Applications of Molecular Cytogenetics Dr Mohammed Alqahtani CSLTCG











- Slides: 38


Applications of Molecular Cytogenetics Dr Mohammed Alqahtani CSLT(CG), CLSp(CG), RT, MBA, Ph. D Genomic Medicine Unit Founder & Director Center of Excellence in Genomic Medicine Research Founder & Director

Lecture Objectives • Understand how molecular cytogenetic techniques can be used to identify clinically relevant chromosome abnormalities • Be aware of the different types of molecular techniques that can be used to identify and clarify chromosome rearrangements

• Molecular Cytogenetic Techniques Powerful complement to conventional cytogenetic analysis of: – aneuploidy – structural rearrangements – submicroscopic rearrangements • microdeletions/duplications • subtelomere rearrangements

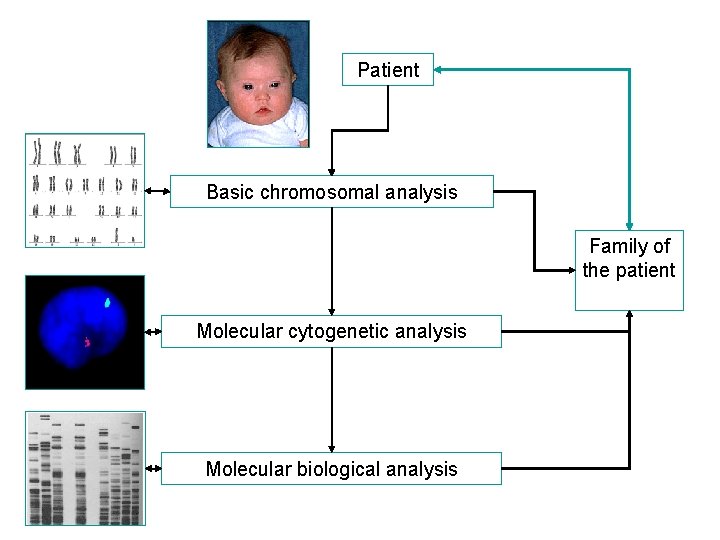
Patient Basic chromosomal analysis Family of the patient Molecular cytogenetic analysis Molecular biological analysis

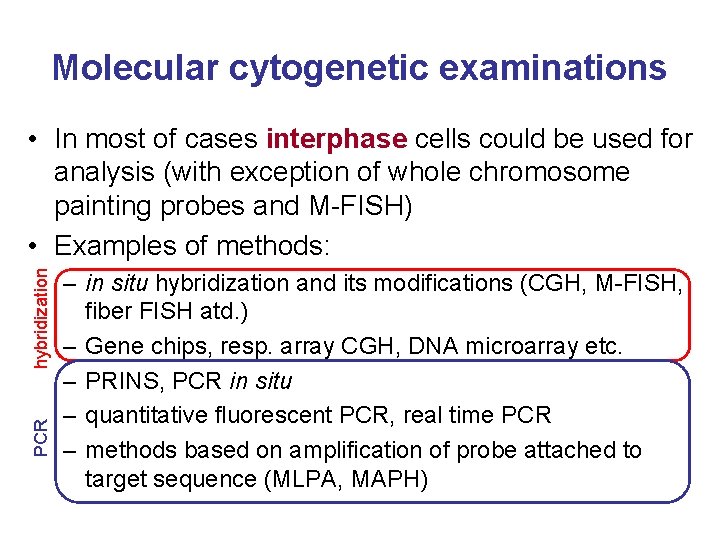
Molecular cytogenetic examinations PCR hybridization • In most of cases interphase cells could be used for analysis (with exception of whole chromosome painting probes and M-FISH) • Examples of methods: – in situ hybridization and its modifications (CGH, M-FISH, fiber FISH atd. ) – Gene chips, resp. array CGH, DNA microarray etc. – PRINS, PCR in situ – quantitative fluorescent PCR, real time PCR – methods based on amplification of probe attached to target sequence (MLPA, MAPH)

