Chapter 10 Lecture Outline See separate Power Point




- Slides: 34

Chapter 10 Lecture Outline See separate Power. Point slides for all figures and tables preinserted into Power. Point without notes. Copyright © Mc. Graw-Hill Education. Permission required for reproduction or display. 1

A Glimpse of History § 1870 s: Bacteria classified by shape (Ferdinand Cohn) § 1908: Physiology rather than morphology (Sigurd Orla. Jensen) § 1930 s: Classification based on evolutionary relationships (Albert Kluyver, C. B. van Niel) § 1970: Relationships determined by comparing physical traits, nucleotide sequences (Roger Stanier) § Late 1970 s: Prokaryotes divided into two major groups based upon ribosomal RNA sequences (Carl Woese) • Led to current three domain system: Bacteria, Archaea, Eukarya

10. 1. Principles of Taxonomy § Taxonomy is the science that studies organisms to arrange them into groups, or taxa § Three separate but interrelated areas: • Identification • Process of characterizing in order to group • Classification • Arranging organisms into similar or related groups • Nomenclature • System of assigning names

10. 1. Principles of Taxonomy § Taxonomic Hierarchies • Species is basic unit: group of morphologically similar organisms capable of producing fertile offspring • Definition problematic for prokaryotes • Species is group of closely related isolates or strains • Informal groupings also used • May be genetically unrelated – Lactic acid bacteria – Anoxygenic phototrophs – Endospore-formers – Sulfate reducers • Kingdoms for prokaryotes still in state of flux

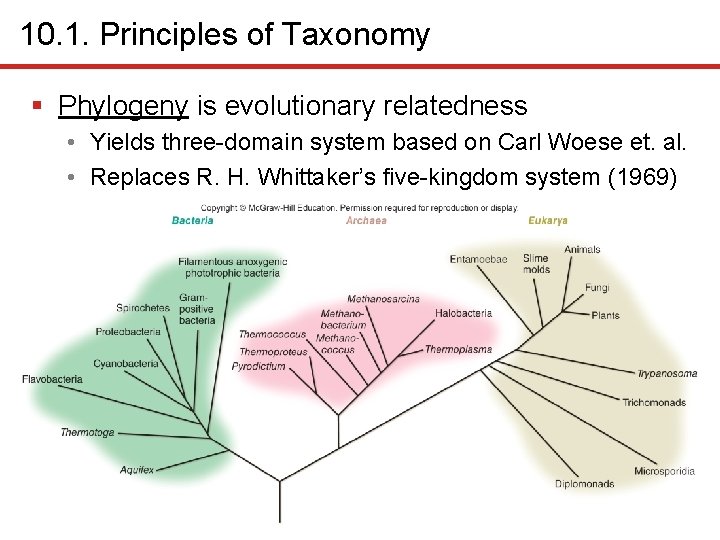
10. 1. Principles of Taxonomy § Phylogeny is evolutionary relatedness • Yields three-domain system based on Carl Woese et. al. • Replaces R. H. Whittaker’s five-kingdom system (1969)

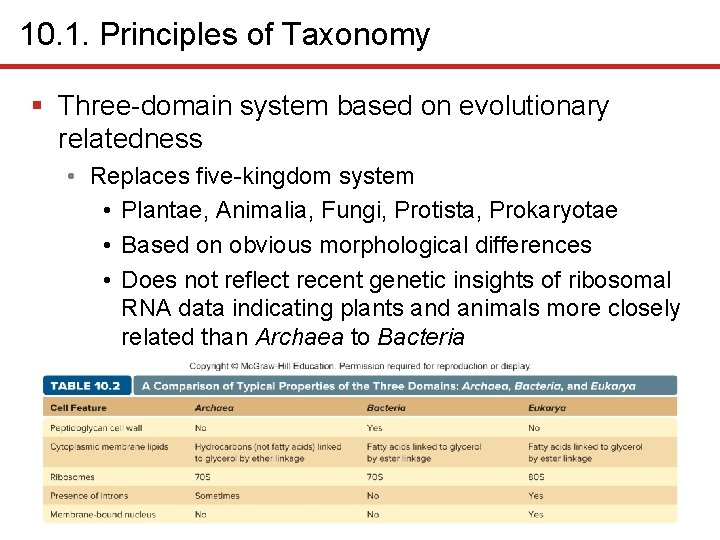
10. 1. Principles of Taxonomy § Three-domain system based on evolutionary relatedness • Replaces five-kingdom system • Plantae, Animalia, Fungi, Protista, Prokaryotae • Based on obvious morphological differences • Does not reflect recent genetic insights of ribosomal RNA data indicating plants and animals more closely related than Archaea to Bacteria

