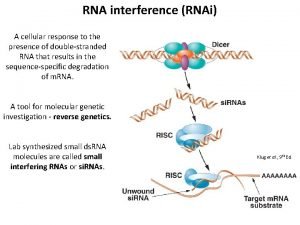
RNAi and chromatin silencing Heterochromatin condensed regions during









- Slides: 24

RNAi and chromatin silencing

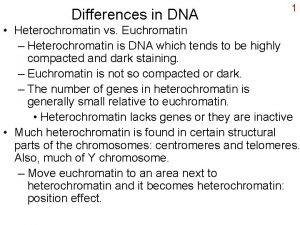
Heterochromatin - condensed regions during cell cycle (E. Heitz, 1928) - transcriptionally repressed and highly condensed structure - recombination suppression, chromosome segregation, gene silencing, - DNA methylation and histone modifications control diverse chromatin states and architectures. Recent work indicates that this highly condensed chromosomal material is transcribed, and rapidly silenced, by an orchestrated sequence of events directed by RNA interference (RNAi). transcription and noncoding RNAs

convergent view of the molecular mechanisms that underlie heterochromatin assembly - centromeric heterochromatin formation in fission yeast - RNA-directed DNA methylation in plants - X chromosome inactivation (XCI) and imprinting in mammals.



Science 297, 1833 -1837, 2002 Regulation of Heterochromatic Silencing and Histone H 3 Lysine-9 Methylation by RNAi Thomas A. Volpe, Catherine Kidner, Ira M. Hall, Grace Teng, Shiv I. S. Grewal, Robert A. Martienssen 1 Eukaryotic heterochromatin is characterized by a high density of repeats and transposons, as well as by modified histones, and influences both gene expression and chromosome segregation. In the fission yeast Schizosaccharomyces pombe, we deleted the argonaute, dicer, and RNA-dependent RNA polymerase gene homologs, which encode part of the machinery responsible for RNA interference (RNAi). Deletion results in the aberrant accumulation of complementary transcripts from centromeric heterochromatic repeats. This is accompanied by transcriptional derepression of transgenes integrated at the centromere, loss of histone H 3 lysine-9 methylation, and impairment of centromere function. We propose that doublestranded RNA arising from centromeric repeats targets formation and maintenance of heterochromatin through RNAi.

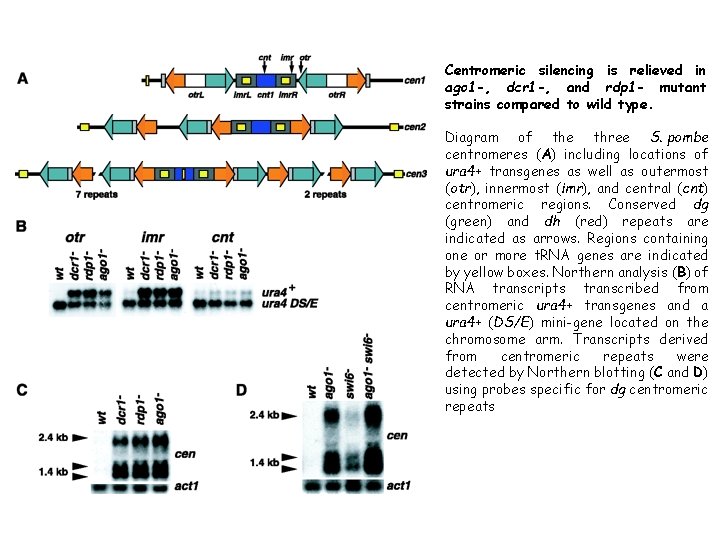
Centromeric silencing is relieved in ago 1 -, dcr 1 -, and rdp 1 - mutant strains compared to wild type. Diagram of the three S. pombe centromeres (A) including locations of ura 4+ transgenes as well as outermost (otr), innermost (imr), and central (cnt) centromeric regions. Conserved dg (green) and dh (red) repeats are indicated as arrows. Regions containing one or more t. RNA genes are indicated by yellow boxes. Northern analysis (B) of RNA transcripts transcribed from centromeric ura 4+ transgenes and a ura 4+ (DS/E) mini-gene located on the chromosome arm. Transcripts derived from centromeric repeats were detected by Northern blotting (C and D) using probes specific for dg centromeric repeats

