Supervised and unsupervised analysis of gene expression data




- Slides: 24

Supervised and unsupervised analysis of gene expression data Bing Zhang Department of Biomedical Informatics Vanderbilt University bing. zhang@vanderbilt. edu

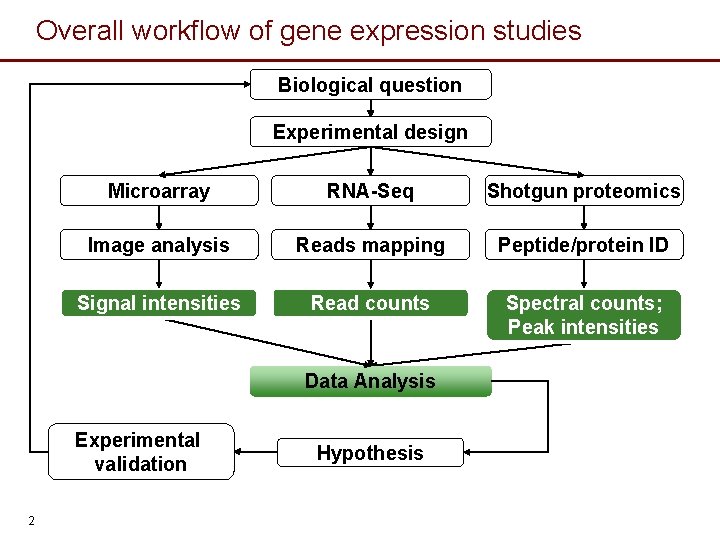
Overall workflow of gene expression studies Biological question Experimental design Microarray RNA-Seq Shotgun proteomics Image analysis Reads mapping Peptide/protein ID Signal intensities Read counts Spectral counts; Peak intensities Data Analysis Experimental validation 2 Hypothesis

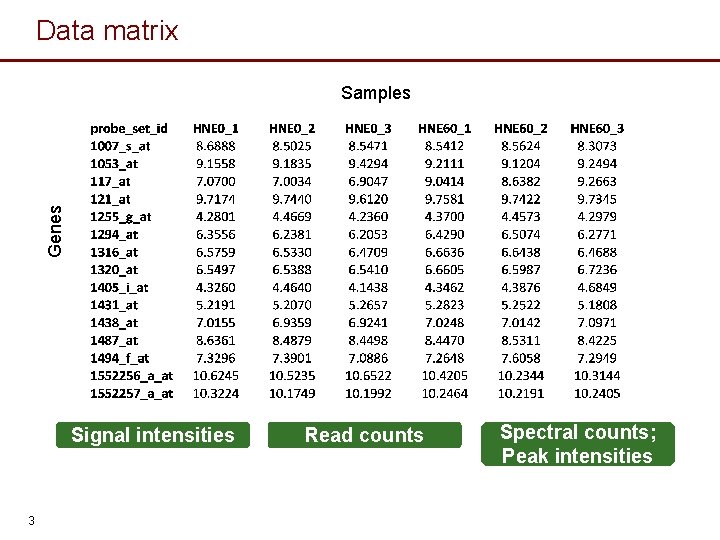
Data matrix Genes Samples Signal intensities 3 Read counts Spectral counts; Peak intensities

Three major goals of gene expression studies n n n 4 Differential expression (supervised analysis) q Input: gene expression data, class label of the samples q Output: differentially expressed genes q e. g. disease biomarker discovery Clustering (unsupervised analysis) q Input: gene expression data q Output: groups of similar samples or genes q e. g. disease subtype identification Classification (machine learning) q Input: gene expression data, class label of the samples (training data) q Output: prediction model q e. g. disease diagnosis and prognosis