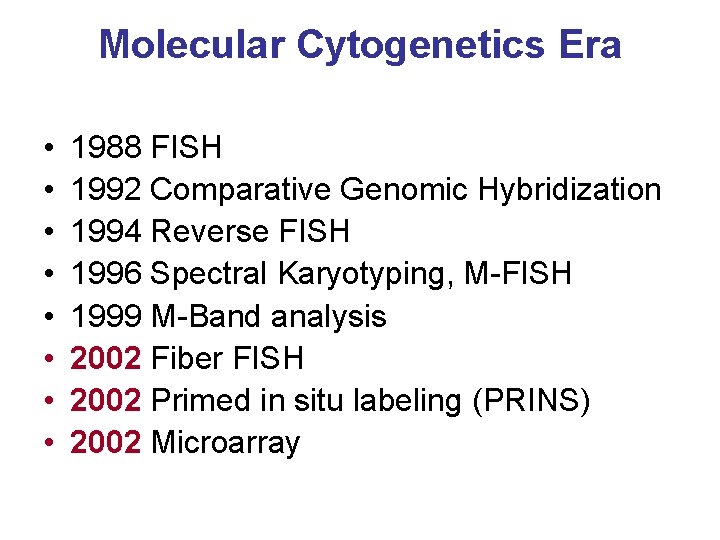
Molecular Cytogenetics Era • • 1988 FISH 1992 Comparative Genomic Hybridization 1994 Reverse FISH 1996 Spectral Karyotyping, M-FISH 1999 M-Band analysis 2002 Fiber FISH 2002 Primed in situ labeling (PRINS) 2002 Microarray

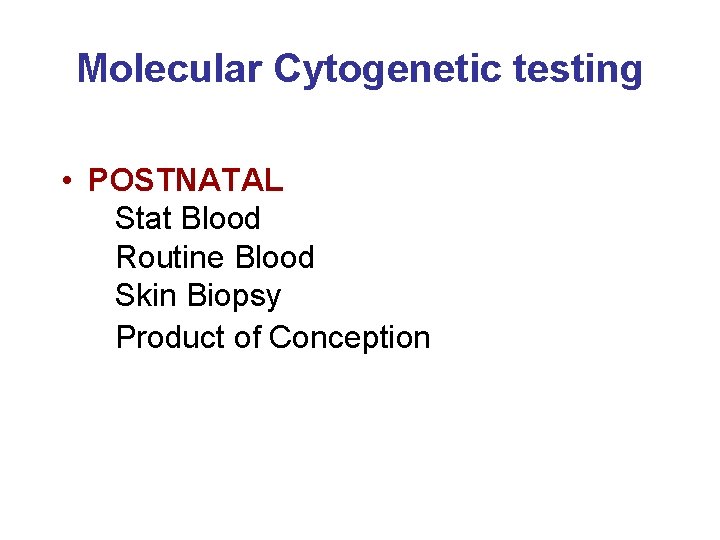
Molecular Cytogenetic testing • POSTNATAL Stat Blood Routine Blood Skin Biopsy Product of Conception

• PRENATAL Amniotic Fluid Chorionic Villus Sampling Fetal Cord Blood • CANCER GENETICS Bone Marrow Oncology Blood Solid Tumor Lymph Node Pleural Effusion Core Biopsy

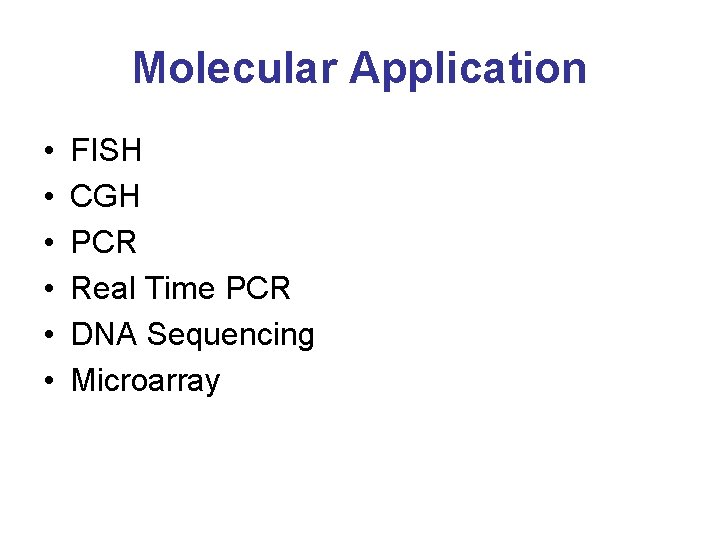
Molecular Application • • • FISH CGH PCR Real Time PCR DNA Sequencing Microarray