The RNAi machinery is required for the initiation and maintenance of the heterochromatic state of centromeric repeats. Reverse strand centromeric transcription occurs in wild-type cells and is degraded posttranscriptionally by the RNAi machinery. Low-level transcription from the forward strand and/or amplification by Rdp 1 results in generation of ds. RNA, which is converted to si. RNA by RNAi. Rdp 1, bound to the chromatin, promotes targeting of histone modifications to specific sequences via si. RNA, resulting in maintenance of the heterochromatic state (HMT, histone methyl transferase).

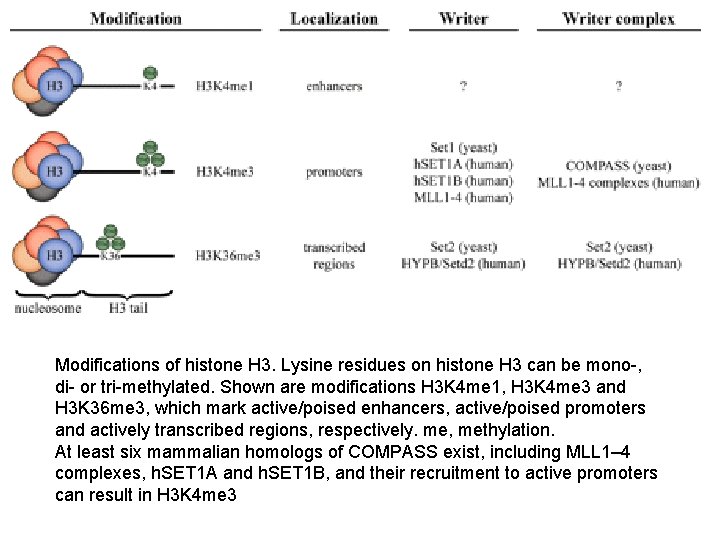
Modifications of histone H 3. Lysine residues on histone H 3 can be mono-, di- or tri-methylated. Shown are modifications H 3 K 4 me 1, H 3 K 4 me 3 and H 3 K 36 me 3, which mark active/poised enhancers, active/poised promoters and actively transcribed regions, respectively. me, methylation. At least six mammalian homologs of COMPASS exist, including MLL 1– 4 complexes, h. SET 1 A and h. SET 1 B, and their recruitment to active promoters can result in H 3 K 4 me 3

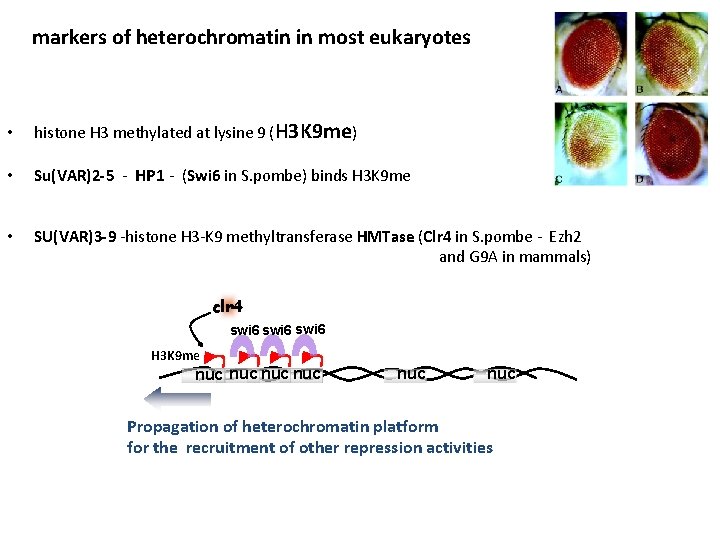
markers of heterochromatin in most eukaryotes • histone H 3 methylated at lysine 9 (H 3 K 9 me) • Su(VAR)2 -5 - HP 1 - (Swi 6 in S. pombe) binds H 3 K 9 me • SU(VAR)3 -9 -histone H 3 -K 9 methyltransferase HMTase (Clr 4 in S. pombe - Ezh 2 and G 9 A in mammals) clr 4 swi 6 H 3 K 9 me nuc nuc nuc Propagation of heterochromatin platform for the recruitment of other repression activities