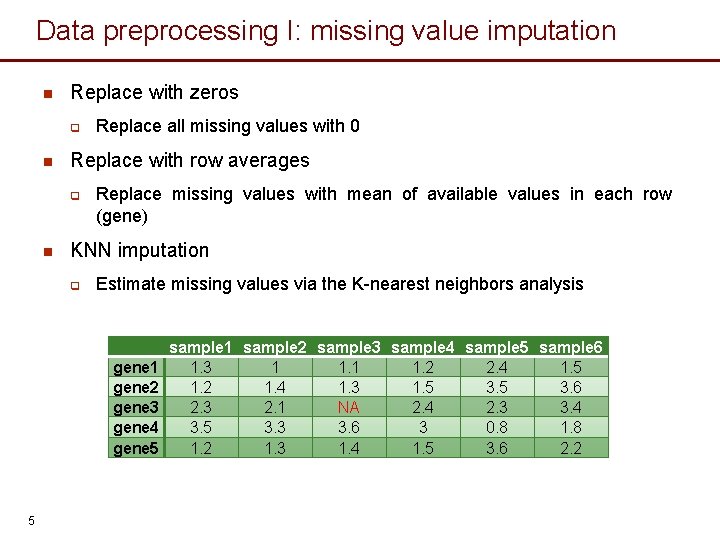
Data preprocessing I: missing value imputation n Replace with zeros q n Replace with row averages q n Replace all missing values with 0 Replace missing values with mean of available values in each row (gene) KNN imputation q Estimate missing values via the K-nearest neighbors analysis gene 1 gene 2 gene 3 gene 4 gene 5 5 sample 1 sample 2 sample 3 sample 4 sample 5 sample 6 1. 3 1 1. 2 2. 4 1. 5 1. 2 1. 4 1. 3 1. 5 3. 6 2. 3 2. 1 NA 2. 4 2. 3 3. 4 3. 5 3. 3 3. 6 3 0. 8 1. 2 1. 3 1. 4 1. 5 3. 6 2. 2

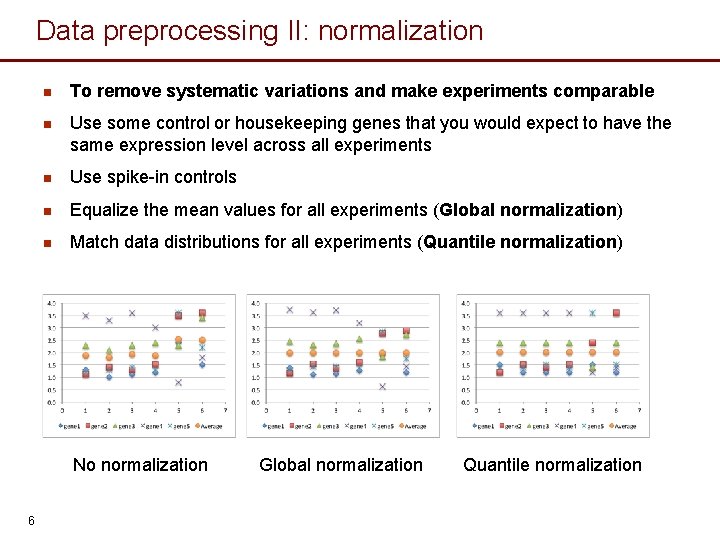
Data preprocessing II: normalization n To remove systematic variations and make experiments comparable n Use some control or housekeeping genes that you would expect to have the same expression level across all experiments n Use spike-in controls n Equalize the mean values for all experiments (Global normalization) n Match data distributions for all experiments (Quantile normalization) No normalization 6 Global normalization Quantile normalization

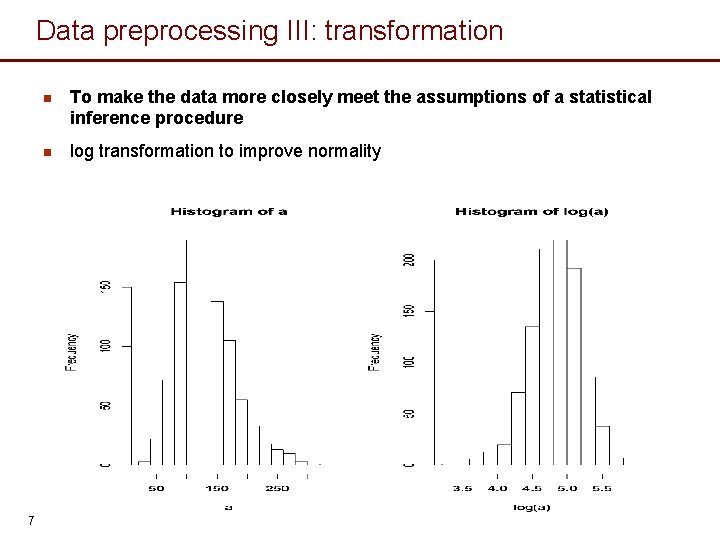
Data preprocessing III: transformation 7 n To make the data more closely meet the assumptions of a statistical inference procedure n log transformation to improve normality

