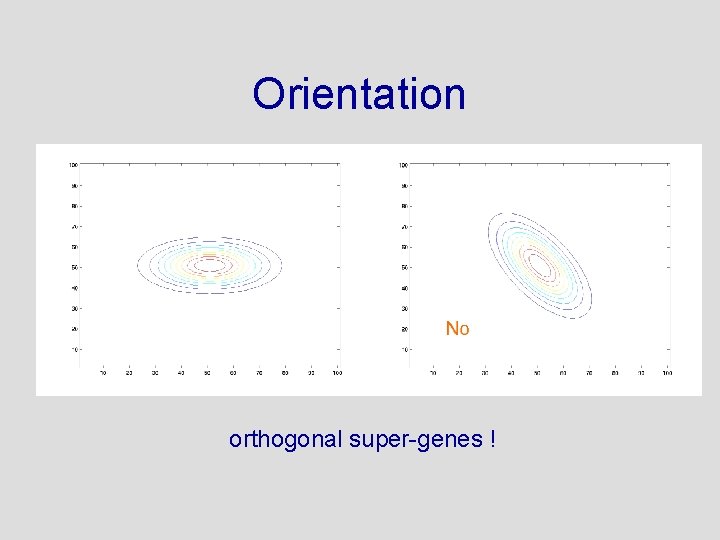
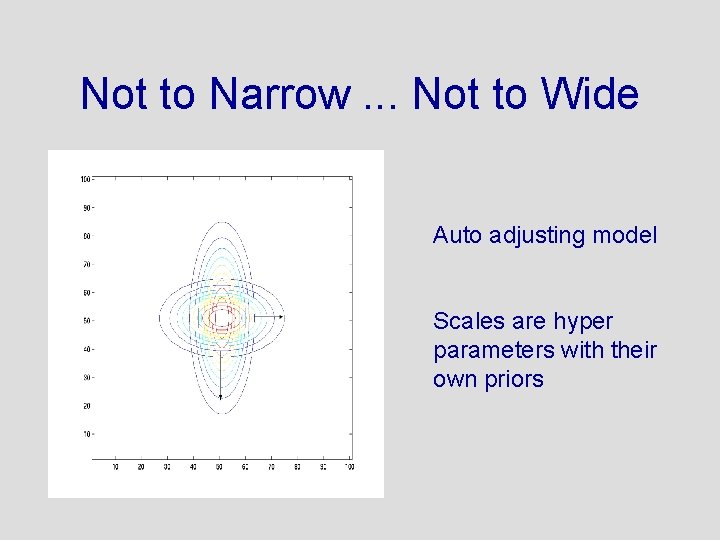
The Prior Needs to Be Designed in 49 Dimensions • • Shape? Center? Orientation? Not to narrow. . . not to wide

Shape multidimensional normal for simplicity

Center Assumptions on the model correspond to assumptions on the diagnosis

Orientation orthogonal super-genes !

Not to Narrow. . . Not to Wide Auto adjusting model Scales are hyper parameters with their own priors

What are the additional assumptions that came in by the prior? • The model can not be dominated by only a few super-genes ( genes! ) • The diagnosis is done based on global changes in the expression profiles influenced by many genes • The assumptions are neutral with respect to the individual diagnosis


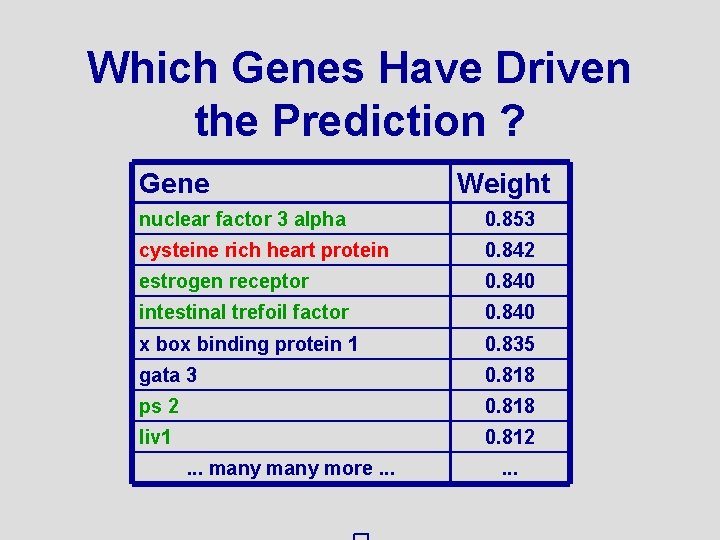
Which Genes Have Driven the Prediction ? Gene Weight nuclear factor 3 alpha 0. 853 cysteine rich heart protein 0. 842 estrogen receptor 0. 840 intestinal trefoil factor 0. 840 x box binding protein 1 0. 835 gata 3 0. 818 ps 2 0. 818 liv 1 0. 812. . . many more. . .

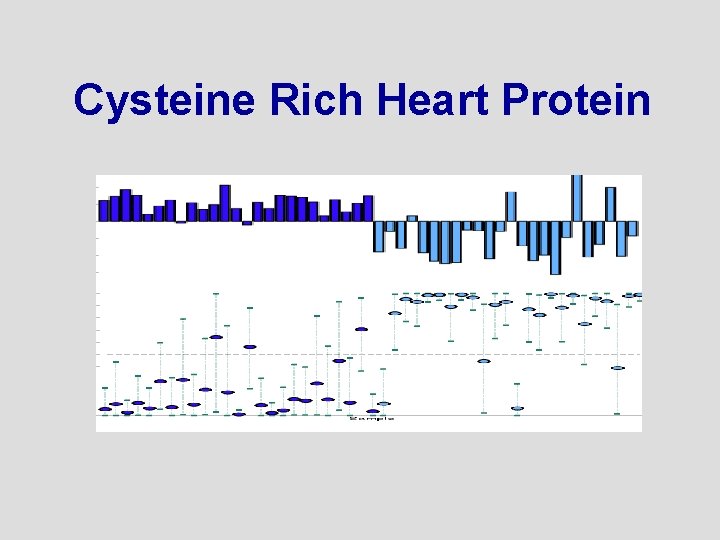
Cysteine Rich Heart Protein

Summary. . . so far • We have solved a relatively simple computational diagnostics problem (ER-status in human breast cancers) • Probit model • Overfitting is a problem • Additional model assumptions do the trick

A Common Problem With Expression Profiles • We do not have enough samples to answer a certain question • A possible strategy: Introduce additional model assumptions

Differential Expression I Setup: Two conditions ( healthy vs sick ), some repetitions, 10 000 genes Which genes are up or down regulated ? The most basic question Good because it is a hypothesis free approach

Differential Expression II 10 000 degrees of freedom A very bad multiple testing problem It is possible in principal, but might require many replications depending on signal to noise ratios SAM: regularized t-statistic + permutation based false positive rates Hard to improve the analysis because it is a hypothesis free approach

Clustering of Genes • Setup: many different conditions - time series - multiple knock-outs • 100% explorative analysis • Essentially it is rearranging the data • Good for finding hypotheses but not for verifying them

Clustering of Profiles (Patients) • Maybe we can find new disease types or refine existing ones • Completely different results when different sets of genes are used • No predictive analysis

Think About Data Analysis Ahead of Time Collect possible questions on the data Which of them are easy ? - Biologists and Bioinformaticians might have a different take on that - Compare: number of samples vs. degrees of freedom It is possible to compensate lack of data with model assumptions: Which assumptions make sense ? More complex question can be the easier ones if they allow for an appropriate model