Fluorescence In Situ Hybridization (FISH)

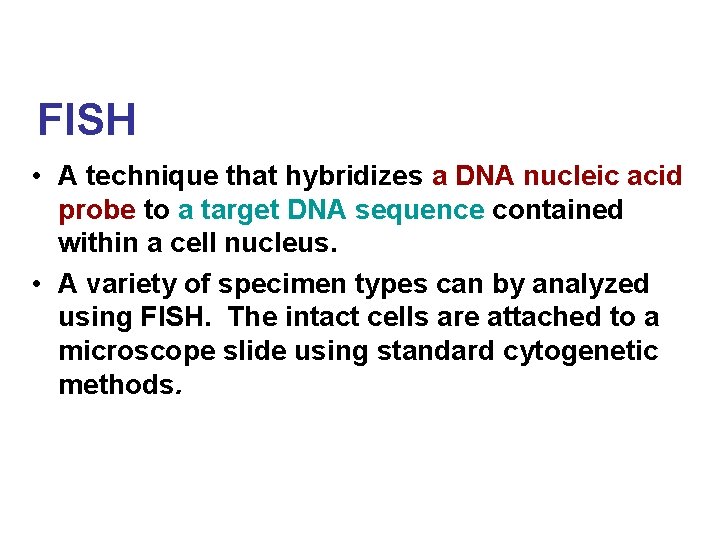
FISH • A technique that hybridizes a DNA nucleic acid probe to a target DNA sequence contained within a cell nucleus. • A variety of specimen types can by analyzed using FISH. The intact cells are attached to a microscope slide using standard cytogenetic methods.

(FISH) TO RULE OUT: Ø Chromosome Microdeletion Detection Ø Interphase Chromosome Enumeration Ø Gene Rearrangements (ie, bcr/abl, PML/RARA) Ø Cryptic Chromosomal Rearrangements Ø Marker Chromosome Identification Ø Chromosome Breakpoint Mapping

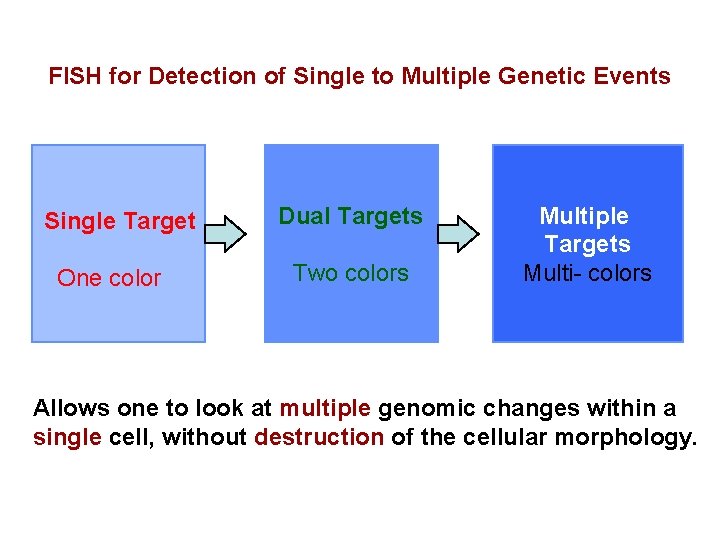
FISH for Detection of Single to Multiple Genetic Events Single Target One color Dual Targets Two colors Multiple Targets Multi- colors Allows one to look at multiple genomic changes within a single cell, without destruction of the cellular morphology.