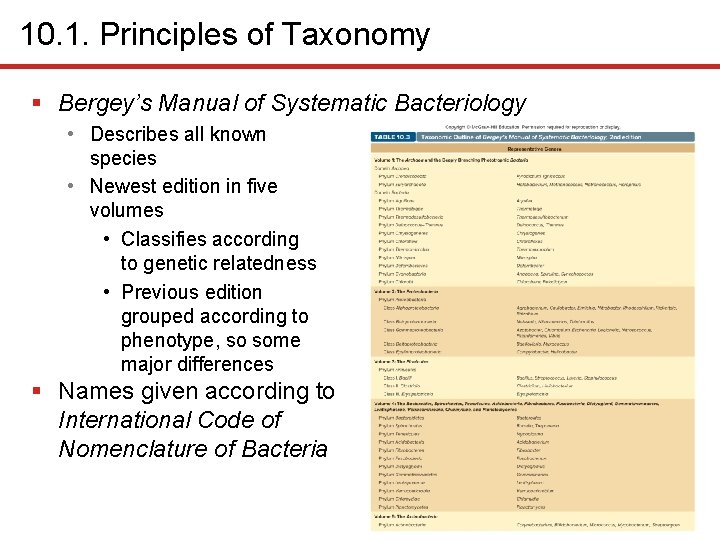
10. 1. Principles of Taxonomy § Bergey’s Manual of Systematic Bacteriology • Describes all known species • Newest edition in five volumes • Classifies according to genetic relatedness • Previous edition grouped according to phenotype, so some major differences § Names given according to International Code of Nomenclature of Bacteria

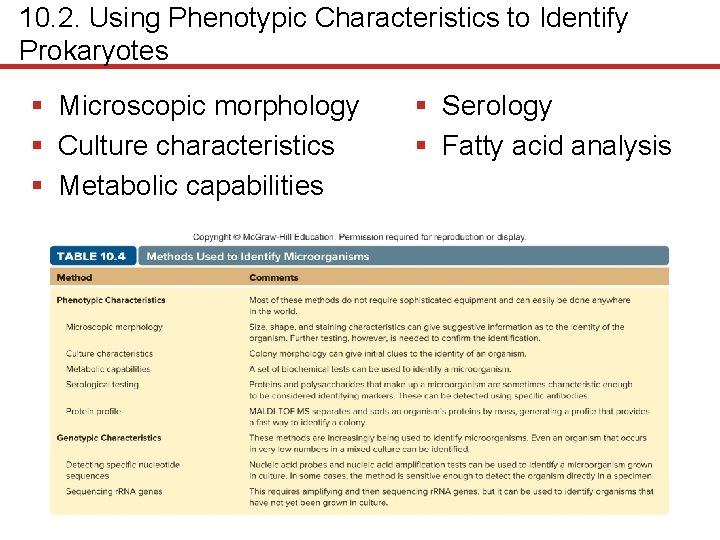
10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Microscopic morphology § Culture characteristics § Metabolic capabilities § Serology § Fatty acid analysis

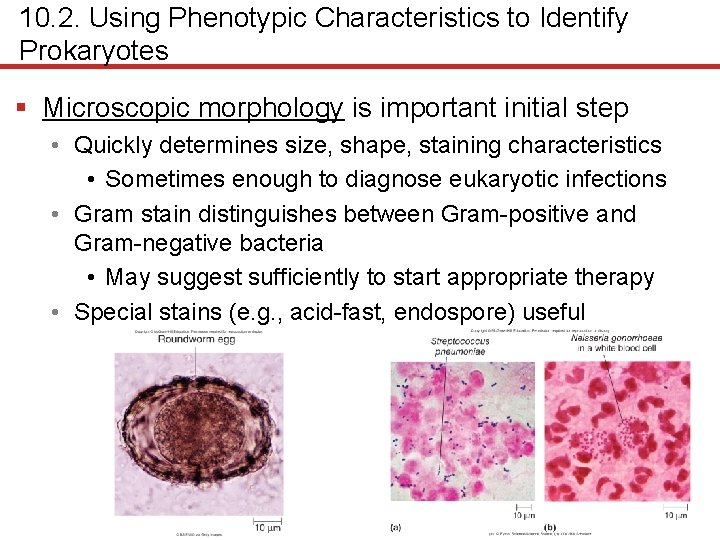
10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Microscopic morphology is important initial step • Quickly determines size, shape, staining characteristics • Sometimes enough to diagnose eukaryotic infections • Gram stain distinguishes between Gram-positive and Gram-negative bacteria • May suggest sufficiently to start appropriate therapy • Special stains (e. g. , acid-fast, endospore) useful

