Chapter 17 Protein From Gene to Protein Synthesis



- Slides: 45

Chapter 17~ Protein From Gene to

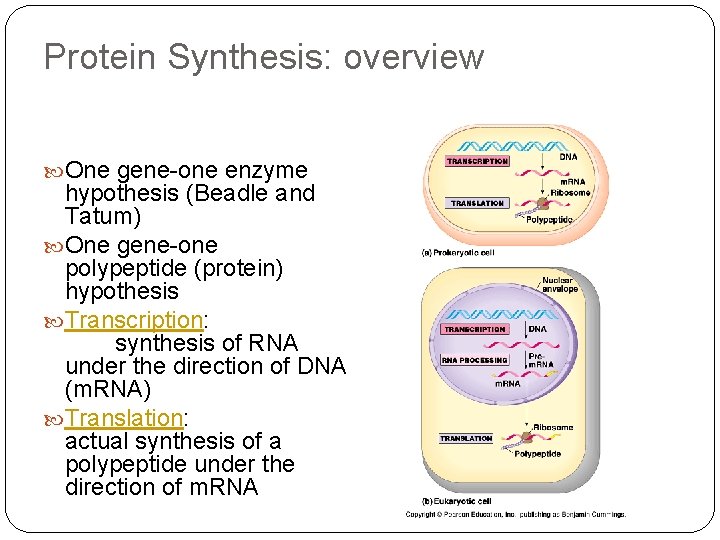
Protein Synthesis: overview One gene-one enzyme hypothesis (Beadle and Tatum) One gene-one polypeptide (protein) hypothesis Transcription: synthesis of RNA under the direction of DNA (m. RNA) Translation: actual synthesis of a polypeptide under the direction of m. RNA

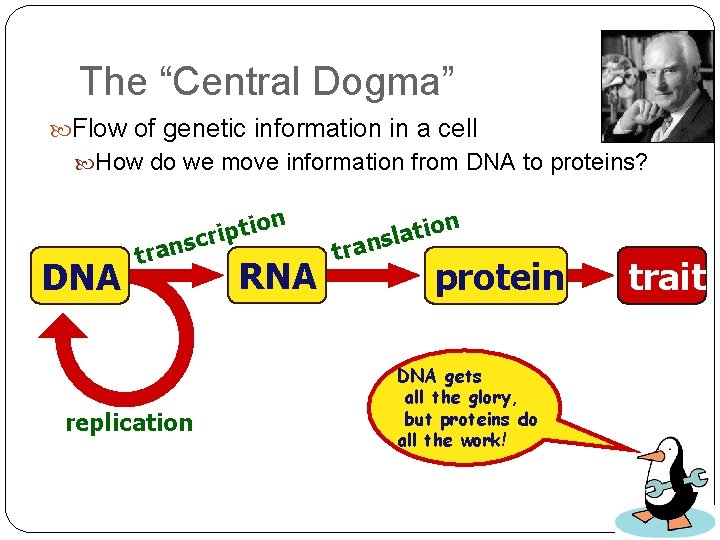
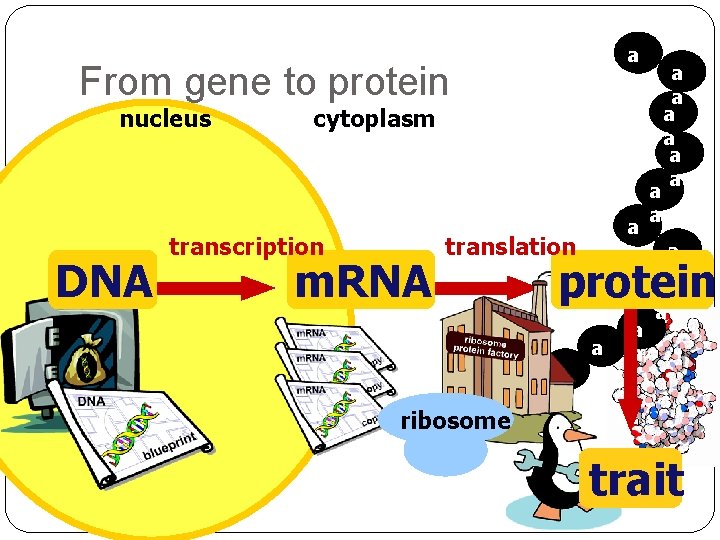
The “Central Dogma” Flow of genetic information in a cell How do we move information from DNA to proteins? DNA on i t p i nscr tra replication RNA tra n o i t a nsl protein DNA gets all the glory, but proteins do all the work! trait

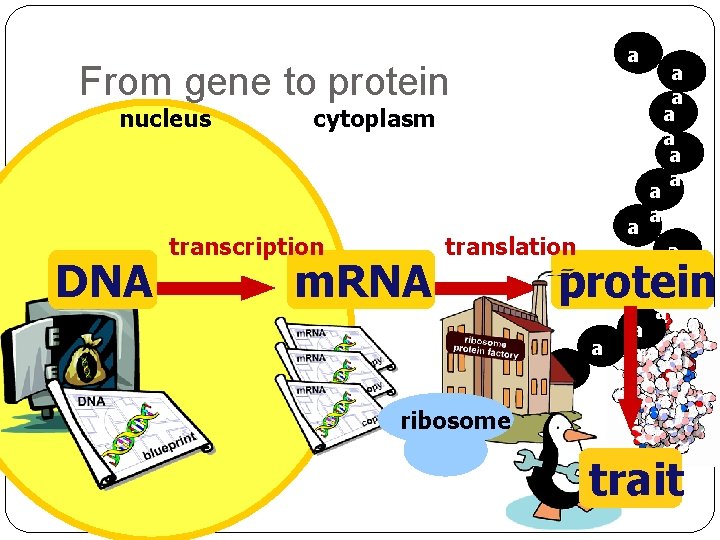
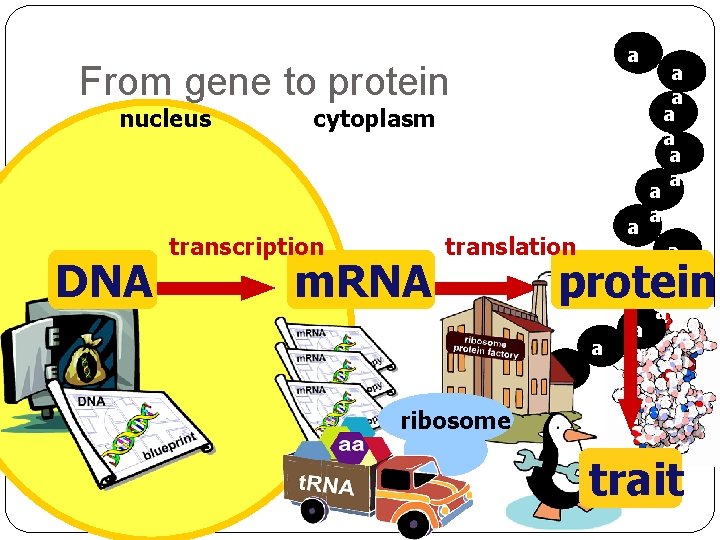
a a From gene to protein nucleus DNA cytoplasm transcription m. RNA a a translation ribosome a a a protein a a a a trait