Probes • Probe is a nucleic acid that – can be labeled with a marker which allows identification and quantitation – will hybridize to another nucleic acid on the basis of base complementarity

Probes Types of labeling • Direct & Indirect • Radioactive (32 P, 35 S, 14 C, 3 H) • Fluorescent • FISH: fluorescent in situ hybridization • Biotinylated (avidin-streptavidin)

Probe • A part of DNA (or RNA) that is complementary to certain sequence on target DNA (i. e. DNA of the patient) • Plasmid, phage DNA, cosmid (or combination of phage and plasmid DNA), YAC • PCR-product (amplification of certain segment of chromosomal DNA)

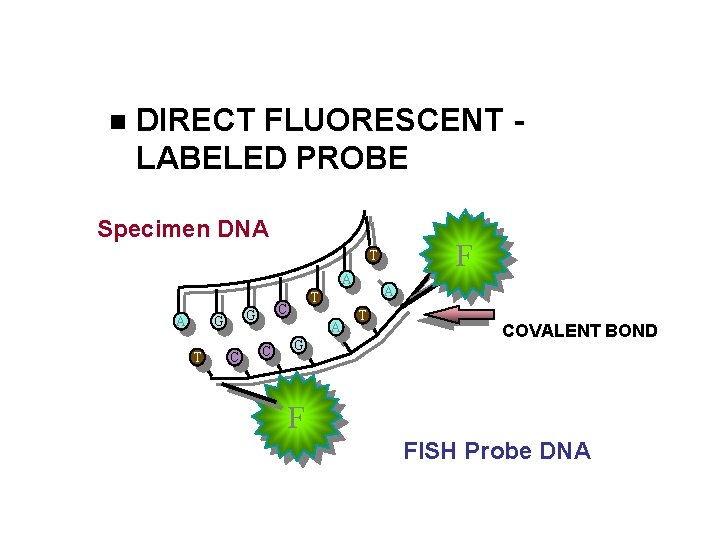
DIRECT FLUORESCENT LABELED PROBE Specimen DNA F T A A T C G G C A T G T COVALENT BOND F FISH Probe DNA


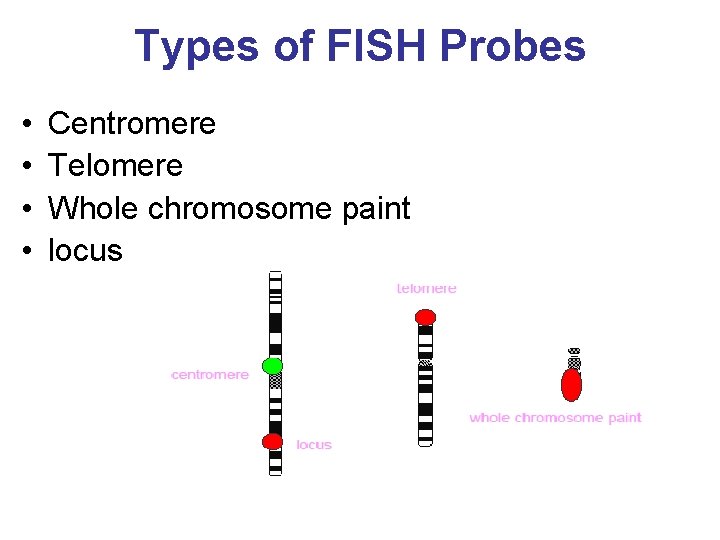
Types of FISH Probes • • Centromere Telomere Whole chromosome paint locus

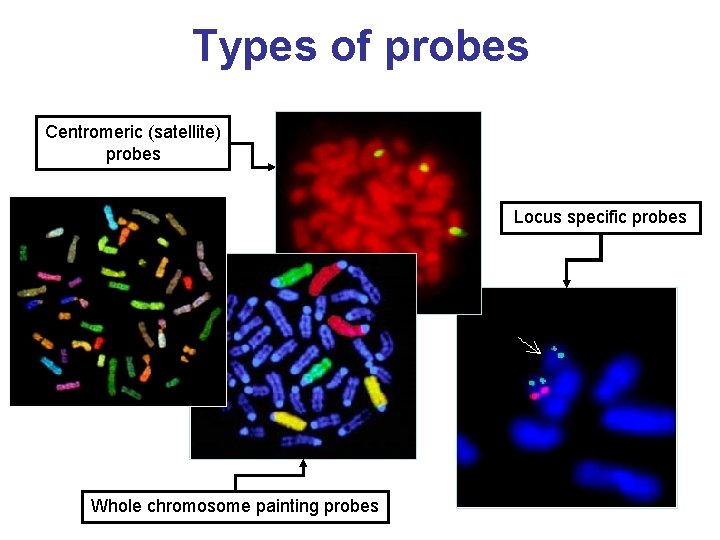
Types of probes Centromeric (satellite) probes Locus specific probes Whole chromosome painting probes