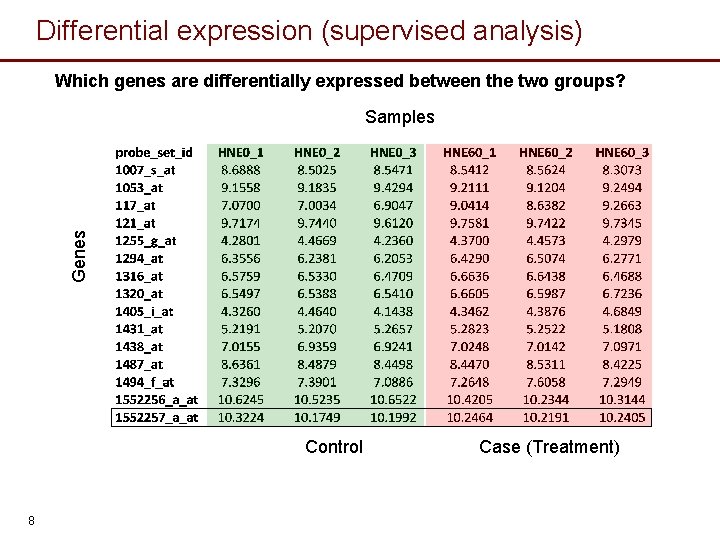
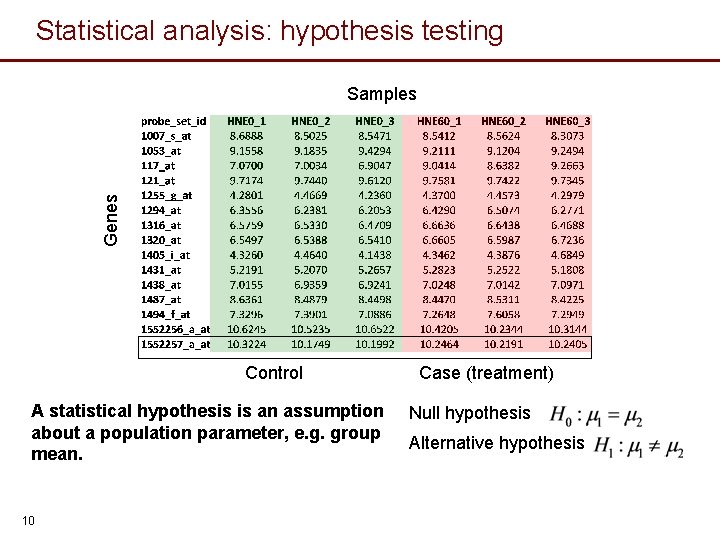
Differential expression (supervised analysis) Which genes are differentially expressed between the two groups? Genes Samples Control 8 Case (Treatment)