Transcription from DNA nucleic acid language to RNA nucleic acid language

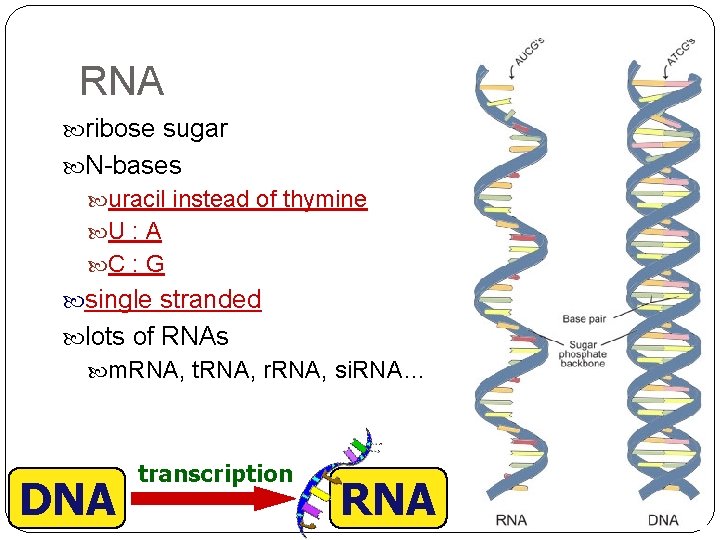
RNA ribose sugar N-bases uracil instead of thymine U : A C : G single stranded lots of RNAs m. RNA, t. RNA, r. RNA, si. RNA… DNA transcription RNA

Transcription Making m. RNA transcribed DNA strand = template strand untranscribed DNA strand = coding strand same sequence as RNA synthesis of complementary RNA strand transcription bubble enzyme RNA polymerase 5 DNA C G 3 build RNA 5 3 A G T A T C T A rewinding m. RNA 5 coding strand G C A T C G T T A 3 G C A U C G T A G C A T RNA polymerase C A G C T G A T 3 5 unwinding template strand

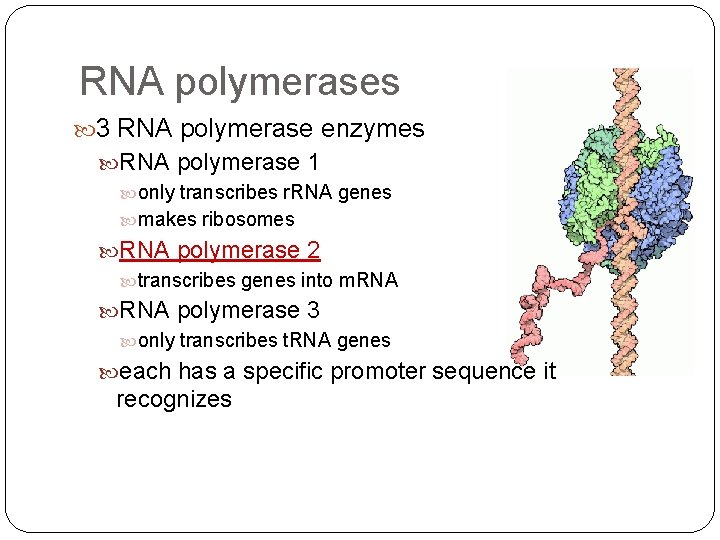
RNA polymerases 3 RNA polymerase enzymes RNA polymerase 1 only transcribes r. RNA genes makes ribosomes RNA polymerase 2 transcribes genes into m. RNA polymerase 3 only transcribes t. RNA genes each has a specific promoter sequence it recognizes

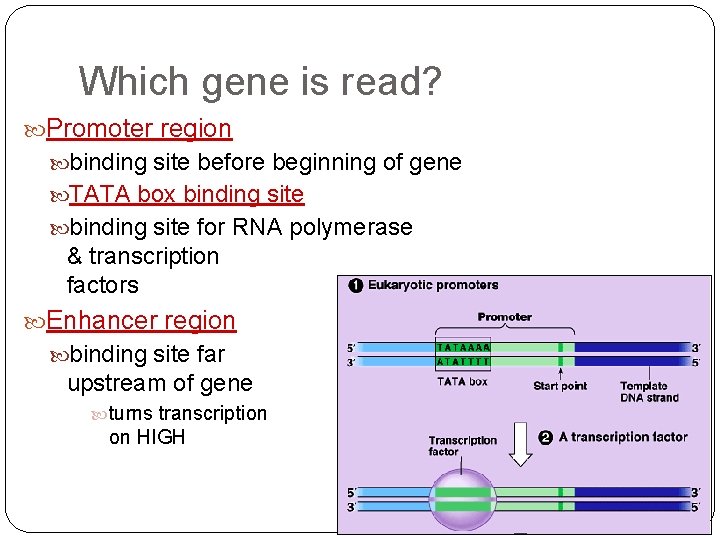
Which gene is read? Promoter region binding site before beginning of gene TATA box binding site for RNA polymerase & transcription factors Enhancer region binding site far upstream of gene turns transcription on HIGH

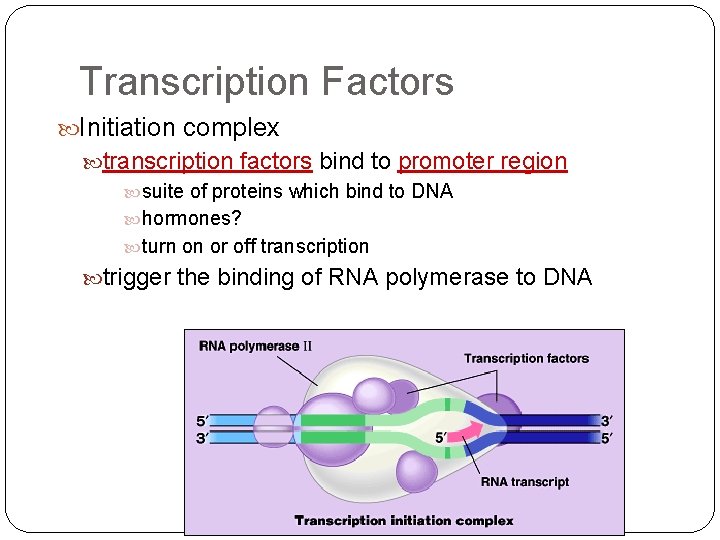
Transcription Factors Initiation complex transcription factors bind to promoter region suite of proteins which bind to DNA hormones? turn on or off transcription trigger the binding of RNA polymerase to DNA

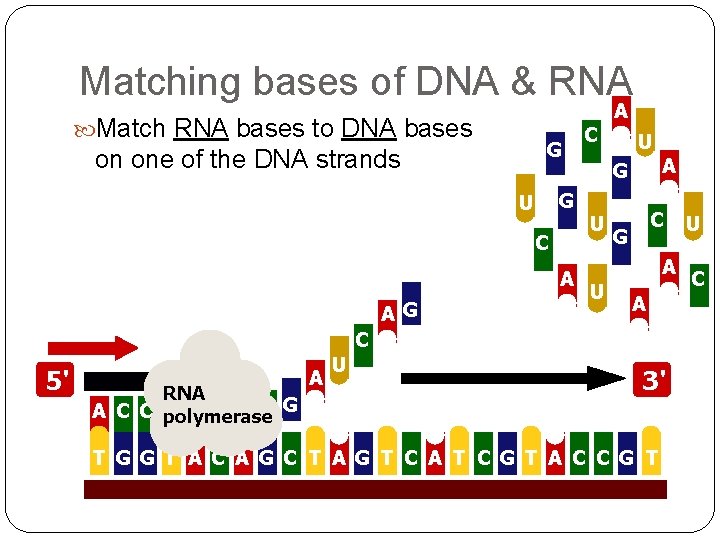
Matching bases of DNA & RNA Match RNA bases to DNA bases G on one of the DNA strands G U C A A G C A U G U A C G A U A C 5' RNA A C C polymerase G A U 3' T G G T A C A G C T A G T C A T C G T A C C G T U C