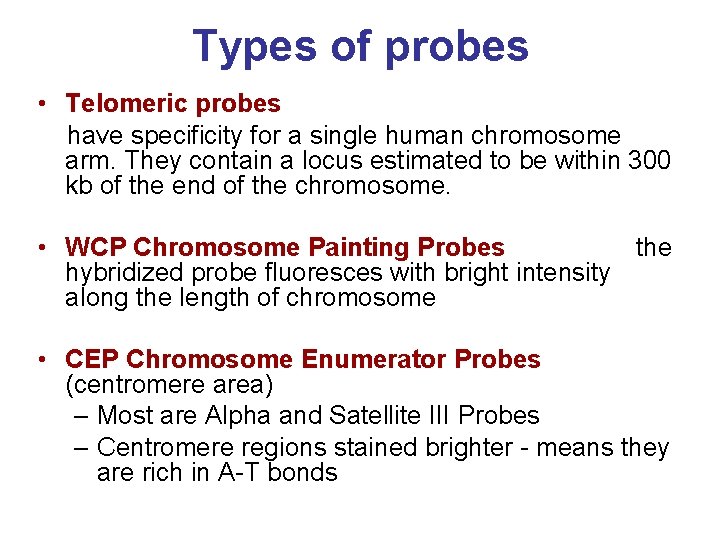
Types of probes • Telomeric probes have specificity for a single human chromosome arm. They contain a locus estimated to be within 300 kb of the end of the chromosome. • WCP Chromosome Painting Probes the hybridized probe fluoresces with bright intensity along the length of chromosome • CEP Chromosome Enumerator Probes (centromere area) – Most are Alpha and Satellite III Probes – Centromere regions stained brighter - means they are rich in A-T bonds

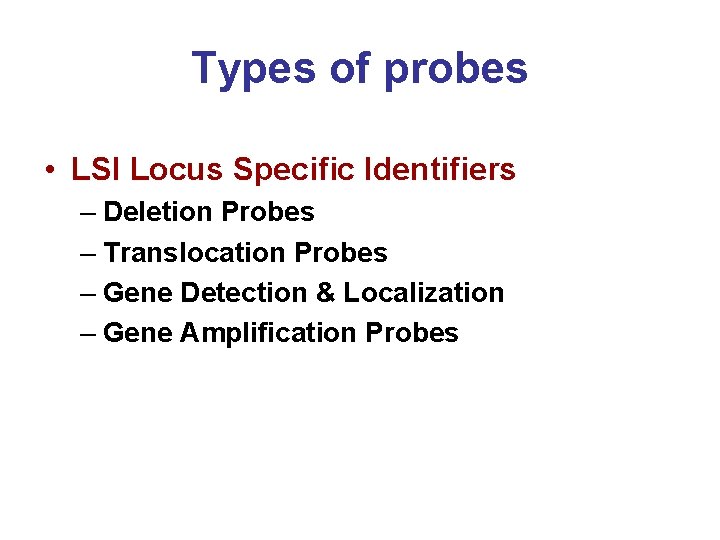
Types of probes • LSI Locus Specific Identifiers – Deletion Probes – Translocation Probes – Gene Detection & Localization – Gene Amplification Probes

In which conditions we have to indicate FISH analysis? • The material doesn't contain metaphase chromosomes – Unsuccessful cultivation – It isn't possible to cultivate the tissue from patient (preimplantation analysis, rapid prenatal examinations, examinations of solid tumors or autopsy material) • Analysis of complicated chromosomal rearrangements

In which conditions we have to indicate FISH analysis? • Identification of marker chromosomes • Analysis of low-frequency mosaic • Diagnosis of submicroscopic (cryptic) chromosomal rearrangements – Microdeletion syndromes – Amplification of oncogenes and microdeletion of tumor-suppressor genes in malignancies

Multi Color FISH • Multicolor FISH can provide “colorized” information relative to chromosome rearrangements, especially useful in specimens where chromosome preparations are less than optimal for standard cytogenetic banding analysis.


