Cell proliferation and control Prof Dr Asuman SUNGUROLU














- Slides: 33

Cell proliferation and control Prof. Dr. Asuman SUNGUROĞLU (Department of Medical Biology)

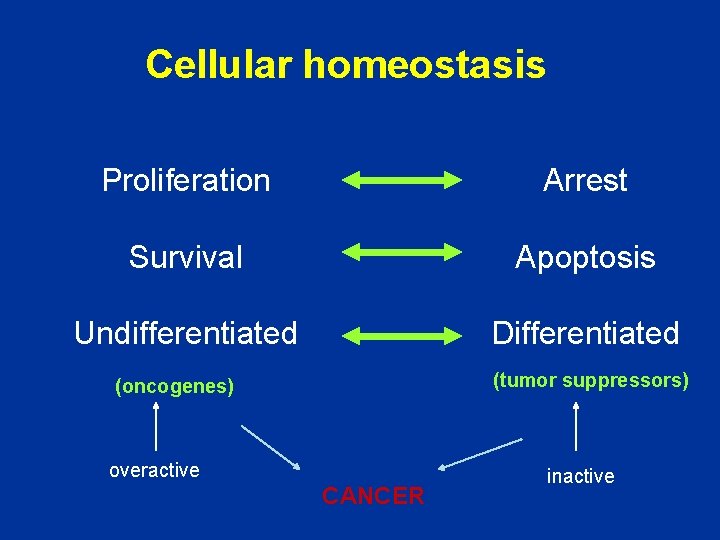
Cellular homeostasis Proliferation Arrest Survival Apoptosis Undifferentiated Differentiated (tumor suppressors) (oncogenes) overactive CANCER inactive

Disruption of homeostasis Activation of oncogenes and/or Inhibition of tumor suppressors CANCER

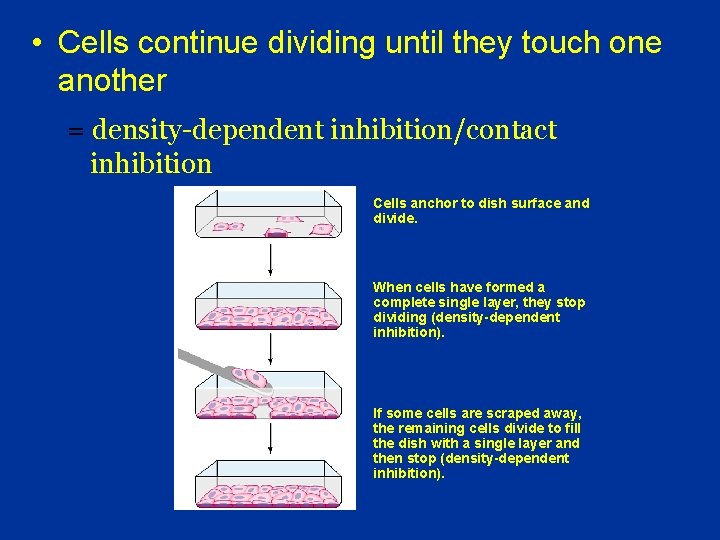
• Cells continue dividing until they touch one another = density-dependent inhibition/contact inhibition Cells anchor to dish surface and divide. When cells have formed a complete single layer, they stop dividing (density-dependent inhibition). If some cells are scraped away, the remaining cells divide to fill the dish with a single layer and then stop (density-dependent inhibition).

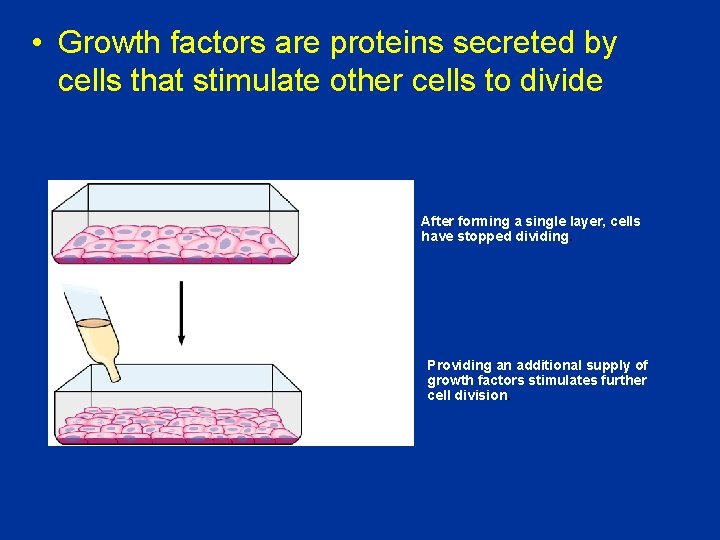
• Growth factors are proteins secreted by cells that stimulate other cells to divide After forming a single layer, cells have stopped dividing. Providing an additional supply of growth factors stimulates further cell division.