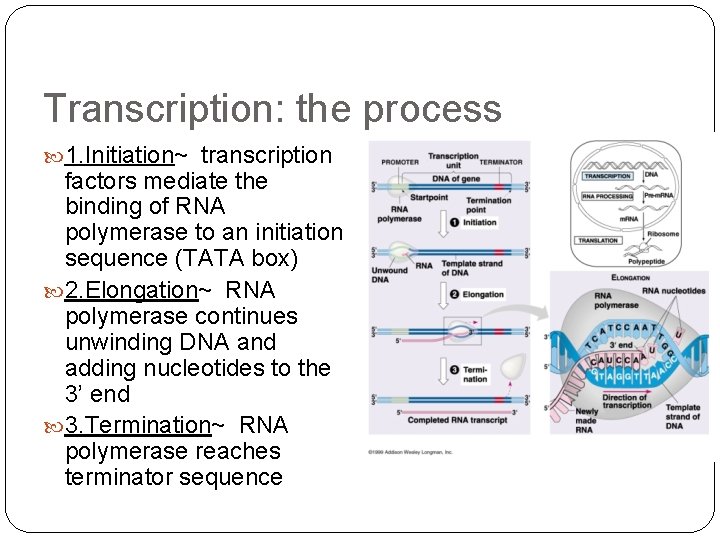
Transcription: the process 1. Initiation~ transcription factors mediate the binding of RNA polymerase to an initiation sequence (TATA box) 2. Elongation~ RNA polymerase continues unwinding DNA and adding nucleotides to the 3’ end 3. Termination~ RNA polymerase reaches terminator sequence

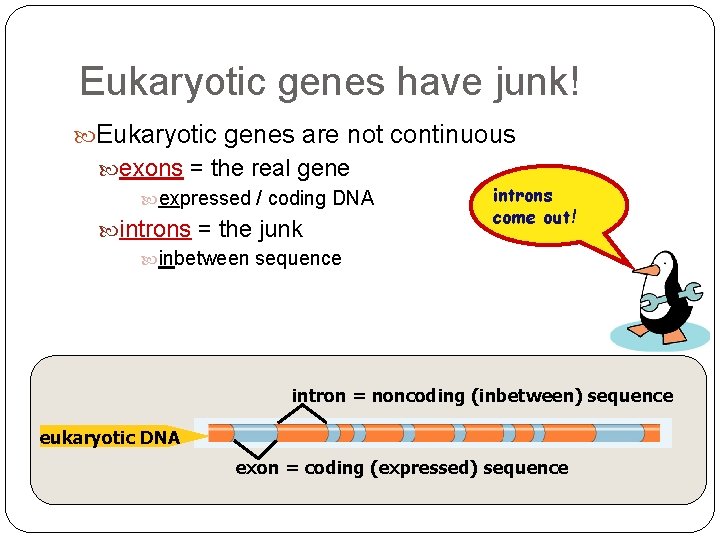
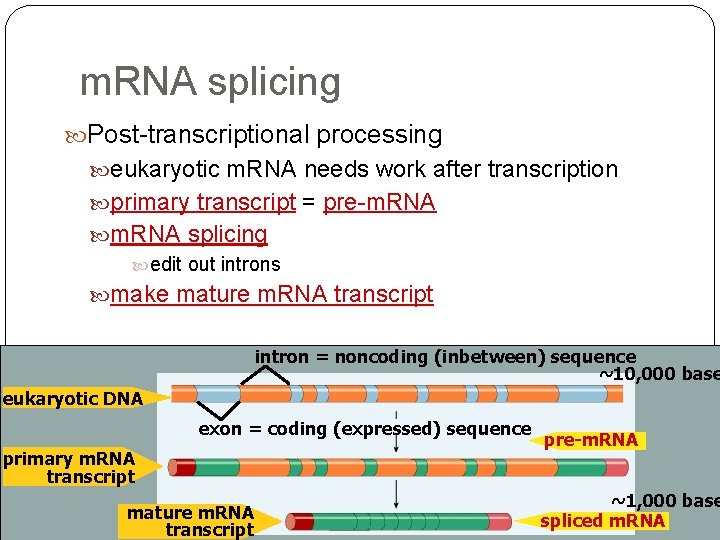
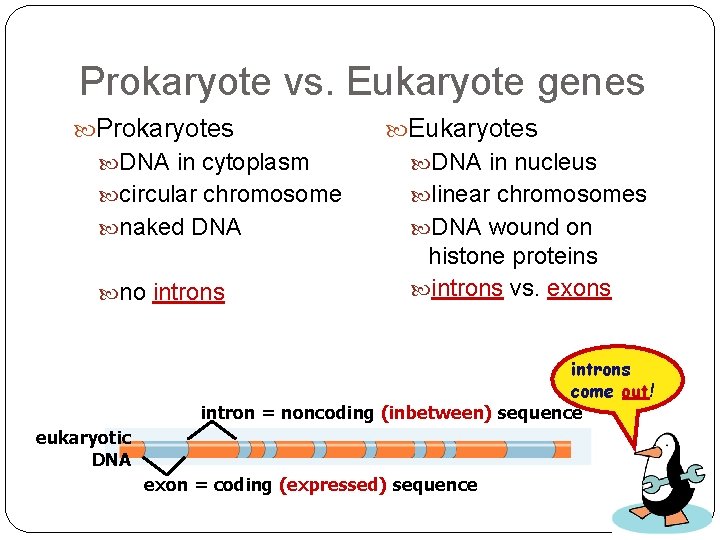
Eukaryotic genes have junk! Eukaryotic genes are not continuous exons = the real gene expressed / coding DNA introns = the junk inbetween sequence introns come out! intron = noncoding (inbetween) sequence eukaryotic DNA exon = coding (expressed) sequence

m. RNA splicing Post-transcriptional processing eukaryotic m. RNA needs work after transcription primary transcript = pre-m. RNA splicing edit out introns make mature m. RNA transcript intron = noncoding (inbetween) sequence ~10, 000 base eukaryotic DNA exon = coding (expressed) sequence primary m. RNA transcript mature m. RNA transcript pre-m. RNA ~1, 000 base spliced m. RNA

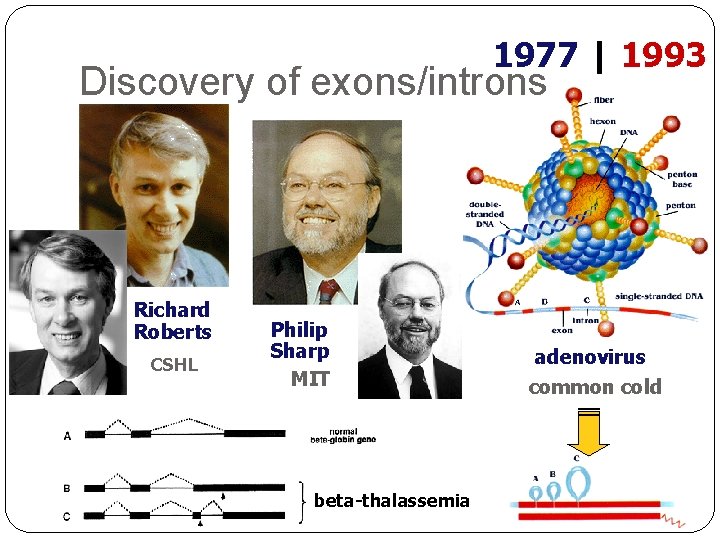
1977 | 1993 Discovery of exons/introns Richard Roberts CSHL Philip Sharp MIT beta-thalassemia adenovirus common cold