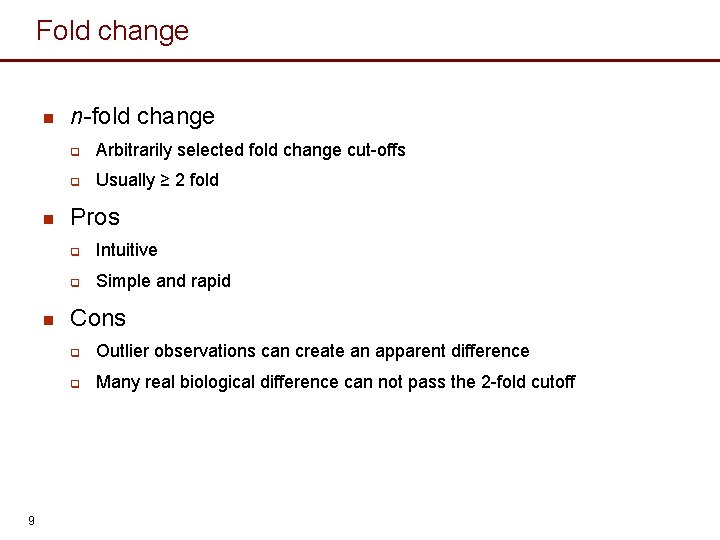
Fold change n n n 9 n-fold change q Arbitrarily selected fold change cut-offs q Usually ≥ 2 fold Pros q Intuitive q Simple and rapid Cons q Outlier observations can create an apparent difference q Many real biological difference can not pass the 2 -fold cutoff

Statistical analysis: hypothesis testing Genes Samples Control A statistical hypothesis is an assumption about a population parameter, e. g. group mean. 10 Case (treatment) Null hypothesis Alternative hypothesis

t-test graph courtesy of www. socialresearchmethods. net 11

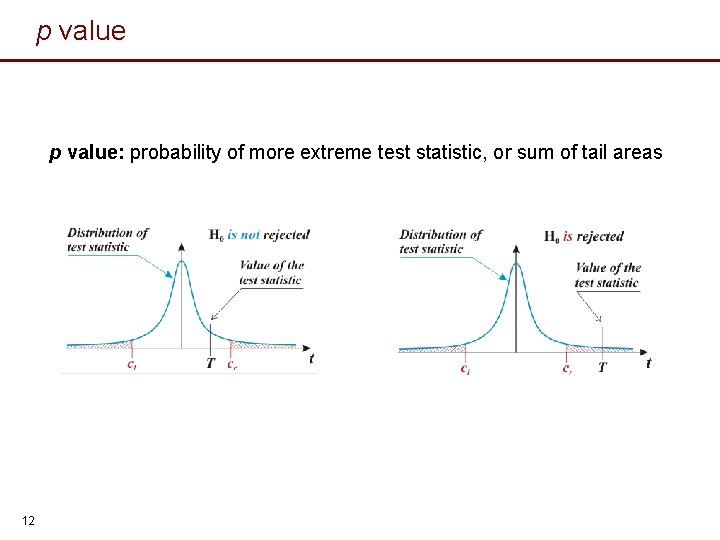
p value: probability of more extreme test statistic, or sum of tail areas 12

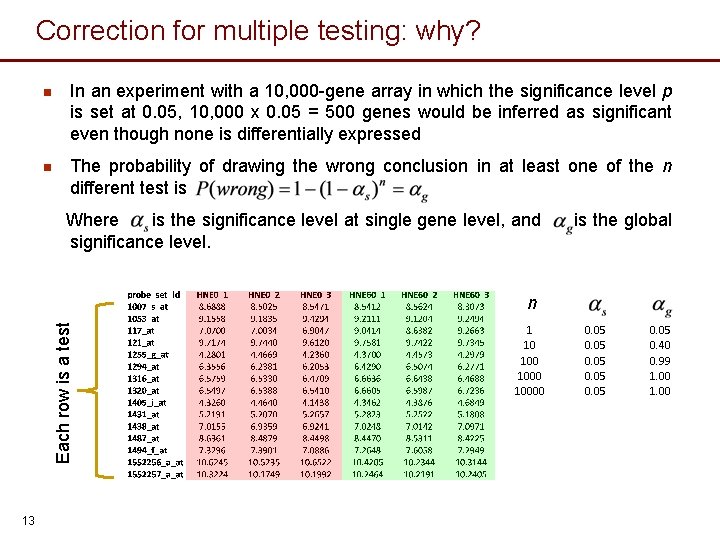
Correction for multiple testing: why? n In an experiment with a 10, 000 -gene array in which the significance level p is set at 0. 05, 10, 000 x 0. 05 = 500 genes would be inferred as significant even though none is differentially expressed n The probability of drawing the wrong conclusion in at least one of the n different test is Where is the significance level at single gene level, and significance level. is the global Each row is a test n 13 1 10 10000 0. 05 0. 40 0. 99 1. 00