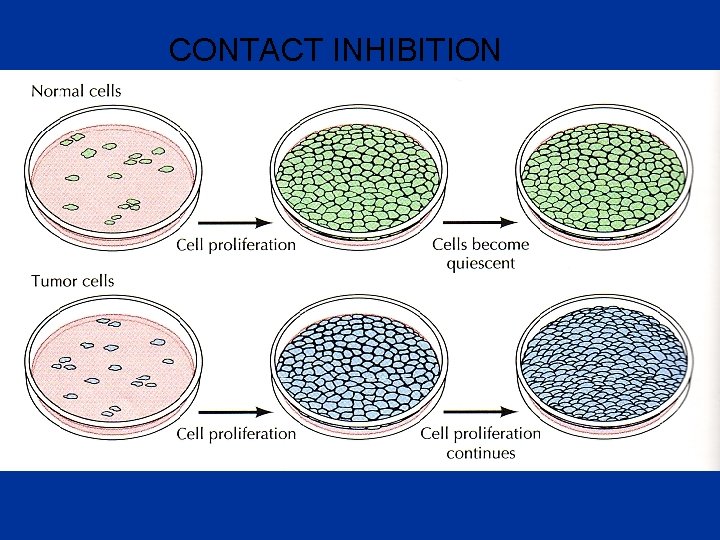
CONTACT INHIBITION

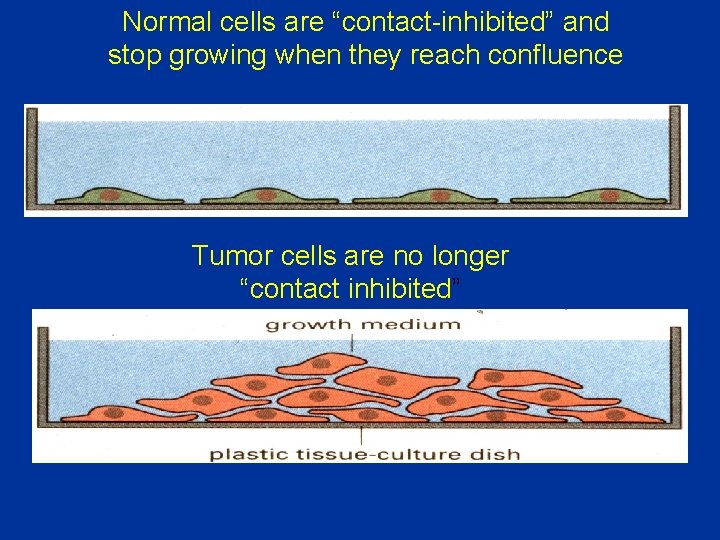
• Loss of contact inhibition by cancer cells in cell culture. • Most normal cells stop proliferating once they have carpeted the dish with a single layer of cells: proliferation seems to depend on contact with the dish, and to be inhibited by contacts with other cells—a phenomenon known as “contact inhibition. ” • Cancer cells, in contrast, usually disregard these restraints and continue to grow, so that they pile up on top of one another

Normal cells are “contact-inhibited” and stop growing when they reach confluence Tumor cells are no longer “contact inhibited”

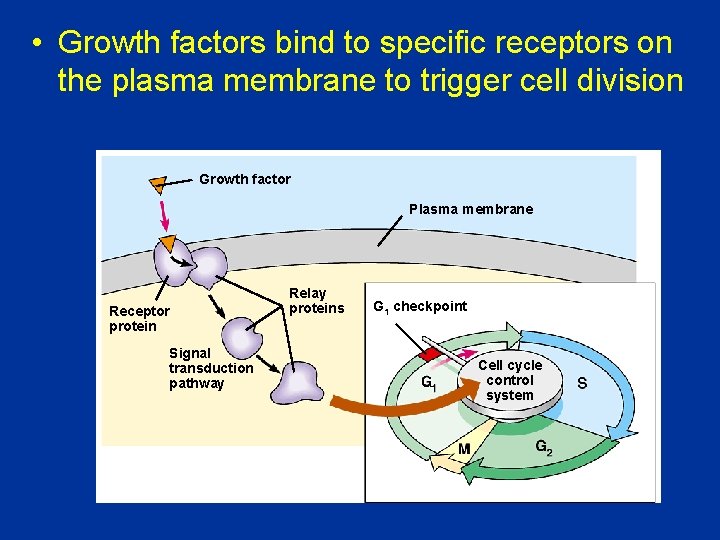
• Growth factors bind to specific receptors on the plasma membrane to trigger cell division Growth factor Plasma membrane Receptor protein Signal transduction pathway Relay proteins G 1 checkpoint Cell cycle control system