Splicing must be accurate No room for mistakes! a single base added or lost throws off the reading frame AUGCGGCTATGGGUCCGAUAAGGGCCAU AUGCGGUCCGAUAAGGGCCAU AUG|CGG|UCC|GAU|AAG|GGC|CAU Met|Arg|Ser|Asp|Lys|Gly|His AUGCGGCTATGGGUCCGAUAAGGGCCAU AUGCGGGUCCGAUAAGGGCCAU AUG|CGG|GUC|CGA|UAA|GGG|CCA|U Met|Arg|Val|Arg|STOP|

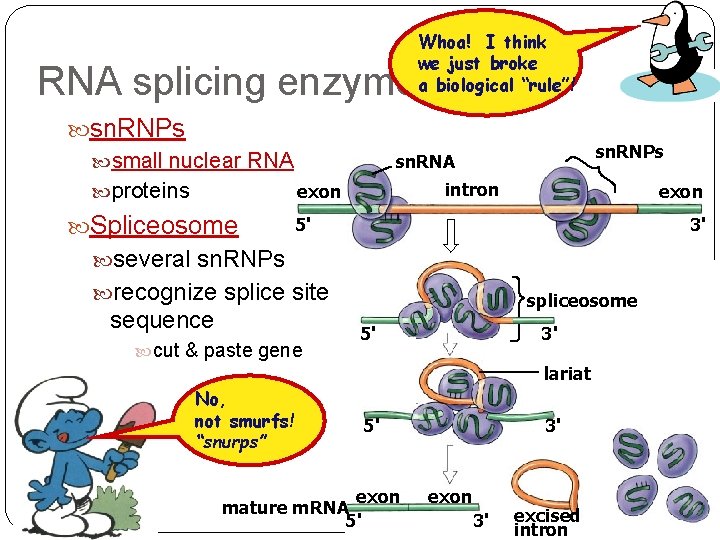
Whoa! I think we just broke a biological “rule”! RNA splicing enzymes sn. RNPs small nuclear RNA exon proteins Spliceosome sn. RNPs sn. RNA intron exon 5' 3' several sn. RNPs recognize splice site sequence cut & paste gene spliceosome 5' 3' lariat No, not smurfs! “snurps” 5' exon mature m. RNA 5' 3' exon 3' excised intron

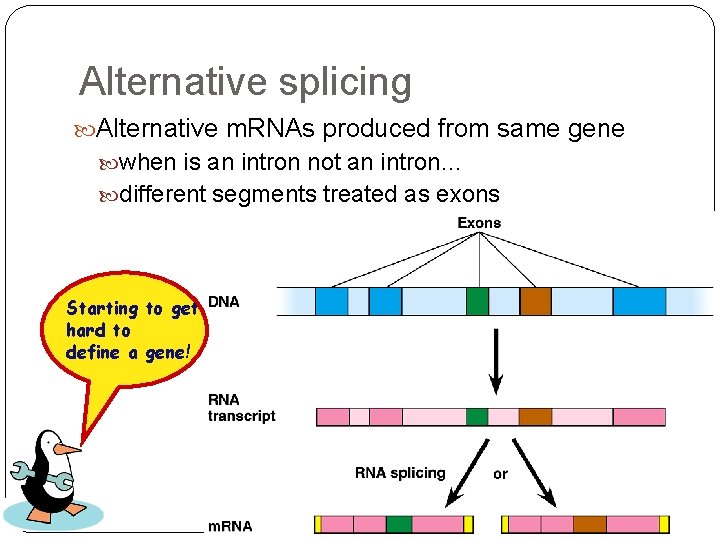
Alternative splicing Alternative m. RNAs produced from same gene when is an intron not an intron… different segments treated as exons Starting to get hard to define a gene!

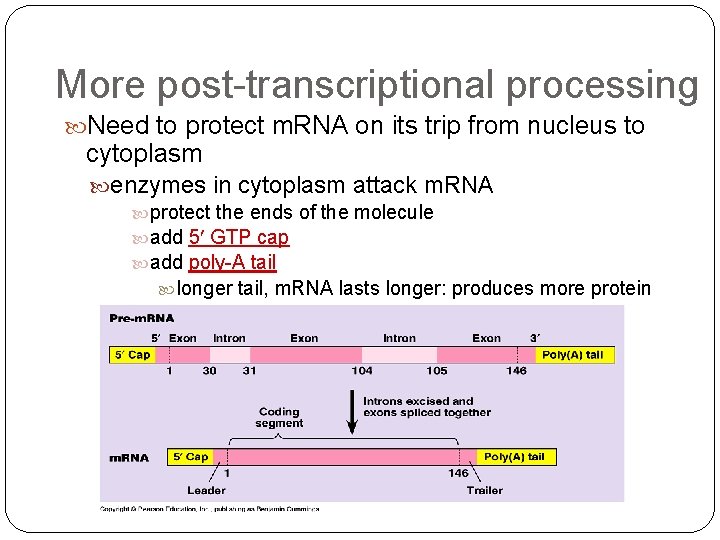
More post-transcriptional processing Need to protect m. RNA on its trip from nucleus to cytoplasm enzymes in cytoplasm attack m. RNA protect the ends of the molecule add 5 GTP cap add poly-A tail longer tail, m. RNA lasts longer: produces more protein

a a From gene to protein nucleus DNA cytoplasm transcription m. RNA a a translation ribosome a a a protein a a a a trait

Translation from nucleic acid language to amino acid language

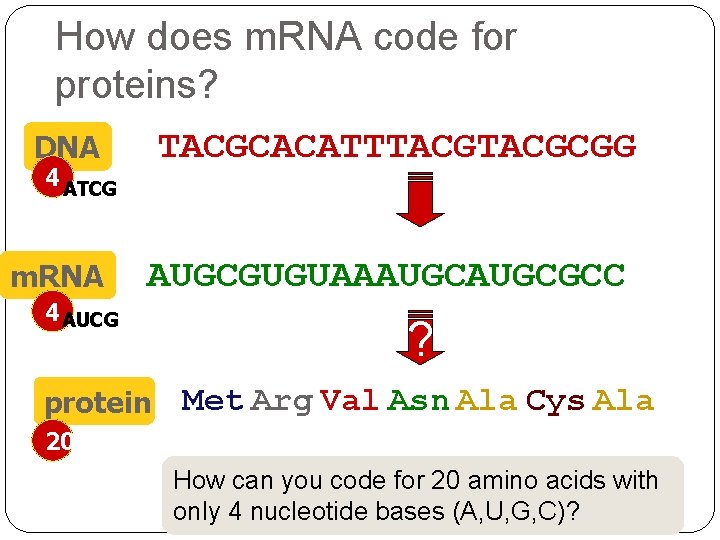
How does m. RNA code for proteins? TACGCACATTTACGCGG DNA 4 ATCG m. RNA 4 AUCG AUGCGUGUAAAUGCGCC protein ? Met Arg Val Asn Ala Cys Ala 20 How can you code for 20 amino acids with only 4 nucleotide bases (A, U, G, C)?