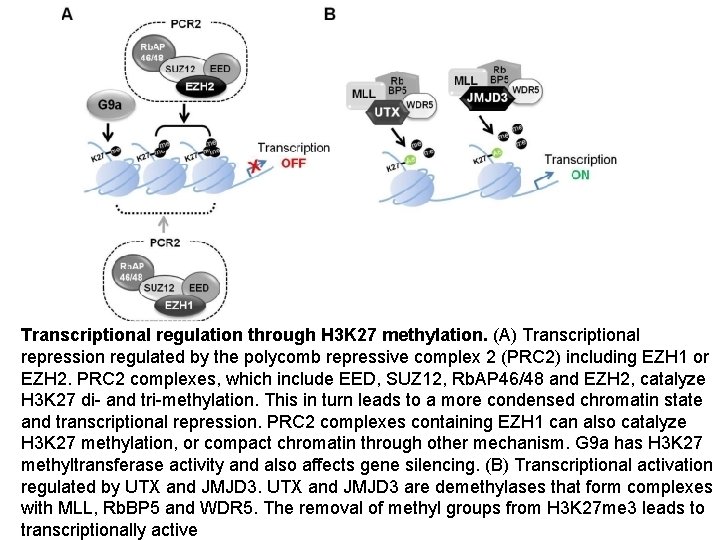
Transcriptional regulation through H 3 K 27 methylation. (A) Transcriptional repression regulated by the polycomb repressive complex 2 (PRC 2) including EZH 1 or EZH 2. PRC 2 complexes, which include EED, SUZ 12, Rb. AP 46/48 and EZH 2, catalyze H 3 K 27 di- and tri-methylation. This in turn leads to a more condensed chromatin state and transcriptional repression. PRC 2 complexes containing EZH 1 can also catalyze H 3 K 27 methylation, or compact chromatin through other mechanism. G 9 a has H 3 K 27 methyltransferase activity and also affects gene silencing. (B) Transcriptional activation regulated by UTX and JMJD 3 are demethylases that form complexes with MLL, Rb. BP 5 and WDR 5. The removal of methyl groups from H 3 K 27 me 3 leads to transcriptionally active

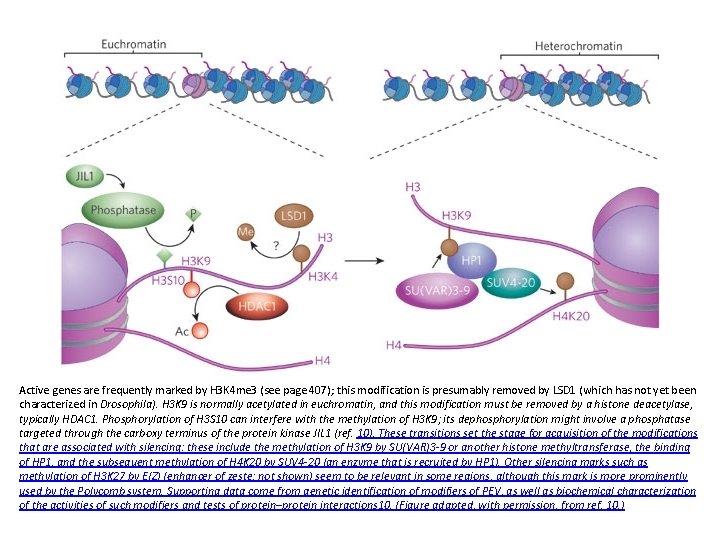
Active genes are frequently marked by H 3 K 4 me 3 (see page 407); this modification is presumably removed by LSD 1 (which has not yet been characterized in Drosophila). H 3 K 9 is normally acetylated in euchromatin, and this modification must be removed by a histone deacetylase, typically HDAC 1. Phosphorylation of H 3 S 10 can interfere with the methylation of H 3 K 9; its dephosphorylation might involve a phosphatase targeted through the carboxy terminus of the protein kinase JIL 1 (ref. 10). These transitions set the stage for acquisition of the modifications that are associated with silencing: these include the methylation of H 3 K 9 by SU(VAR)3 -9 or another histone methyltransferase, the binding of HP 1, and the subsequent methylation of H 4 K 20 by SUV 4 -20 (an enzyme that is recruited by HP 1). Other silencing marks such as methylation of H 3 K 27 by E(Z) (enhancer of zeste; not shown) seem to be relevant in some regions, although this mark is more prominently used by the Polycomb system. Supporting data come from genetic identification of modifiers of PEV, as well as biochemical characterization of the activities of such modifiers and tests of protein–protein interactions 10. (Figure adapted, with permission, from ref. 10. )