10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Culture characteristics can give clues • Streptococci colonies generally fairly small • Serratia marcescens colonies often red at 22°C • Pseudomonas aeruginosa often produces green pigment • Cultures also have distinct fruity odor • Differential media aids in identification • Streptococcus pyogenes (strep throat) yields β -hemolytic colonies on blood agar • E. coli (urinary tract infection) ferments lactose, forms pink colonies on Mac. Conkey agar

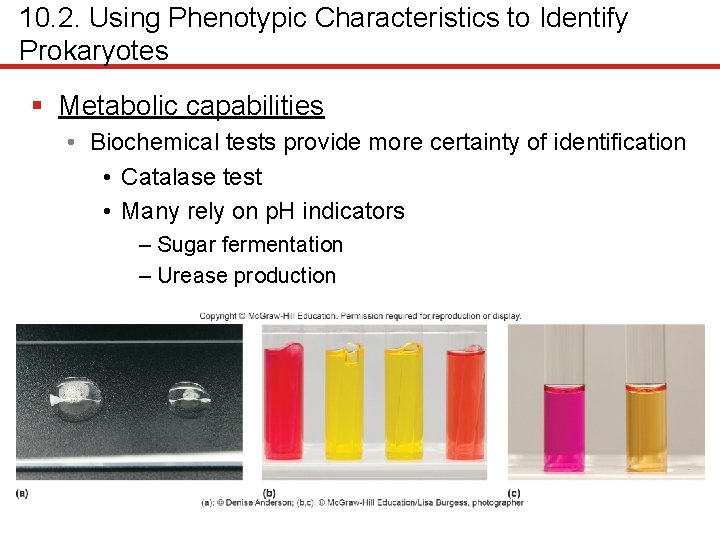
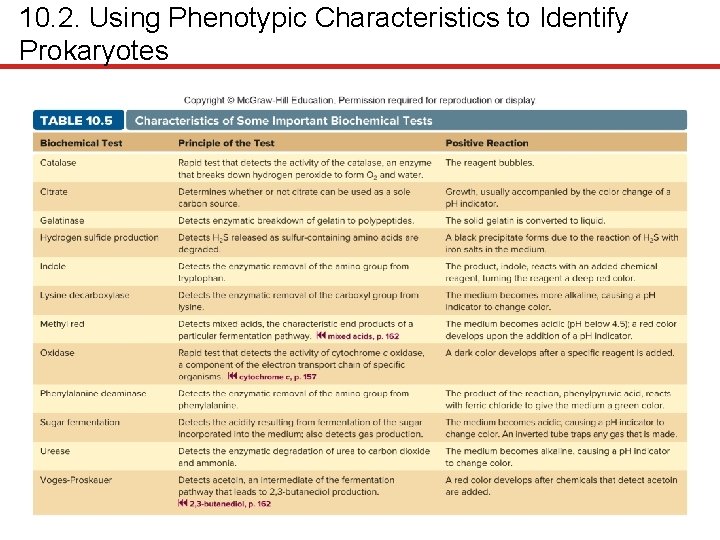
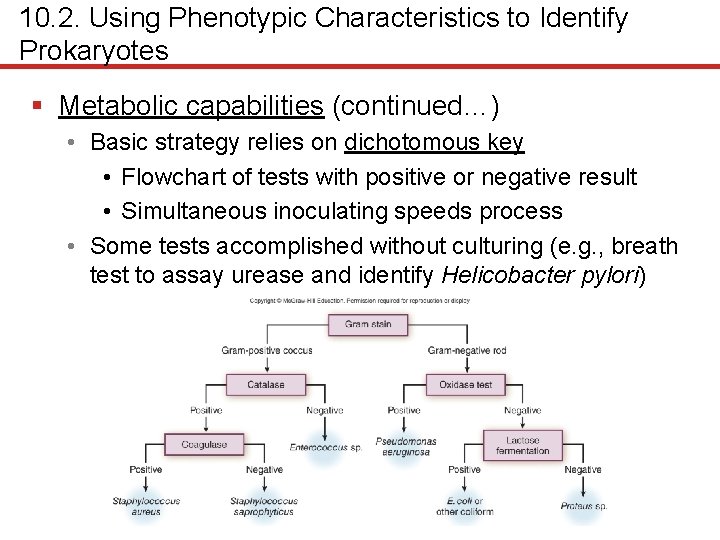
10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Metabolic capabilities • Biochemical tests provide more certainty of identification • Catalase test • Many rely on p. H indicators – Sugar fermentation – Urease production