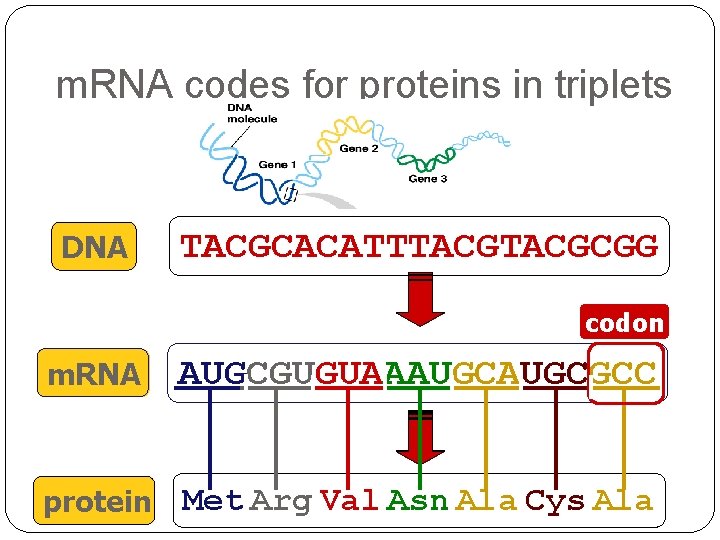
m. RNA codes for proteins in triplets DNA TACGCACATTTACGCGG codon m. RNA AUGCGUGUAAAUGCGCC ? protein Met Arg Val Asn Ala Cys Ala

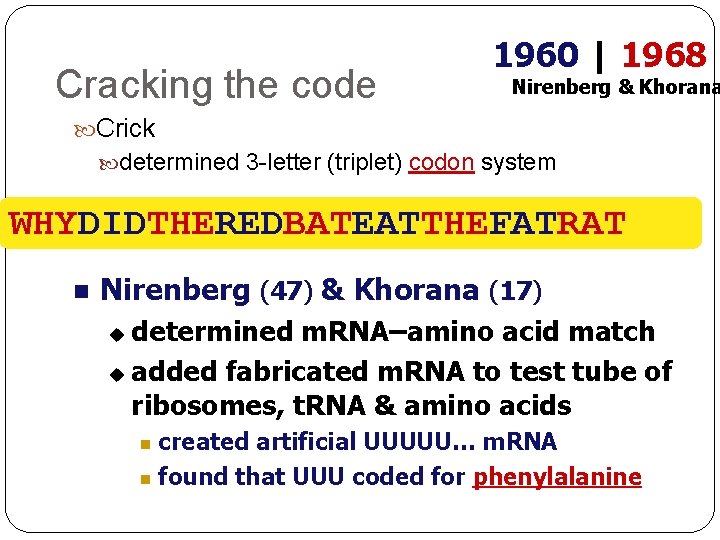
Cracking the code 1960 | 1968 Nirenberg & Khorana Crick determined 3 -letter (triplet) codon system WHYDIDTHEREDBATEATTHEFATRAT Nirenberg (47) & Khorana (17) determined m. RNA–amino acid match u added fabricated m. RNA to test tube of ribosomes, t. RNA & amino acids u created artificial UUUUU… m. RNA found that UUU coded for phenylalanine

Marshall Nirenberg 1960 | 1968 Har Khorana

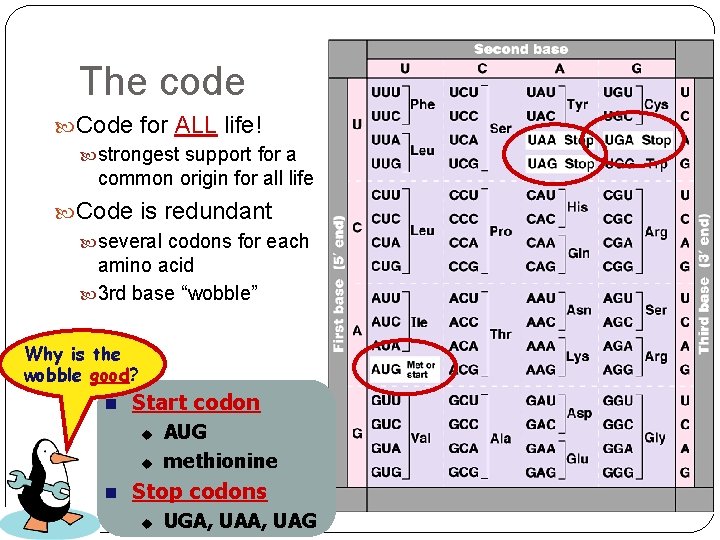
The code Code for ALL life! strongest support for a common origin for all life Code is redundant several codons for each amino acid 3 rd base “wobble” Why is the wobble good? Start codon u u AUG methionine Stop codons u UGA, UAG

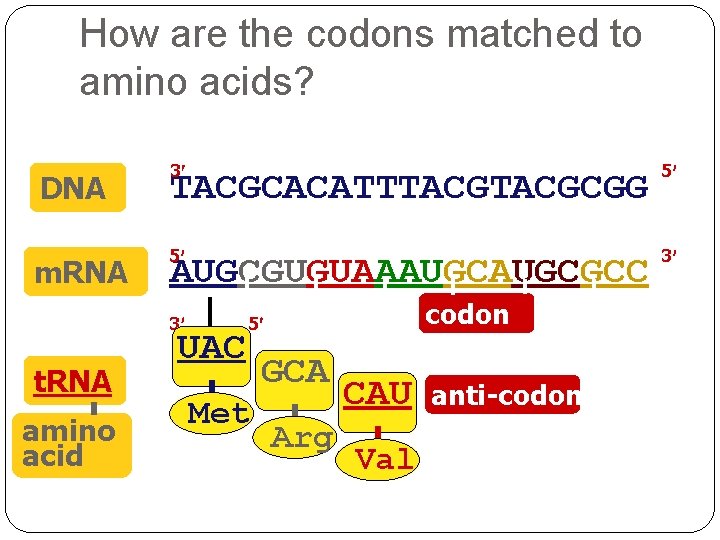
How are the codons matched to amino acids? DNA m. RNA 3 TACGCACATTTACGCGG 5 5 3 AUGCGUGUAAAUGCGCC 3 t. RNA amino acid UAC codon 5 Met GCA Arg CAU Val anti-codon

a a From gene to protein nucleus DNA cytoplasm transcription m. RNA a a translation ribosome a a a protein a a a a trait

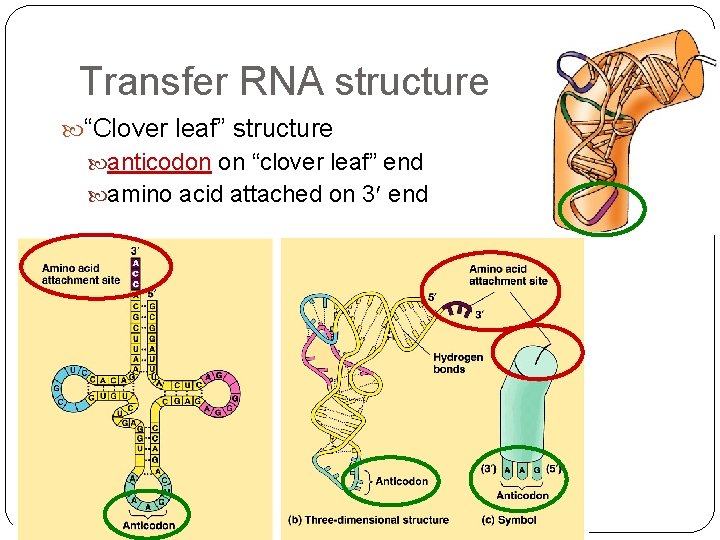
Transfer RNA structure “Clover leaf” structure anticodon on “clover leaf” end amino acid attached on 3 end