Cancer cells have • 1. Oncogene activation: self-sufficiency in growth signal • 2. Inactivation of tumor suppressor genes: insensitivity to growth-inhibitory signals • 3. Evasion of programmed cell death • 4. Defect in DNA repair: can’t maintain genomic integrity • 5. Expression of telomerase: limitless replicative potential • 6. Sustained angiogenesis • 7. Ability to invade and metastasize

Cancer cells: • keep dividing while disregarding incoming intra and extracellular signals • are indifferentiated cells • are genetically and epigenetically instable • avoid Apoptosis, • avoid replicative senecence, either by producing telomerase or by acquiring another way of stabilizing their telomeres. • capable of invasion • have metastase ability Metastase is the settlement of the primer tumor derived cells to the secondary organs by the blood cells.


• Telomerase is a RNA-dependent DNA polymerase, reverse transcriptase, an enzyme that transcribes single-stranded RNA into TTAGGG repeats and thereby elongates the telomeres (Blackburn, 2005).


• Telomerase is a ribonucleoprotein complex that maintains chromosome ends. It is a cellular reverse transcriptase composed of both RNA and proteins that employs its internal RNA component as a template for the synthesis of telomeric DNA. • It stabilizes telomere length by adding hexameric (TTAGGG)n repeats onto telomeric ends of chromosomes. After adding six bases, the enzyme is believed to pause while it repositions (translocates) the template RNA in order to synthesize the subsequent 6 bp repeat. • This extension of the 3′ DNA template end in turn allows replication of the 5′ end of the lagging strand. It thus compensates for the continued erosion of telomeres and has been referred to as a ‘cellular immortalizing enzyme. ’

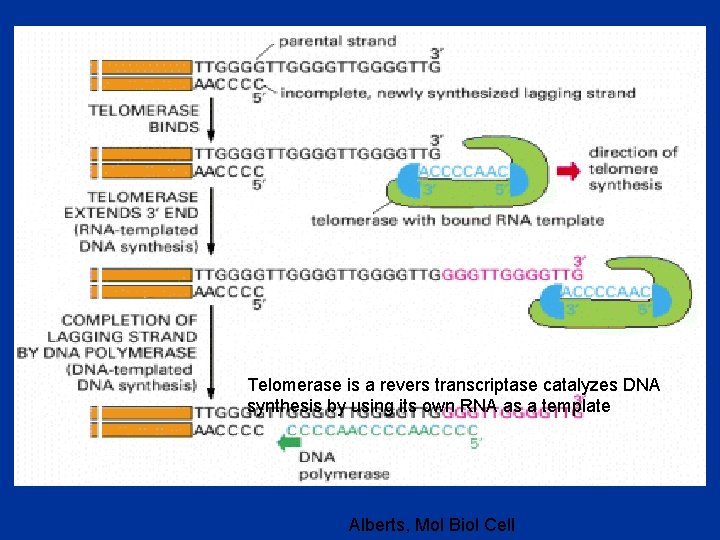
Telomerase is a revers transcriptase catalyzes DNA synthesis by using its own RNA as a template Alberts, Mol Biol Cell

Risk Factors § § § Old age Unhealthy lifestyle , Poor diet, lack of physical activity, or being overweight. Environmental factors, defined broadly to include tobacco use, diet, sunlight and infectious agents(Viruse and bacteria). Carcinogens –Alcohol-Chemicals and other substance Radiation Heredity, Family history of cancer (Genetic susceptibility)