10. 2. Using Phenotypic Characteristics to Identify Prokaryotes

10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Metabolic capabilities (continued…) • Basic strategy relies on dichotomous key • Flowchart of tests with positive or negative result • Simultaneous inoculating speeds process • Some tests accomplished without culturing (e. g. , breath test to assay urease and identify Helicobacter pylori)

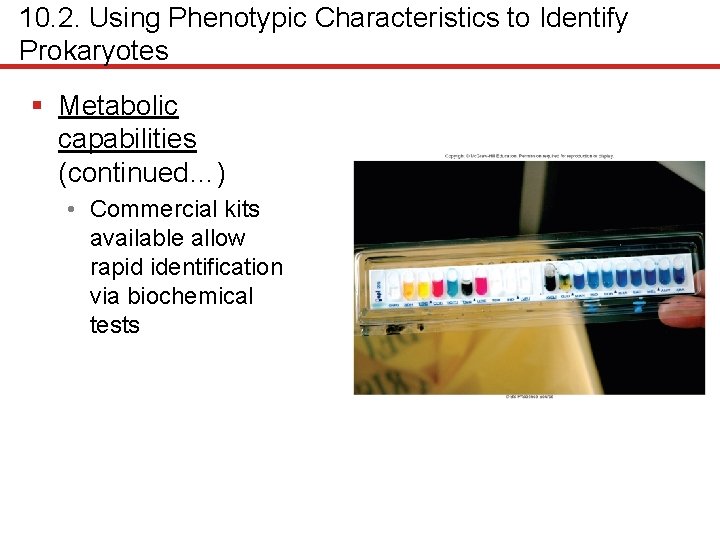
10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Metabolic capabilities (continued…) • Commercial kits available allow rapid identification via biochemical tests

10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § Serology • Proteins, polysaccharides of prokaryotic cells can serve as identifying markers • Most useful include surface structures of cell wall, capsule, flagella, pili • Some Streptococcus species contain unique carbohydrate in cell wall • Serological tests use antibodies for detection (Chapter 18)

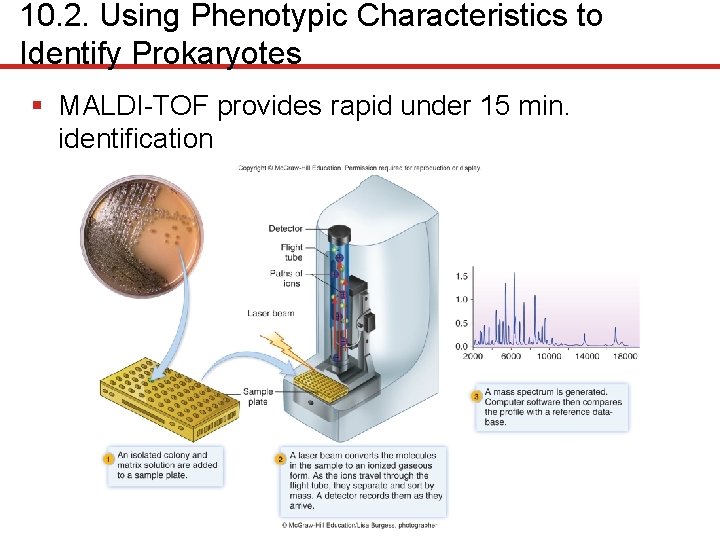
10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § MALDI-TOF ( matrix- assisted laser desorption ionization time of flight mass spectrometry) • Measure the masses of various components using mass spectrophotometer • Sample spotted on sample plate with matrix • Laser beam vaporizes and ionizes sample • Time of flight: small ions travel faster than larger ones in tube • Mass spectrum a “fingerprint” or profile of the proteins and other macromolecules in the cell

10. 2. Using Phenotypic Characteristics to Identify Prokaryotes § MALDI-TOF provides rapid under 15 min. identification

10. 3. Using Genotypic Characteristics to Identify Prokaryotes § Detecting Specific Nucleotide Sequences • Tests can identify sequences unique to species or group • Nucleic acid probes • Nucleic acid amplification tests (NAATs) • Limitation is each detects only single possibility • Need to run multiple probes if organism being tested could be one of multiple different species or related groups