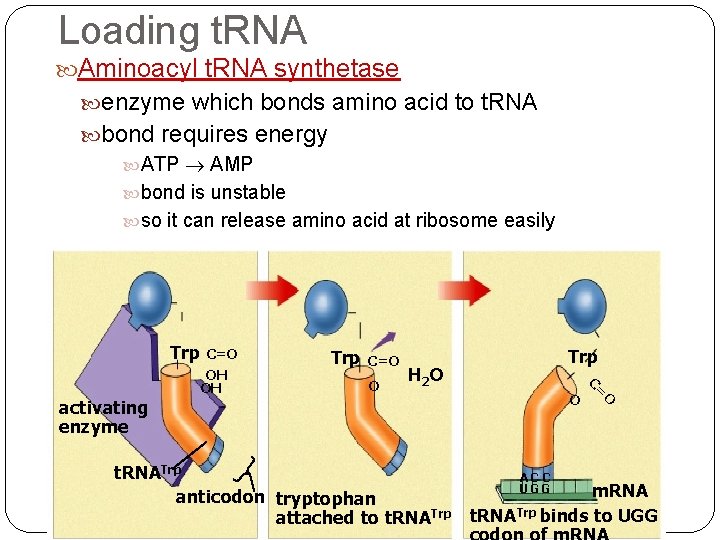
Loading t. RNA Aminoacyl t. RNA synthetase enzyme which bonds amino acid to t. RNA bond requires energy ATP AMP bond is unstable so it can release amino acid at ribosome easily Trp C=O O Trp H 2 O AC C UGG O t. RNATrp anticodon tryptophan attached to t. RNATrp O C= activating enzyme C=O OH OH m. RNA t. RNATrp binds to UGG

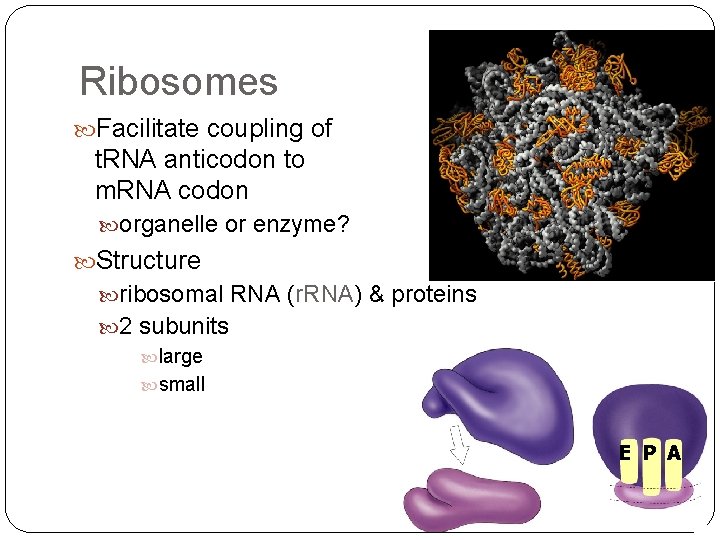
Ribosomes Facilitate coupling of t. RNA anticodon to m. RNA codon organelle or enzyme? Structure ribosomal RNA (r. RNA) & proteins 2 subunits large small E P A

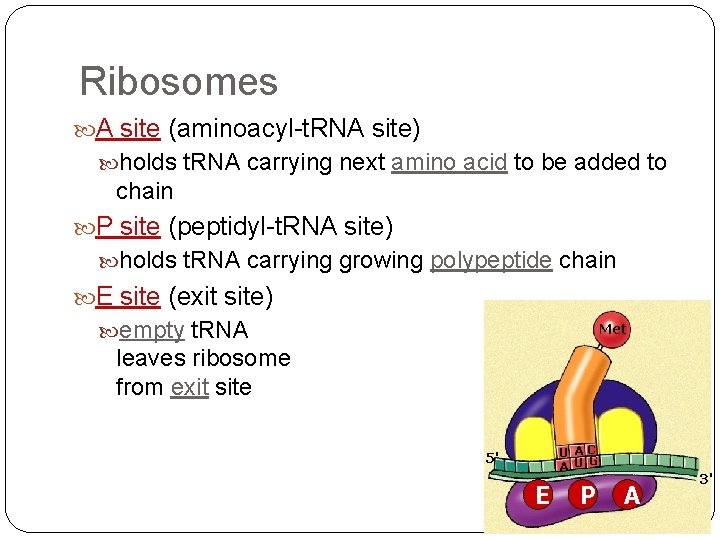
Ribosomes A site (aminoacyl-t. RNA site) holds t. RNA carrying next amino acid to be added to chain P site (peptidyl-t. RNA site) holds t. RNA carrying growing polypeptide chain E site (exit site) empty t. RNA Met leaves ribosome from exit site U A C A U G 5' E P A 3'

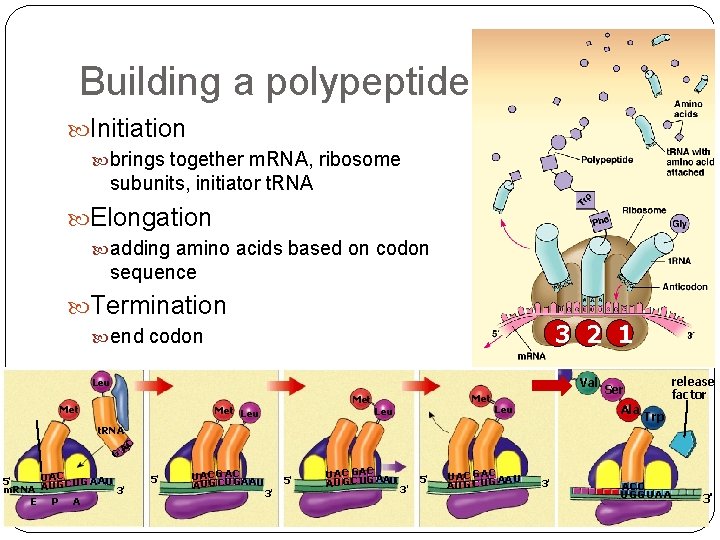
Building a polypeptide Initiation brings together m. RNA, ribosome subunits, initiator t. RNA Elongation adding amino acids based on codon sequence Termination 3 2 1 end codon Val Leu Met Met Leu Ala Leu release factor Ser Trp A C t. RNA G UAC 5' C UG A A U m. RNA A U G 3' E P A 5' UA C G A C A A UG C U G AU 5' 3' U A C GA C A U G C UG AA U 3' 5' U AC G A C AA U G C UG 3' A CC U GG U A A 3'