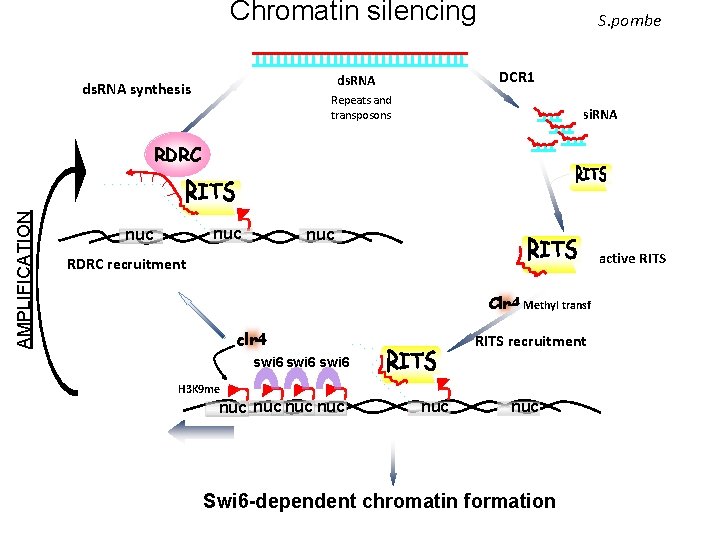
Chromatin silencing DCR 1 ds. RNA synthesis S. pombe Repeats and transposons si. RNA AMPLIFICATION RDRC nuc nuc active RITS RDRC recruitment Clr 4 Methyl transf clr 4 RITS recruitment swi 6 H 3 K 9 me nuc nuc nuc Swi 6 -dependent chromatin formation

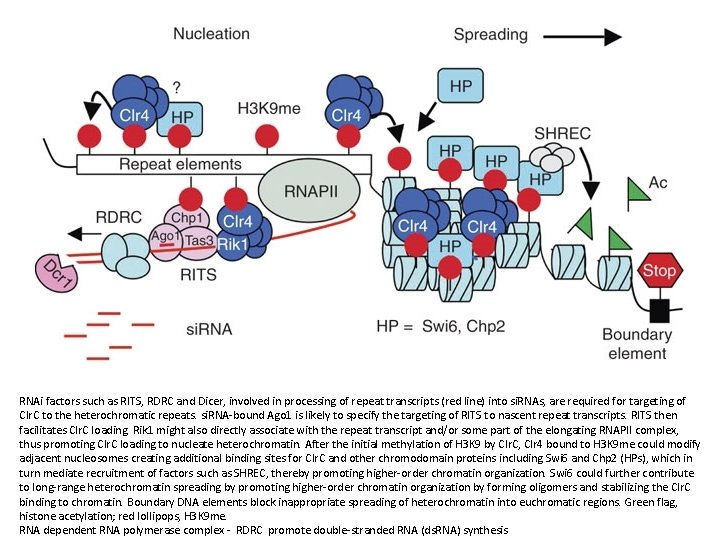
RNAi factors such as RITS, RDRC and Dicer, involved in processing of repeat transcripts (red line) into si. RNAs, are required for targeting of Clr. C to the heterochromatic repeats. si. RNA-bound Ago 1 is likely to specify the targeting of RITS to nascent repeat transcripts. RITS then facilitates Clr. C loading. Rik 1 might also directly associate with the repeat transcript and/or some part of the elongating RNAPII complex, thus promoting Clr. C loading to nucleate heterochromatin. After the initial methylation of H 3 K 9 by Clr. C, Clr 4 bound to H 3 K 9 me could modify adjacent nucleosomes creating additional binding sites for Clr. C and other chromodomain proteins including Swi 6 and Chp 2 (HPs), which in turn mediate recruitment of factors such as SHREC, thereby promoting higher-order chromatin organization. Swi 6 could further contribute to long-range heterochromatin spreading by promoting higher-order chromatin organization by forming oligomers and stabilizing the Clr. C binding to chromatin. Boundary DNA elements block inappropriate spreading of heterochromatin into euchromatic regions. Green flag, histone acetylation; red lollipops, H 3 K 9 me. RNA dependent RNA polymerase complex - RDRC promote double-stranded RNA (ds. RNA) synthesis