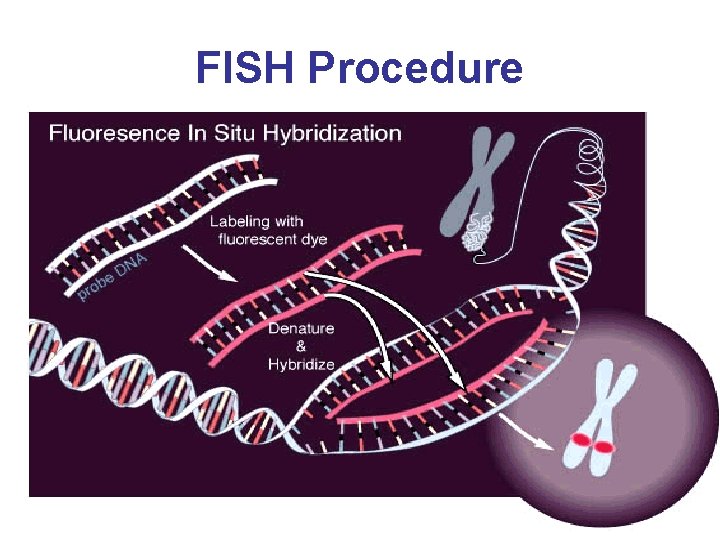
FISH Procedure • • • Denature the chromosomes Denature the probe Hybridization Fluorescence staining Examine slides or store in the dark

FISH Procedure

Direct Label FISH Technology

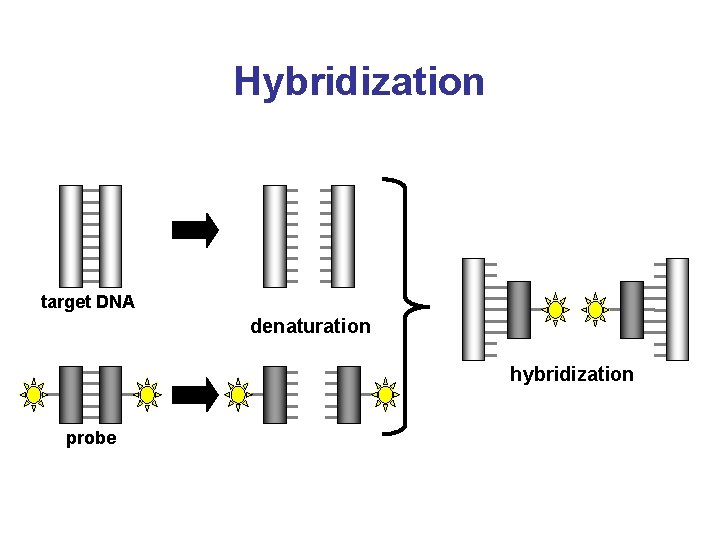
Hybridization target DNA denaturation hybridization probe

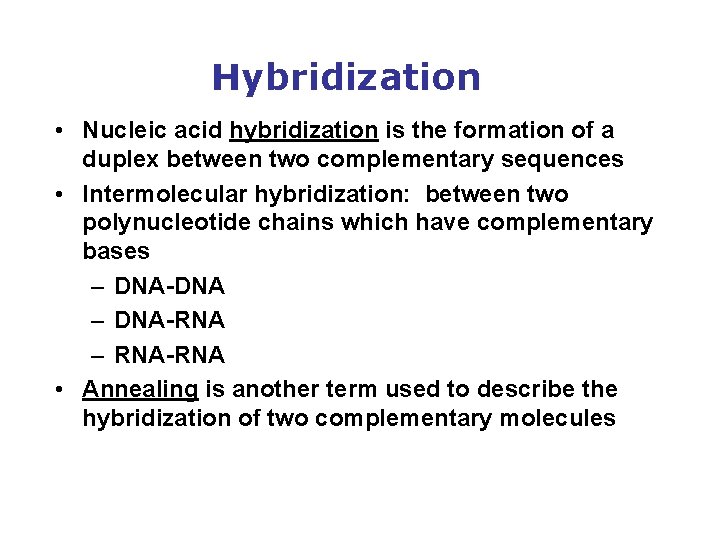
Hybridization • Nucleic acid hybridization is the formation of a duplex between two complementary sequences • Intermolecular hybridization: between two polynucleotide chains which have complementary bases – DNA-DNA – DNA-RNA – RNA-RNA • Annealing is another term used to describe the hybridization of two complementary molecules

Automated Hybridization HYBrite™ • The probe and target DNA are denatured together. • Faster, easier, and safer hybridization.