Correction for multiple testing: how? n Control the family-wise error rate (FWER), the probability that there is a single type I error in the entire set (family) of hypotheses tested. e. g. Standard Bonferroni Correction: uncorrected p value x no. of genes tested n Control the false discovery rate (FDR), the expected proportion of false positives among the number of rejected hypotheses. e. g. Benjamini and Hochberg correction. q Ranking all genes according to their p value q Picking a desired FDR level, q (e. g. 5%) q Starting from the top of the list, accept all genes with , where i is the number of genes accepted so far, and m is the total number of genes tested. p 0. 00003 0. 00004 0. 0003 0. 0008 0. 002 0. 01 0. 049 0. 23 0. 55 0. 92 14 Bonferroni 0. 0003 0. 0004 0. 003 0. 008 0. 02 0. 1 0. 49 1 1 1 Rank (i) 1 2 3 4 5 6 7 8 9 10 q 0. 05 0. 05 (i/m)*q 0. 0050 0. 0100 0. 0150 0. 0200 0. 0250 0. 0300 0. 0350 0. 0400 0. 0450 0. 0500 significant? 1 1 1 0 0

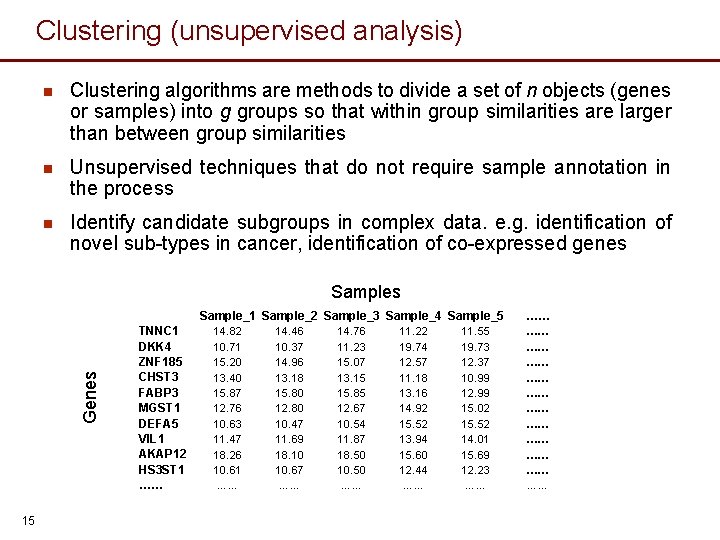
Clustering (unsupervised analysis) n Clustering algorithms are methods to divide a set of n objects (genes or samples) into g groups so that within group similarities are larger than between group similarities n Unsupervised techniques that do not require sample annotation in the process n Identify candidate subgroups in complex data. e. g. identification of novel sub-types in cancer, identification of co-expressed genes Samples Genes Sample_1 Sample_2 Sample_3 Sample_4 Sample_5 15 TNNC 1 DKK 4 ZNF 185 CHST 3 FABP 3 MGST 1 DEFA 5 VIL 1 AKAP 12 HS 3 ST 1 …… 14. 82 10. 71 15. 20 13. 40 15. 87 12. 76 10. 63 11. 47 18. 26 10. 61 …… 14. 46 10. 37 14. 96 13. 18 15. 80 12. 80 10. 47 11. 69 18. 10 10. 67 …… 14. 76 11. 23 15. 07 13. 15 15. 85 12. 67 10. 54 11. 87 18. 50 10. 50 …… 11. 22 19. 74 12. 57 11. 18 13. 16 14. 92 15. 52 13. 94 15. 60 12. 44 …… 11. 55 19. 73 12. 37 10. 99 12. 99 15. 02 15. 52 14. 01 15. 69 12. 23 …… …… …… ……