Marc Buhler – Basel Nat Struct Mol Biol. 2013 Aug; 20(8): 994 -1000 nc. RNA can act as EVICTORS nc. RNAs can counteract the spreading of heterochromatin into neighboring euchromatin BORDERLINE prevents spreading of the HP 1 protein Swi 6 and histone H 3 Lys 9 methylation beyond the pericentromeric repeat region of S. pombe It acts in a locus-dependent manner and isprocessed by Dicer into s. RNAs It evicts HP 1 from the chromatin Borderline acts as an allosteric regulator – Mass spec diff. interctors if SWI-6 alone or bound to the nc. RNA


Role of non coding RNAs and RNAi in DNA methylation and histone modification

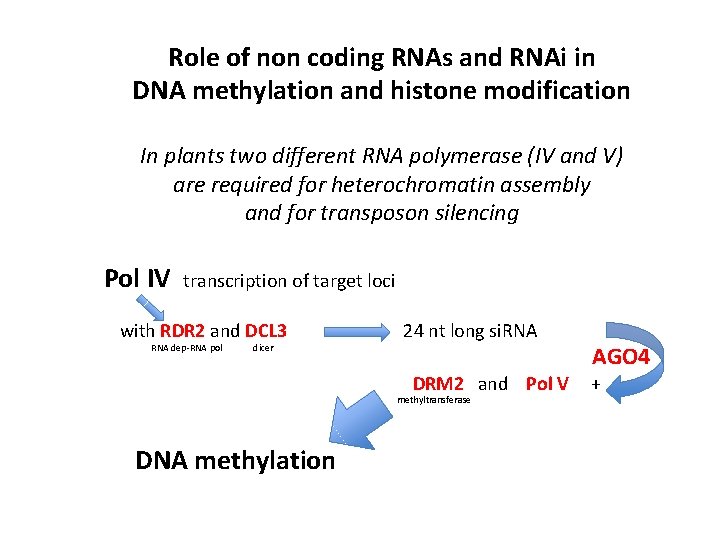
Role of non coding RNAs and RNAi in DNA methylation and histone modification In plants two different RNA polymerase (IV and V) are required for heterochromatin assembly and for transposon silencing Pol IV transcription of target loci with RDR 2 and DCL 3 RNA dep-RNA pol dicer 24 nt long si. RNA DRM 2 and Pol V methyltransferase DNA methylation AGO 4 +

Pol IVa and Pol IVb are players in the Rd. DM pathway RNA polymerase IV silences certain transposons and repetitive DNA in a short interfering RNA pathway involving RNA-dependent RNA polymerase 2 and Dicer-like 3 The existence of this distinct silencing polymerase may explain the paradoxical involvement of an RNA silencing pathway in maintenance of transcriptional silencing. higher plants have five multi-subunit nuclear RNA polymerases: the ubiquitous Pol I, II and III, which are essential for viability; plus two non-essential polymerases, Pol IVa and Pol Ivb which specialize in small RNA-mediated gene silencing pathways In the RNA-directed DNA methylation (Rd. DM) pathway of transcriptional gene silencing double-stranded RNAs generated with the involvement of RDR 2 are cleaved by DCL 3, and the resulting si. RNAs are loaded into AGO 4–RISC and/or AGO 6–RISC complexes that mediate the de novo methylation of cytosines within DNA sequences complementary to the si. RNAs