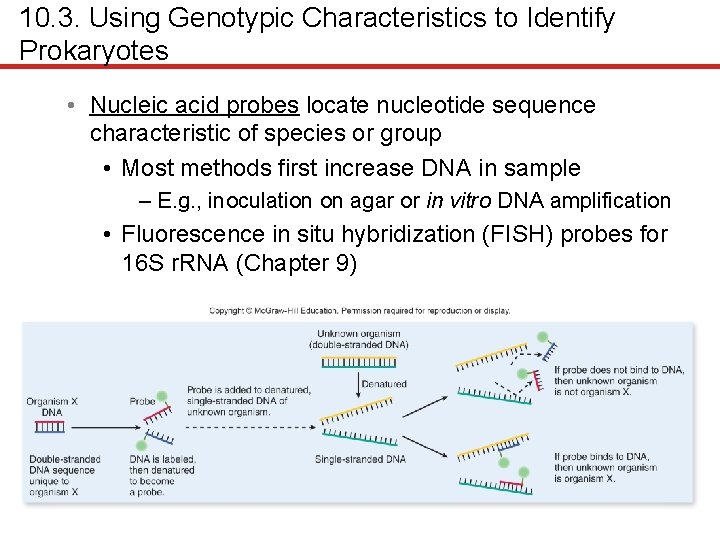
10. 3. Using Genotypic Characteristics to Identify Prokaryotes • Nucleic acid probes locate nucleotide sequence characteristic of species or group • Most methods first increase DNA in sample – E. g. , inoculation on agar or in vitro DNA amplification • Fluorescence in situ hybridization (FISH) probes for 16 S r. RNA (Chapter 9)

10. 3. Using Genotypic Characteristics to Identify Prokaryotes • Nucleic acid amplification tests (NAATs) used to increase number of copies of specific DNA sequences • Allows detection of small numbers of organisms – Often from body fluids, soil, food, water • Detection of organisms that cannot be cultured • Polymerase chain reaction (PCR) common technique (Chapter 9)

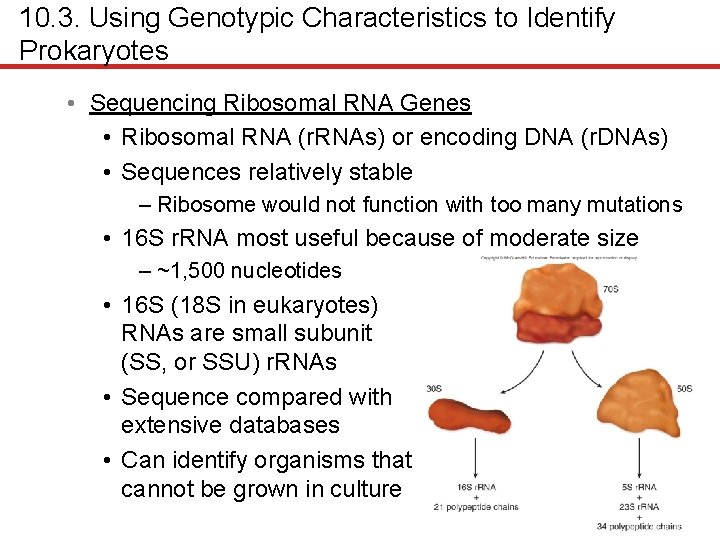
10. 3. Using Genotypic Characteristics to Identify Prokaryotes • Sequencing Ribosomal RNA Genes • Ribosomal RNA (r. RNAs) or encoding DNA (r. DNAs) • Sequences relatively stable – Ribosome would not function with too many mutations • 16 S r. RNA most useful because of moderate size – ~1, 500 nucleotides • 16 S (18 S in eukaryotes) RNAs are small subunit (SS, or SSU) r. RNAs • Sequence compared with extensive databases • Can identify organisms that cannot be grown in culture

10. 4. Characterizing Strain Differences § Characterizing strains important • Foodborne illnesses • Diagnosing certain diseases • Forensic investigations of bioterrorism, biocrimes