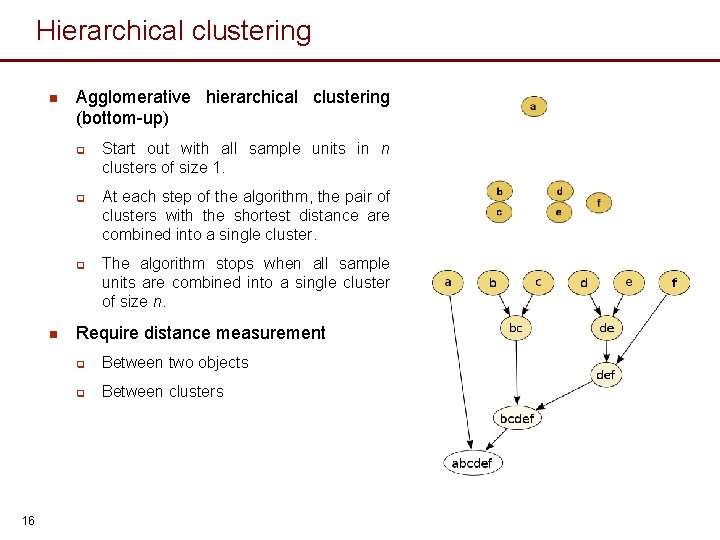
Hierarchical clustering n Agglomerative hierarchical clustering (bottom-up) q q q n 16 Start out with all sample units in n clusters of size 1. At each step of the algorithm, the pair of clusters with the shortest distance are combined into a single cluster. The algorithm stops when all sample units are combined into a single cluster of size n. Require distance measurement q Between two objects q Between clusters

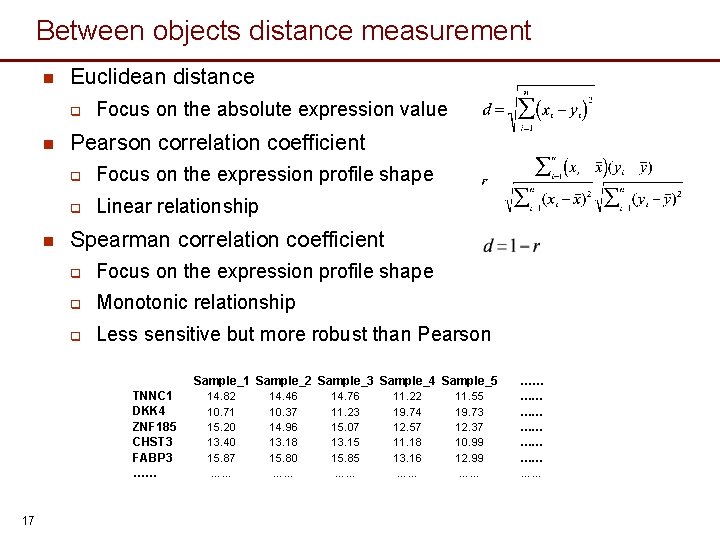
Between objects distance measurement n Euclidean distance q n n Focus on the absolute expression value Pearson correlation coefficient q Focus on the expression profile shape q Linear relationship Spearman correlation coefficient q Focus on the expression profile shape q Monotonic relationship q Less sensitive but more robust than Pearson Sample_1 Sample_2 Sample_3 Sample_4 Sample_5 TNNC 1 DKK 4 ZNF 185 CHST 3 FABP 3 …… 17 14. 82 10. 71 15. 20 13. 40 15. 87 …… 14. 46 10. 37 14. 96 13. 18 15. 80 …… 14. 76 11. 23 15. 07 13. 15 15. 85 …… 11. 22 19. 74 12. 57 11. 18 13. 16 …… 11. 55 19. 73 12. 37 10. 99 12. 99 …… ……