RNA silencing and DNA methylation in Arabidopsis • Cytosine DNA methylation silences harmful DNAs such as transposons and retroviruses • Maintenance DNA methyltransferases propagate pre -existing DNA methylation in the CG sequence context by methylating hemi-methylated sites after DNA replication • RNA Silencing Genes Control de Novo DNA Methylation in Arabidopsis

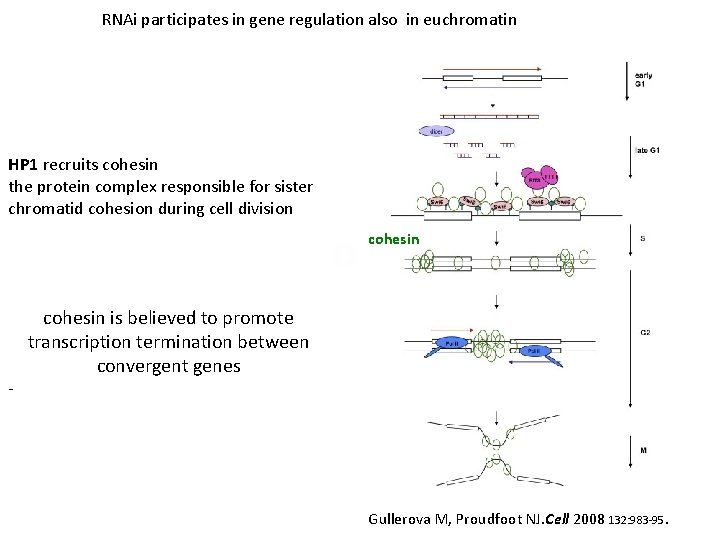
RNAi participates in gene regulation also in euchromatin HP 1 recruits cohesin the protein complex responsible for sister chromatid cohesion during cell division cohesin - cohesin is believed to promote transcription termination between convergent genes Gullerova M, Proudfoot NJ. Cell 2008 132: 983 -95.

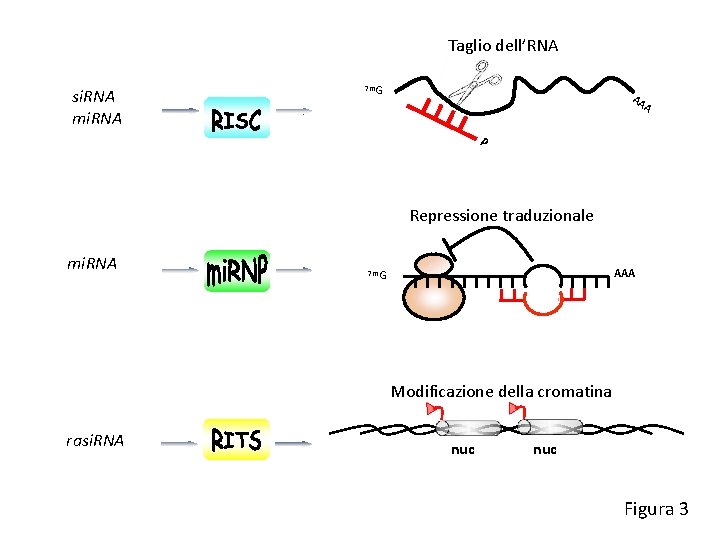
Taglio dell’RNA si. RNA mi. RNA 7 m. G AA A P Repressione traduzionale mi. RNA AAA 7 m. G Modificazione della cromatina rasi. RNA nuc Figura 3