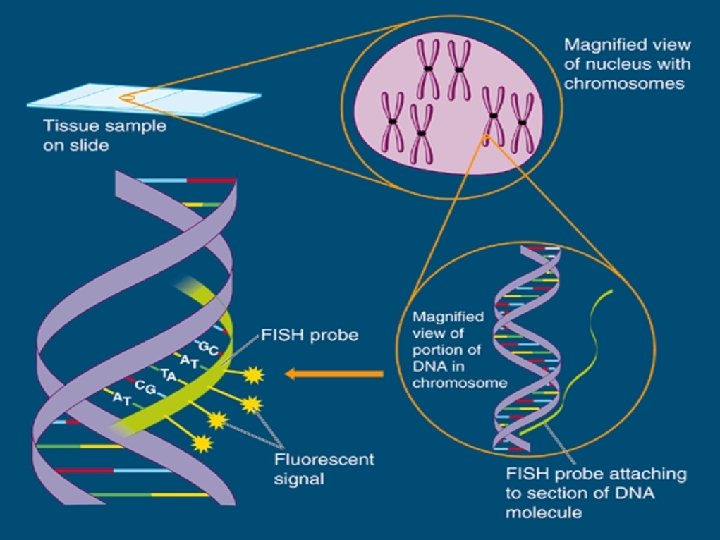
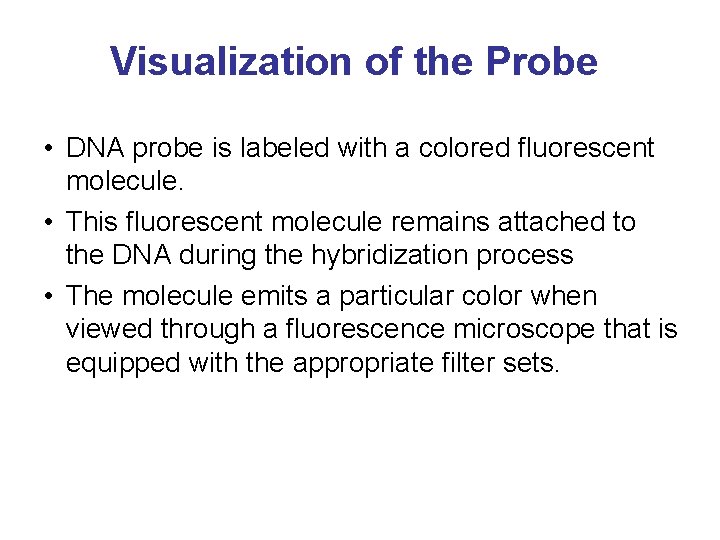
Visualization of the Probe • DNA probe is labeled with a colored fluorescent molecule. • This fluorescent molecule remains attached to the DNA during the hybridization process • The molecule emits a particular color when viewed through a fluorescence microscope that is equipped with the appropriate filter sets.

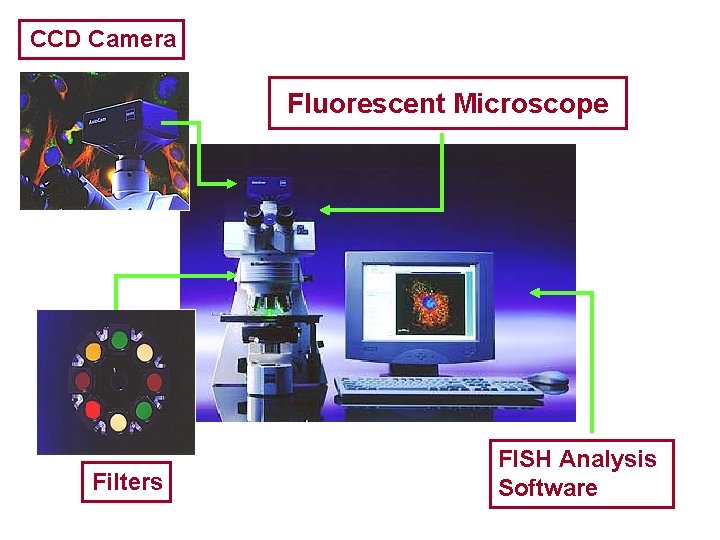
CCD Camera Fluorescent Microscope Filters FISH Analysis Software