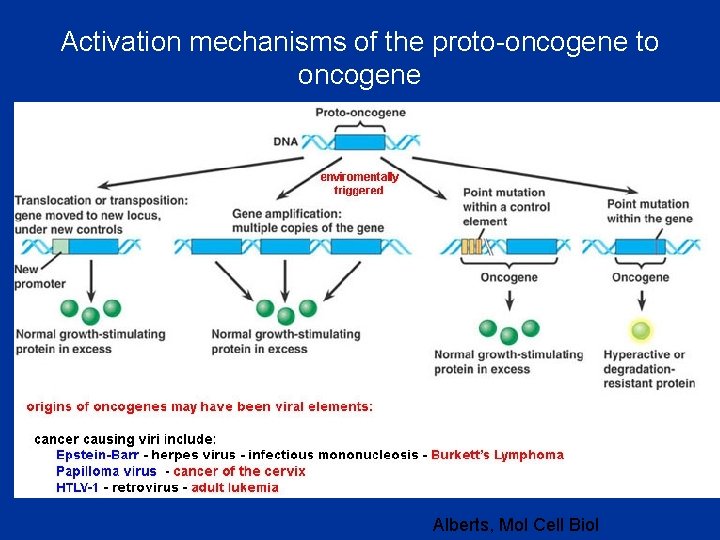
• Factors that cause genetic changes tend to provoke the development of cancer. Thus, carcinogenesis (the generation of cancer) can be linked to mutagenesis (the production of a change in the DNA sequence). This correlation is particularly clear for two classes of external agents: • (1) chemical carcinogens (which typically cause simple local changes in the nucleotide sequence), • (2) radiation such as x-rays (which typically cause chromosome breaks and translocations) or ultraviolet • (UV) light (which causes specific DNA base alterations). • - oncogenic viruses • - bacteria(Helycobacter pylori)

Tumors and Cancer -Cell division is highly controlled in normal differantiated cells and these cells remain in one location -Loss of cellular divison control leads to high cell replication -Hyperplasia(cell structure and function altered slightly, localized mass, mostly encapsulated--Benign tumor -Dysplasia(Cell strucrure and functi 9 on widely alteredinvasion and metastasis (spread to secondary organs to the body through blood vessels------Malign tumor

Tumorigenesis is a Multistep Pathway • Mutation of proto-oncogenes and tumor suppressor genes • Initiation – 1 st mutation – Increased proliferation of a single cell • Progression – Additional mutations – Selection for more aggressive cells • With increase in histopathological abnormalities, there is an increase in the number of mutations at defined genetic loci • Clonal selection!

What mutations cause cancer? 1. Anything that damages DNA *Radiation ----Physical agents *Chemical agents---Carcinogens 2. Anything that stimulates the rate of mitosis *Viruses *Oncogenes *Tumor suppressor genes

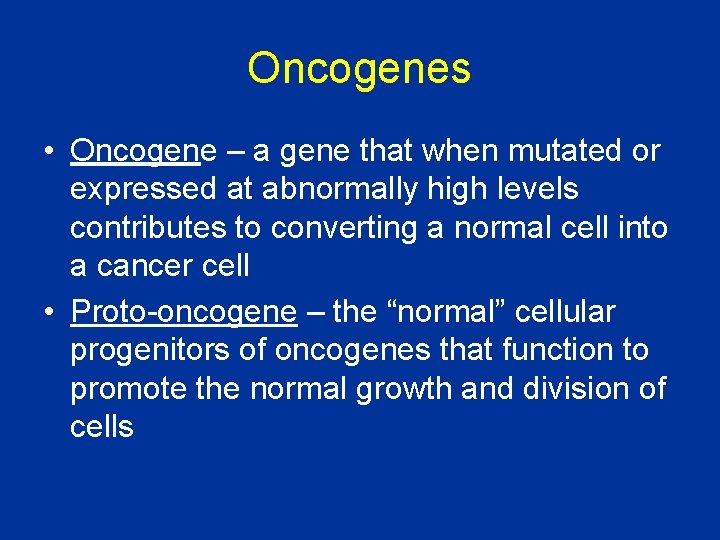
Oncogenes • Oncogene – a gene that when mutated or expressed at abnormally high levels contributes to converting a normal cell into a cancer cell • Proto-oncogene – the “normal” cellular progenitors of oncogenes that function to promote the normal growth and division of cells

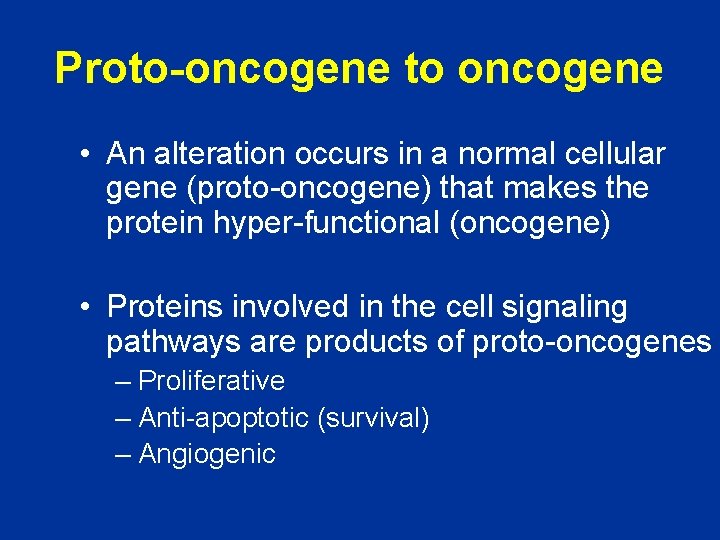
Proto-oncogene to oncogene • An alteration occurs in a normal cellular gene (proto-oncogene) that makes the protein hyper-functional (oncogene) • Proteins involved in the cell signaling pathways are products of proto-oncogenes – Proliferative – Anti-apoptotic (survival) – Angiogenic