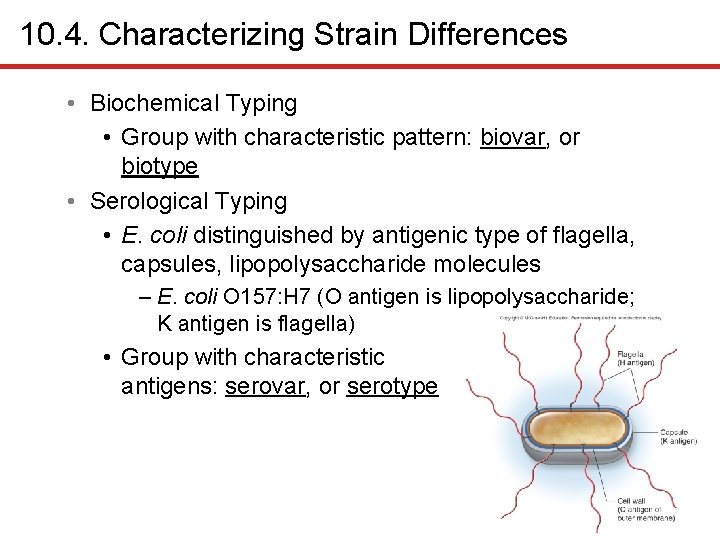
10. 4. Characterizing Strain Differences • Biochemical Typing • Group with characteristic pattern: biovar, or biotype • Serological Typing • E. coli distinguished by antigenic type of flagella, capsules, lipopolysaccharide molecules – E. coli O 157: H 7 (O antigen is lipopolysaccharide; K antigen is flagella) • Group with characteristic antigens: serovar, or serotype

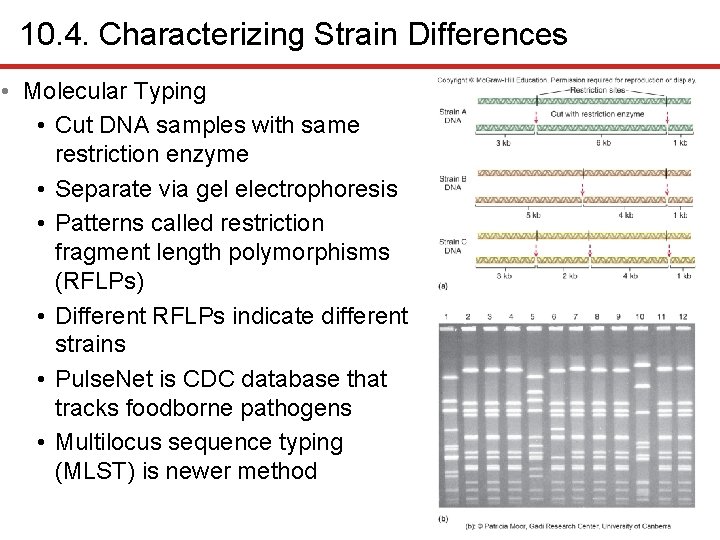
10. 4. Characterizing Strain Differences • Molecular Typing • Cut DNA samples with same restriction enzyme • Separate via gel electrophoresis • Patterns called restriction fragment length polymorphisms (RFLPs) • Different RFLPs indicate different strains • Pulse. Net is CDC database that tracks foodborne pathogens • Multilocus sequence typing (MLST) is newer method

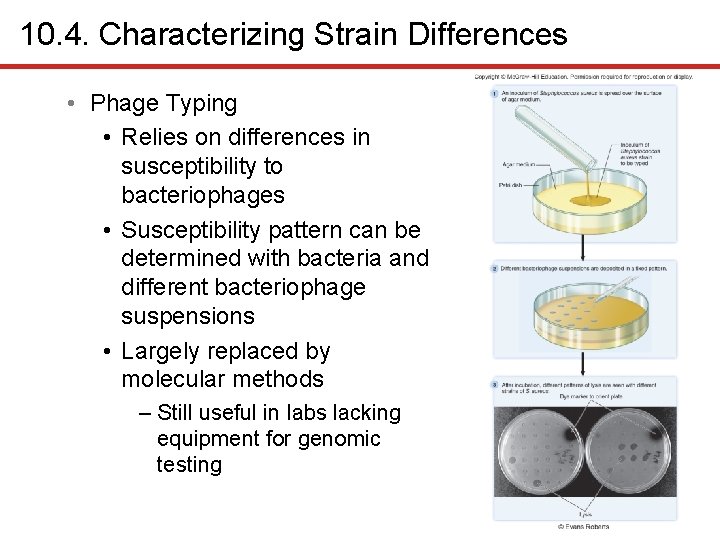
10. 4. Characterizing Strain Differences 1 • Phage Typing • Relies on differences in susceptibility to bacteriophages • Susceptibility pattern can be determined with bacteria and different bacteriophage suspensions • Largely replaced by molecular methods – Still useful in labs lacking equipment for genomic testing