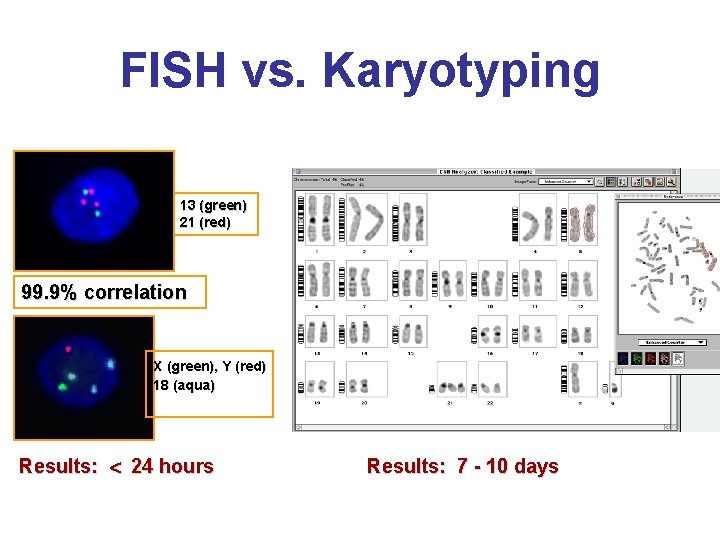
FISH vs. Karyotyping 13 (green) 21 (red) 99. 9% correlation X (green), Y (red) 18 (aqua) Results: 24 hours Results: 7 - 10 days
 Omar alqahtani
Omar alqahtani Cytogenetics
Cytogenetics Cytogenetics
Cytogenetics Naacls accredited cytogenetics education program
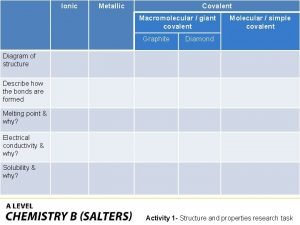
Naacls accredited cytogenetics education program Ionic covalent metallic
Ionic covalent metallic Zinc oxide + nitric acid → zinc nitrate + water
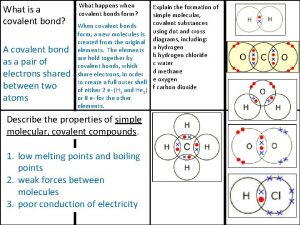
Zinc oxide + nitric acid → zinc nitrate + water Covalent bond melting point
Covalent bond melting point T tubules
T tubules Mohammed el husseiny
Mohammed el husseiny Dr mohammed tarrabain
Dr mohammed tarrabain Dr samah mohammed
Dr samah mohammed Mohammed yusuf (boko haram)
Mohammed yusuf (boko haram) Professor mohammed arif
Professor mohammed arif Auditory pathway ecolima
Auditory pathway ecolima Mohammed mona md
Mohammed mona md Mohammed
Mohammed Faisal mohammed
Faisal mohammed Dr reshma mohammed
Dr reshma mohammed Mohammed al kamali
Mohammed al kamali Natisod in english
Natisod in english Mohammed airaj
Mohammed airaj Dr mohammed shaker
Dr mohammed shaker Armoghan mohammed
Armoghan mohammed Mohammed j zaki
Mohammed j zaki Dr samah mohammed
Dr samah mohammed Prof mohammed arif
Prof mohammed arif Pneumococci
Pneumococci Prophet soliman
Prophet soliman Dr mohammed shaker
Dr mohammed shaker Dr mohammed ahmed
Dr mohammed ahmed Public ralations
Public ralations Rahaf mohammed video
Rahaf mohammed video Mohammed ashfaq ahmed
Mohammed ashfaq ahmed Aleem mohammed
Aleem mohammed Mohammed's scimitar
Mohammed's scimitar Dr samah mohammed
Dr samah mohammed Mohammed airaj
Mohammed airaj Mohammed aledhari
Mohammed aledhari Dr mohammed ahmed
Dr mohammed ahmed