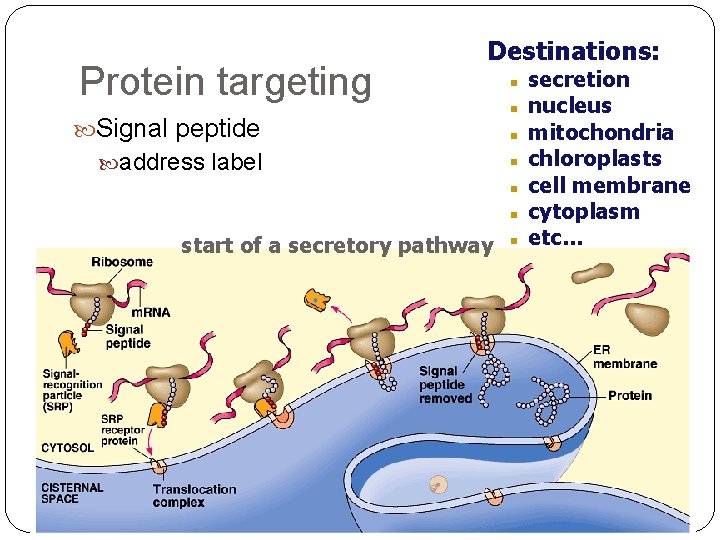
Protein targeting Destinations: Signal peptide address label start of a secretory pathway secretion nucleus mitochondria chloroplasts cell membrane cytoplasm etc…

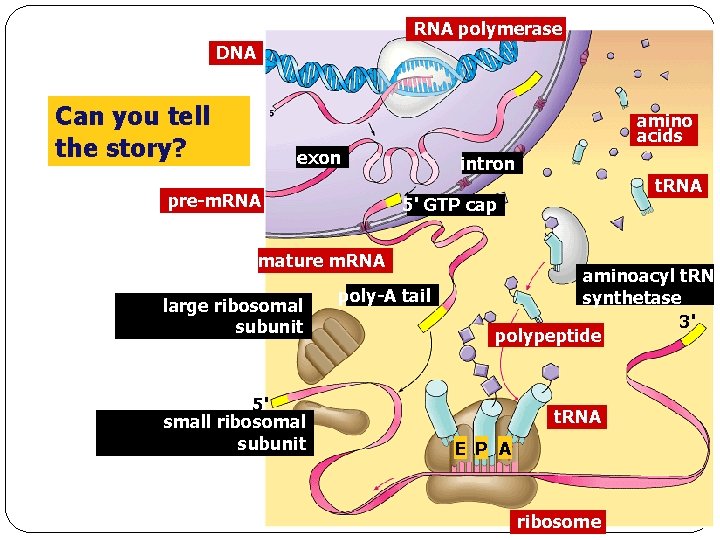
RNA polymerase DNA Can you tell the story? amino acids exon pre-m. RNA intron 5' GTP cap mature m. RNA large ribosomal subunit 5' small ribosomal subunit t. RNA poly-A tail aminoacyl t. RNA synthetase 3' polypeptide t. RNA E P A ribosome

Prokaryote vs. Eukaryote genes Prokaryotes Eukaryotes DNA in cytoplasm DNA in nucleus circular chromosome linear chromosomes naked DNA wound on no introns histone proteins introns vs. exons introns come out! intron = noncoding (inbetween) sequence eukaryotic DNA exon = coding (expressed) sequence

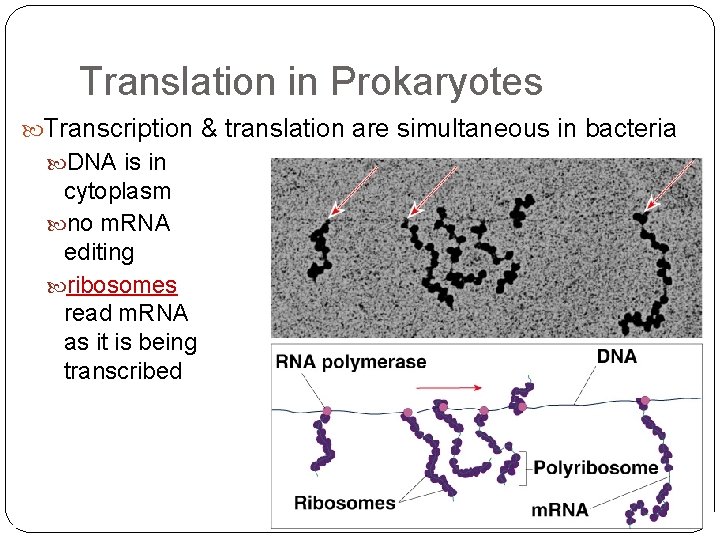
Translation in Prokaryotes Transcription & translation are simultaneous in bacteria DNA is in cytoplasm no m. RNA editing ribosomes read m. RNA as it is being transcribed

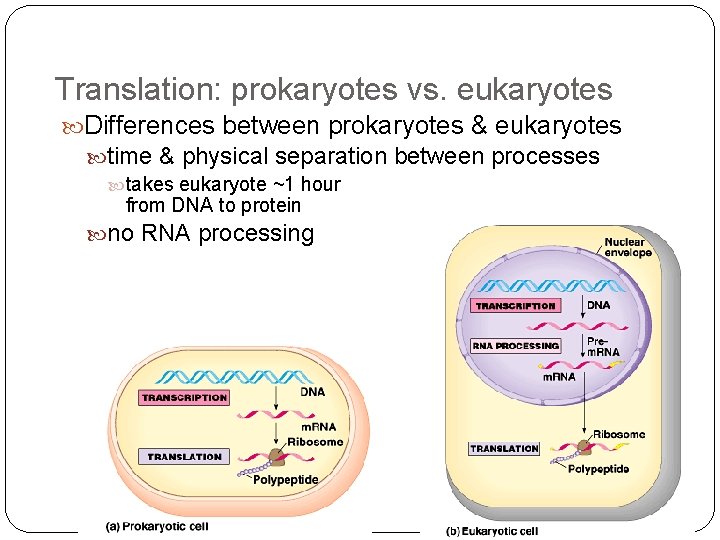
Translation: prokaryotes vs. eukaryotes Differences between prokaryotes & eukaryotes time & physical separation between processes takes eukaryote ~1 hour from DNA to protein no RNA processing

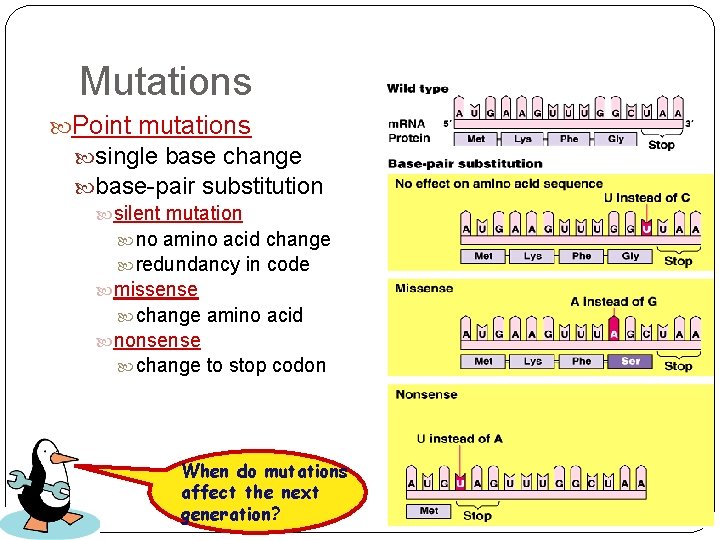
Mutations Point mutations single base change base-pair substitution silent mutation no amino acid change redundancy in code missense change amino acid nonsense change to stop codon When do mutations affect the next generation?