Src – the first oncogene • • The first oncogene was discovered before we understood what oncogenes really are! • Peyton Rous, 1911 – cell-free extract from chicken tumors can cause new tumors when injected into healthy chickens • Rous Sarcoma – a form of cancer which infects chickens • Rous Sarcoma Virus (RSV) – a retrovirus which generates a c. DNA that inserts into the chickens’ DNA • v-Src – the gene which gets expressed by RSV

• Many oncogenes identified in viruses • • But… Most human cancers are not viral in origin • • Cellular versions of oncogenes = protooncogenes • Src expression/activity upregulated in: • • Breast ca • • Pancreatic ca • • Ovarian ca • • Head & Neck ca • • Lung ca • • Gastric ca

Activation mechanisms of the proto-oncogene to oncogene Alberts, Mol Cell Biol

Oncogenes • • • Growth factors Growth factor receptors G proteins Intracellular kinases Transcription factors

Tumor suppressors • Normally function to suppress the formation of cancer – Growth arrest – Apoptosis – DNA repair – Differentiation – Anti-angiogenesis

Alberts Mol Cell Biol

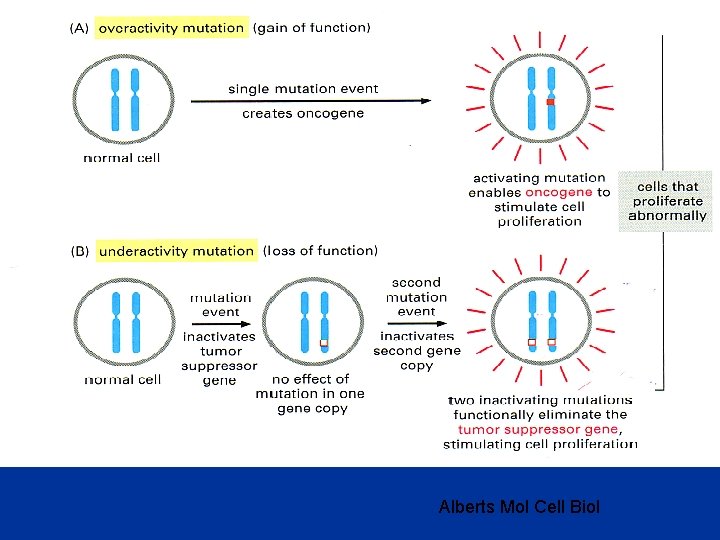
Tumor Suppressor Genes • Normal physiologic function of the wild type gene is to slow cell growth, cause cell differentiation, activate apoptosis or repair DNA. Require mutations (or silencing) of both alleles (Knudson’s two hit hypothesis). • TP 53 tumor suppressor gene TP 53 is mutated in more than half of human tumors One of its normal functions is to prevent replication of DNA which has been damaged, guardian of genome

Mechanisms of TSG inactivation • • Gene deletion • • Oncogenes activation mechanisms • Gene amplification • Insertion of powerful (viral) promoters • Activating mutations • Direct mutation • Loss of Heterozygosity • Epigenetic silencing (promoter methylation) • Promoters rich in the sequence cytosine-guanosine (Cp. G) • Cytosine gets methylated • HDAC protein complexes recognize methyl-Cp. G • HDAC removes histone acetylations • Histones form “closed” DNA conformation • turn off transcription)

Caretaker genes are involved in the maintenance of the genome stability and include genes implicated in DNA repair. CT genes, are not oncogenic, but their loss of function increases the risk of mutations in other genes • Maintain genomic stability by DNA repair or whole chromosome stability. • Caretaker genes, including oncogenes and tumor suppressor genes. (eg BRCA 1 -BRCA 2) • Gatekeeper genes inhibit cell growth or induce apoptosis. (eg p 53)

• Molecular Biology of the Cell • Alberts