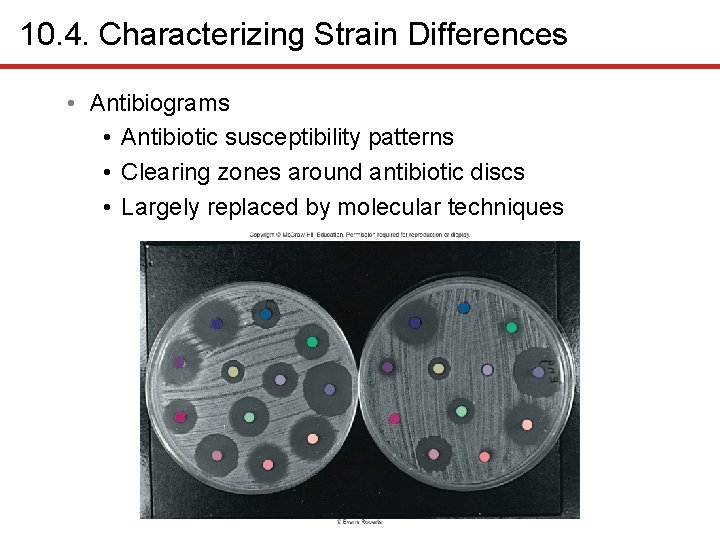
10. 4. Characterizing Strain Differences • Antibiograms • Antibiotic susceptibility patterns • Clearing zones around antibiotic discs • Largely replaced by molecular techniques

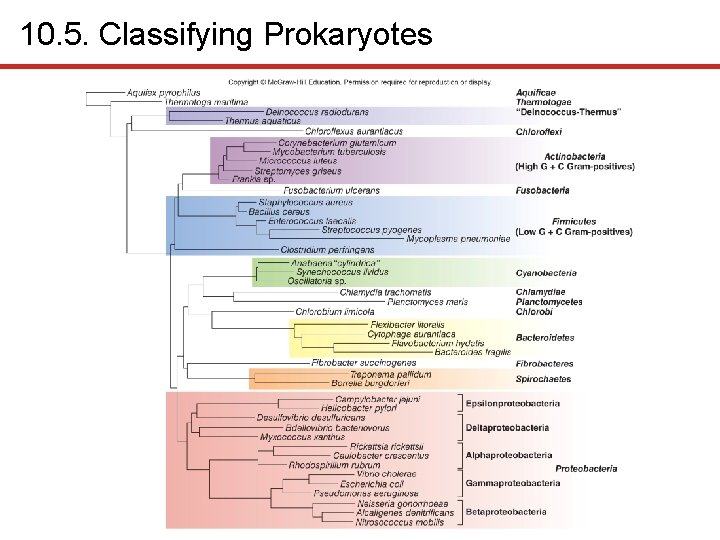
10. 5. Classifying Prokaryotes § Classification historically based on phenotypic traits • Size, shape, staining, metabolic capabilities • But phenotypically similar organisms may be only distantly related; conversely, closely related organisms may appear dissimilar § New molecular techniques more accurate • Provide greater insights into evolutionary relatedness • DNA sequences viewed as evolutionary chronometers • Provide relative measure of time elapsed since divergence from common ancestor • Mutations accumulate over time • DNA sequencing allows construction of phylogenetic tree

10. 5. Classifying Prokaryotes

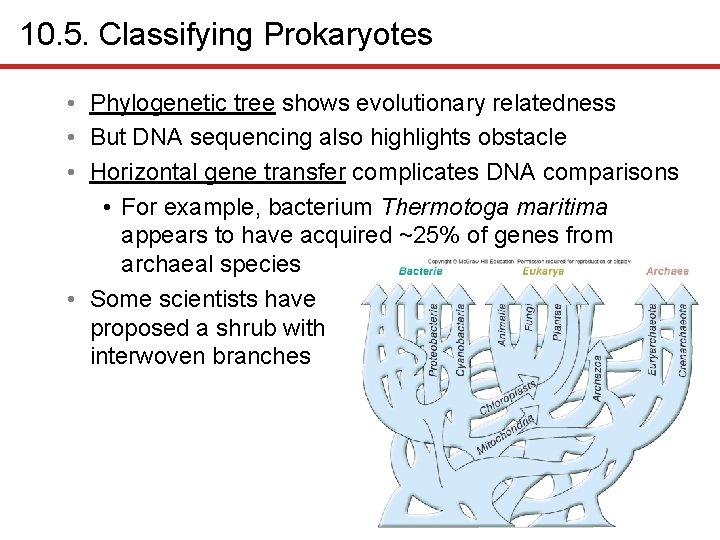
10. 5. Classifying Prokaryotes • Phylogenetic tree shows evolutionary relatedness • But DNA sequencing also highlights obstacle • Horizontal gene transfer complicates DNA comparisons • For example, bacterium Thermotoga maritima appears to have acquired ~25% of genes from archaeal species • Some scientists have proposed a shrub with interwoven branches