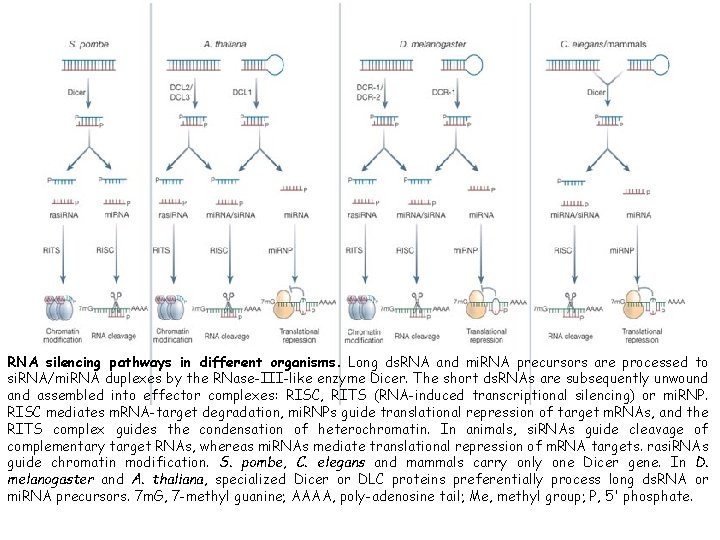
RNA silencing pathways in different organisms. Long ds. RNA and mi. RNA precursors are processed to si. RNA/mi. RNA duplexes by the RNase-III-like enzyme Dicer. The short ds. RNAs are subsequently unwound assembled into effector complexes: RISC, RITS (RNA-induced transcriptional silencing) or mi. RNP. RISC mediates m. RNA-target degradation, mi. RNPs guide translational repression of target m. RNAs, and the RITS complex guides the condensation of heterochromatin. In animals, si. RNAs guide cleavage of complementary target RNAs, whereas mi. RNAs mediate translational repression of m. RNA targets. rasi. RNAs guide chromatin modification. S. pombe, C. elegans and mammals carry only one Dicer gene. In D. melanogaster and A. thaliana, specialized Dicer or DLC proteins preferentially process long ds. RNA or mi. RNA precursors. 7 m. G, 7 -methyl guanine; AAAA, poly-adenosine tail; Me, methyl group; P, 5' phosphate.

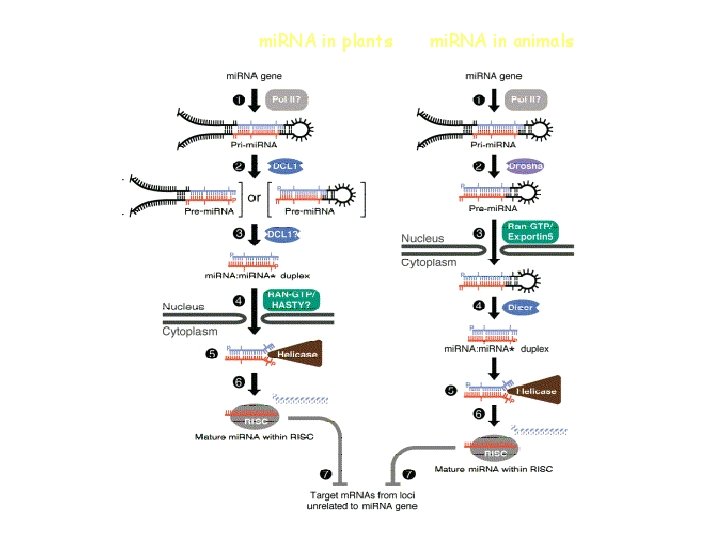
mi. RNA in plants mi. RNA in animals
 Rnai
Rnai Ahringer rnai library
Ahringer rnai library Heterochromatin and euchromatin difference
Heterochromatin and euchromatin difference Cell bio
Cell bio Heterochromatin and euchromatin
Heterochromatin and euchromatin Heterochromatin
Heterochromatin Vertical gene transfer
Vertical gene transfer Mcb 140
Mcb 140 Sex nn
Sex nn Chromatin vs chromozom
Chromatin vs chromozom Chromatin vs chromozom
Chromatin vs chromozom Chromatin vs chromozom
Chromatin vs chromozom Barr body is found in
Barr body is found in Chromatin in a sentence
Chromatin in a sentence Gold standard malaria
Gold standard malaria Primodial germ cell
Primodial germ cell Chromatin
Chromatin Chromatin draws together to create
Chromatin draws together to create Chromatin
Chromatin Phase of mitosis
Phase of mitosis Chromatin states
Chromatin states Chromatin immunoprecipitation
Chromatin immunoprecipitation 반보존적 복제
반보존적 복제 Ohem
Ohem Condensed structural formula of formaldehyde
Condensed structural formula of formaldehyde