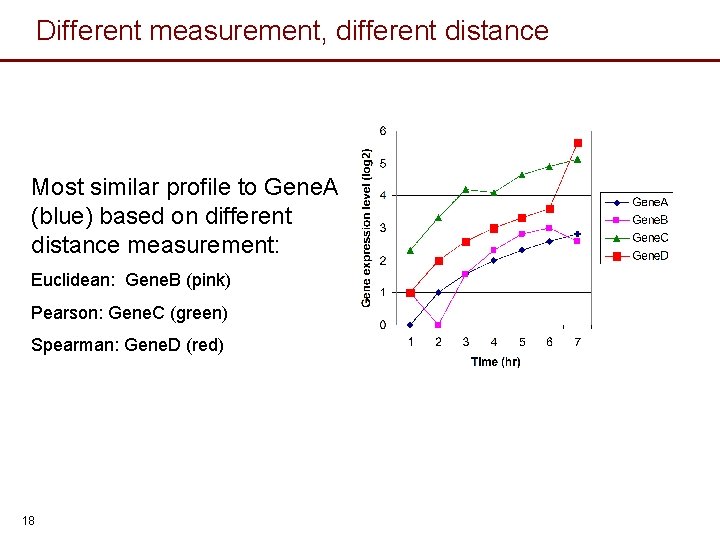
Different measurement, different distance Most similar profile to Gene. A (blue) based on different distance measurement: Euclidean: Gene. B (pink) Pearson: Gene. C (green) Spearman: Gene. D (red) 18

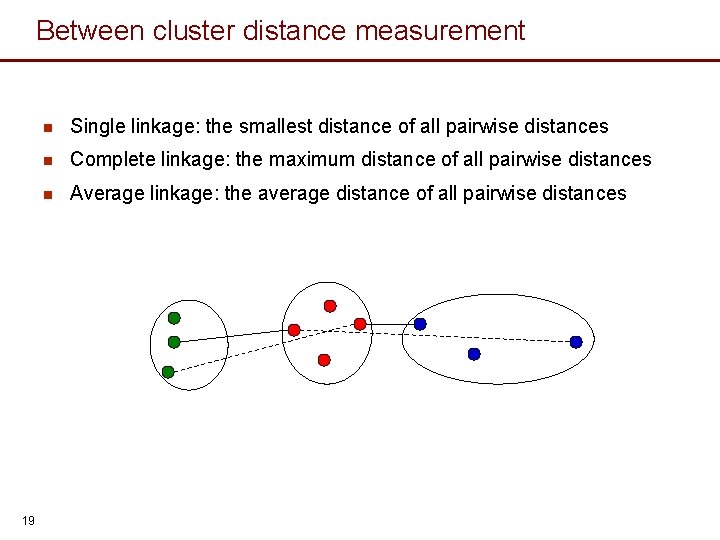
Between cluster distance measurement 19 n Single linkage: the smallest distance of all pairwise distances n Complete linkage: the maximum distance of all pairwise distances n Average linkage: the average distance of all pairwise distances

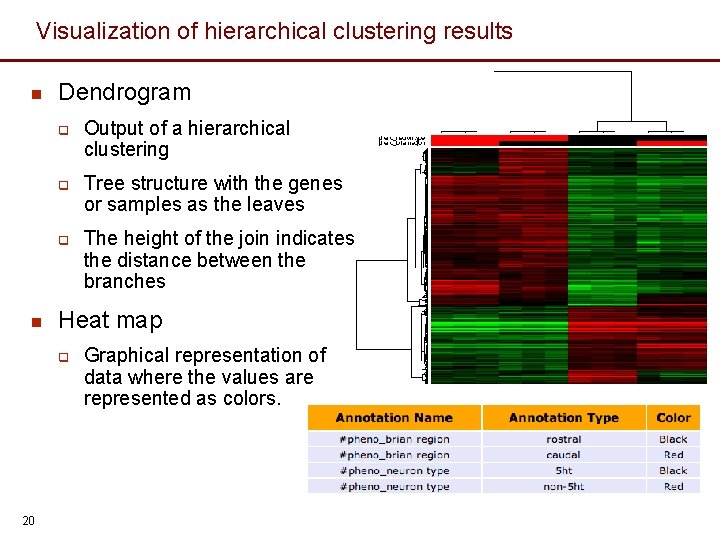
Visualization of hierarchical clustering results n Dendrogram q q q n Tree structure with the genes or samples as the leaves The height of the join indicates the distance between the branches Heat map q 20 Output of a hierarchical clustering Graphical representation of data where the values are represented as colors.