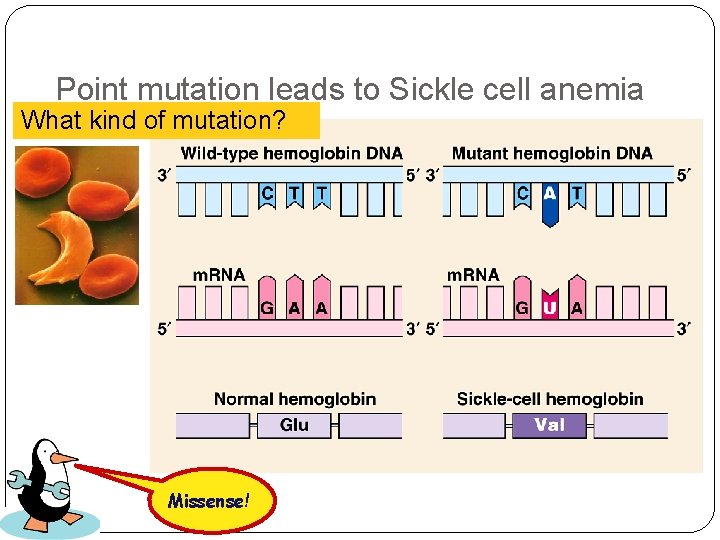
Point mutation leads to Sickle cell anemia What kind of mutation? Missense!

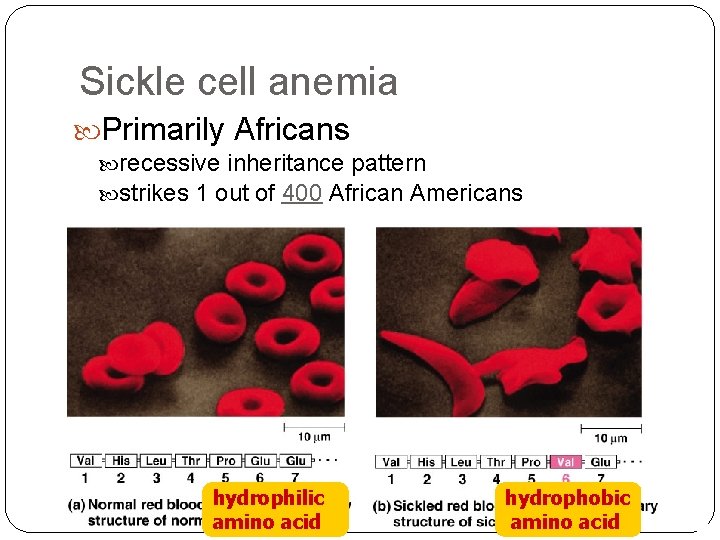
Sickle cell anemia Primarily Africans recessive inheritance pattern strikes 1 out of 400 African Americans hydrophilic amino acid hydrophobic amino acid

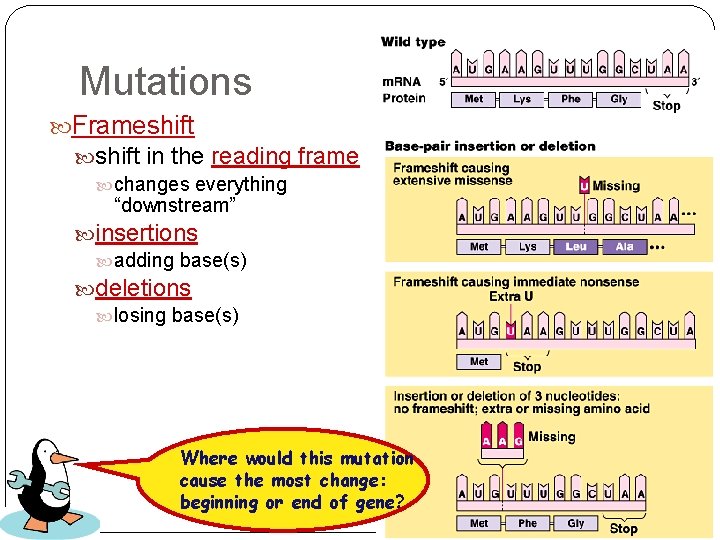
Mutations Frameshift in the reading frame changes everything “downstream” insertions adding base(s) deletions losing base(s) Where would this mutation cause the most change: beginning or end of gene?

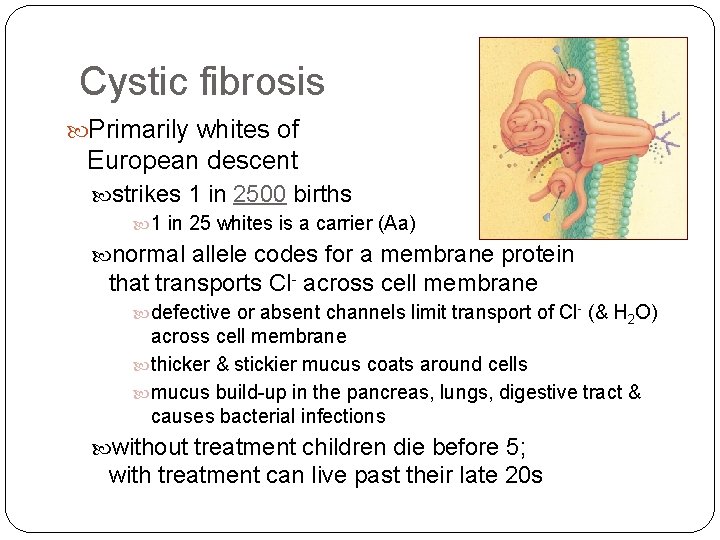
Cystic fibrosis Primarily whites of European descent strikes 1 in 2500 births 1 in 25 whites is a carrier (Aa) normal allele codes for a membrane protein that transports Cl- across cell membrane defective or absent channels limit transport of Cl- (& H 2 O) across cell membrane thicker & stickier mucus coats around cells mucus build-up in the pancreas, lungs, digestive tract & causes bacterial infections without treatment children die before 5; with treatment can live past their late 20 s

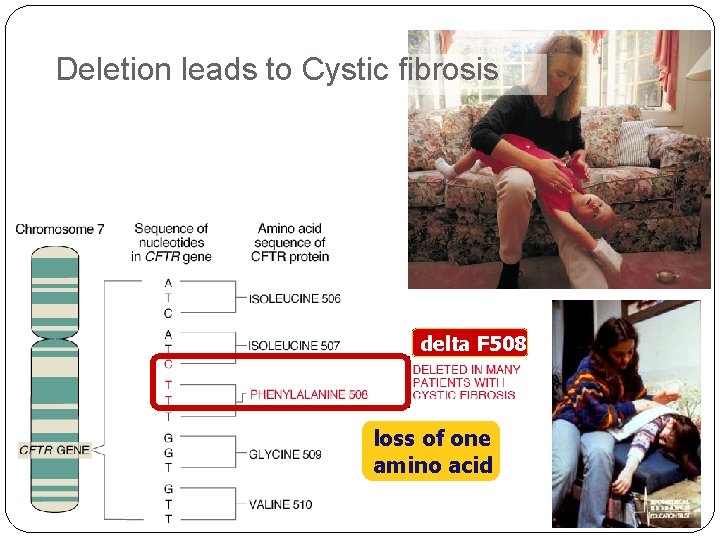
Deletion leads to Cystic fibrosis delta F 508 loss of one amino acid

What’s the value of mutations? 2007 -2008