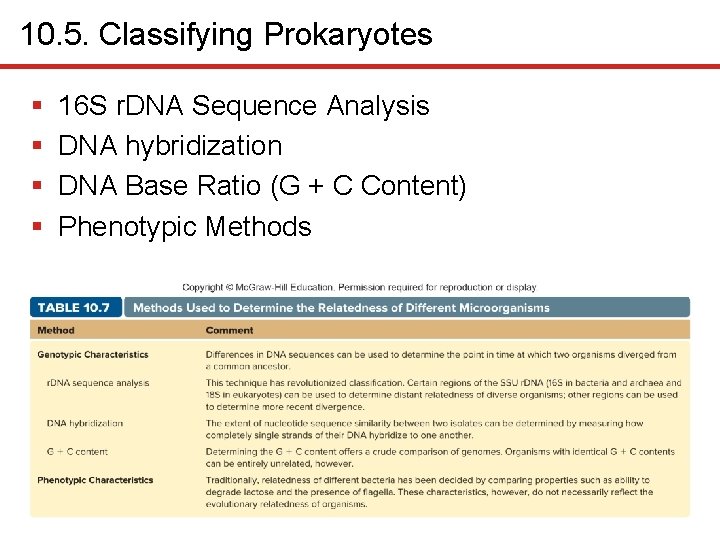
10. 5. Classifying Prokaryotes § § 16 S r. DNA Sequence Analysis DNA hybridization DNA Base Ratio (G + C Content) Phenotypic Methods

10. 5. Classifying Prokaryotes § 16 S r. DNA Sequence Analysis • Comparisons revolutionized classification • Sequences highly conserved since function critical • Lack of mutations allows identification of distant relatedness • Certain regions relatively variable, can determine recent divergence • Horizontal gene transfer appears rare • Culturing not necessary • May not resolve at species level since closely related prokaryotes can have identical 16 S r. DNA sequences – DNA hybridization a better tool in these cases

10. 5. Classifying Prokaryotes § DNA Hybridization • Relatedness of organisms can be determined by similarity of nucleotide sequences • Sequence homology measured by DNA hybridization • Extent of hybridization reflects degree of similarity • Complementary base pairing of single strands • If high percentage, considered related – 70% similarity often considered same species – But Shigella and Escherichia should be grouped in same species based on DNA hybridization

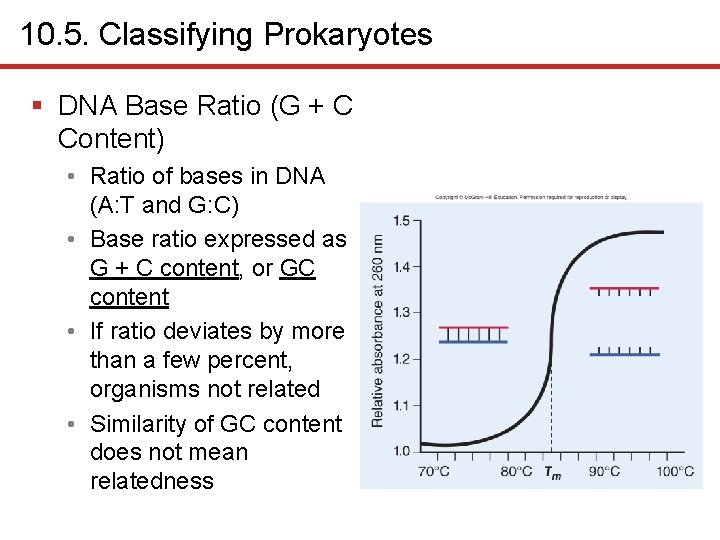
10. 5. Classifying Prokaryotes § DNA Base Ratio (G + C Content) • Ratio of bases in DNA (A: T and G: C) • Base ratio expressed as G + C content, or GC content • If ratio deviates by more than a few percent, organisms not related • Similarity of GC content does not mean relatedness

10. 5. Classifying Prokaryotes § Phenotypic Methods • Have been largely replaced by 16 S ribosomal nucleic acid sequence methods • Some taxonomists believe classification should be based on more than just genotypic traits • Phenotypic methods still important since provide foundation for prokaryotic identification