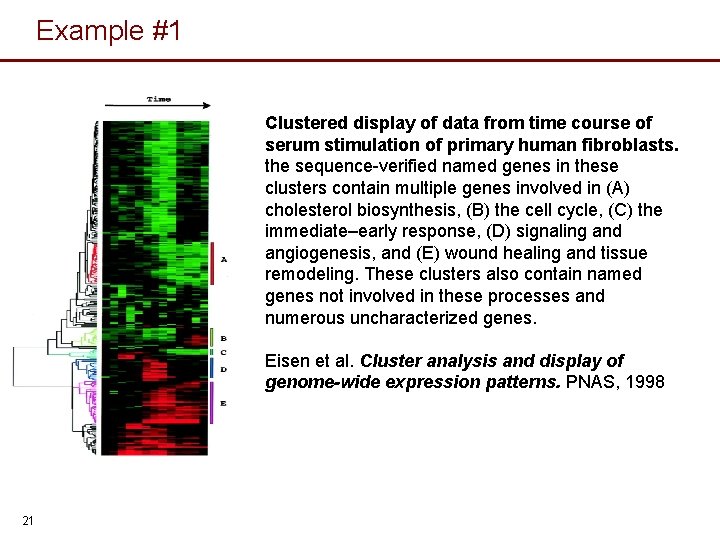
Example #1 Clustered display of data from time course of serum stimulation of primary human fibroblasts. the sequence-verified named genes in these clusters contain multiple genes involved in (A) cholesterol biosynthesis, (B) the cell cycle, (C) the immediate–early response, (D) signaling and angiogenesis, and (E) wound healing and tissue remodeling. These clusters also contain named genes not involved in these processes and numerous uncharacterized genes. Eisen et al. Cluster analysis and display of genome-wide expression patterns. PNAS, 1998 21

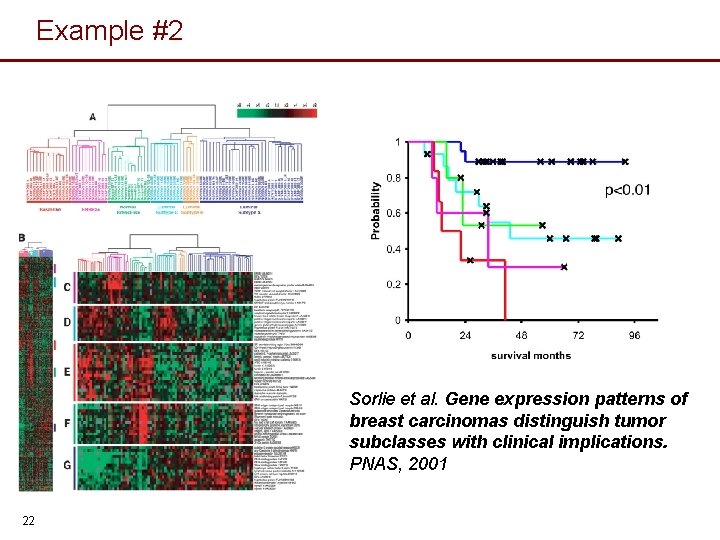
Example #2 Sorlie et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS, 2001 22

Summary n n n 23 Three major goals of gene expression studies n Agglomerative hierarchical clustering q Differential expression (supervised analysis) q Bottom-up q Clustering (unsupervised analysis) q Between objects distance measurement q Classification (machine learning) Gene expression data pre-processing steps q Missing data imputation q Normalization q Transformation q Differential expression analysis q Student’s t-test q Multiple-test adjustment n Control the family-wise error rate (FWER) n Control the false discovery rate (FDR) q n Euclidean distance n Pearson’s correlation coefficient n Spearman’s correlation coefficient Between cluster distance measurement n Single linkage n Complete linkage n Average linkage Visualization n Dendrogram n Heat map

Reading Sabates-Bellver et al. , Mol Cancer Res, 5(12): 1263 -1275, 2007 24